Bio-Rad iQ 5 Optical System Software, Version 2.1 User Manual

iQ™5 Optical System Software
Instruction Manual
Compatible with the MyiQ™2,
MyiQ™, and iQ™5 real-time
PCR detection systems
NOTICE TO PURCHASER
Bio-Rad’s real-time thermal cyclers are licensed real-time thermal cyclers under Applera’s United States Patent 6,814,934 B1 for use in research, human in vitro diagnostics, and for all other fields except the field of veterinary diagnostics. Purchase of this instrument conveys a limited nontransferable immunity from suit for the purchaser’s own internal research and development and for use in applied fields under one or more of U.S. Patents 5,656,493, 5,333,675, 5,475,610 (claims 1, 44, 158, 160–163 and 167 only), and 6,703,236 (claims 1–7 only), or corresponding claims in their non-U.S. counterparts, owned by Applera Corporation. No right is conveyed expressly, by implication or by estoppel under any other patent claim, such as claims to apparatus, reagents, kits, or methods such as 5' nuclease methods. Further information on purchasing licenses may be obtained by contacting the Director of Licensing, Applied Biosystems, 850 Lincoln Centre Drive, Foster City, California 94404, USA.
Bio-Rad’s thermal cyclers and real-time thermal cyclers are covered by one or more of the following U.S. patents or their foreign counterparts owned by Eppendorf AG: U.S. Patent Nos. 6,767,512 and 7,074,367.
SYBR is a trademark of Molecular Probes, Inc. Bio-Rad Laboratories, Inc. is licensed by Molecular Probes, Inc. to sell reagents containing SYBR® Green I for use in real-time PCR, for research purposes only.
Cy is a trademark of GE Healthcare. SYBR and Texas Red are trademarks of Molecular Probes, Inc. HEX, TAMRA, TET, and VIC are trademarks of Applera Corp. Excel, Windows, and Windows XP are trademarks of Microsoft Corporation. Phillips is a trademark of Phillips Screw Company Corporation.
i
Safety Information
Grounding
Always connect the MyiQTM2, MyiQTM, or iQTM5 optics module to a three-prong, grounded AC outlet using the AC power cord provided with the system. Do not use an adaptor to a two-terminal outlet. Always ensure that you set the module power switch to the off position when you connect or disconnect power cords.
Handling
Handle all components of the real-time PCR detection system with care and with clean, dry hands at all times. The optical system contains mirrors and lenses that may shatter if the unit is dropped or struck with great force. If the unit is damaged such that internal components or wires are exposed, contact your local Bio-Rad office immediately. Do not attempt to repair or power on the instrument.
Servicing
The only user-serviceable parts of the optics module are the lamp and filters. Call your local Bio-Rad office for all other optics module and thermal cycler related service. When you replace the lamp or filters, open only the outer casing of the optics module. The camera lamp may get extremely hot during system operation. Do not attempt to remove the lamp without powering off the instrument and allowing the system to cool for at least 15 minutes. To prevent skin burns and fire hazards, do not attempt to operate the real-time PCR detection system while the camera case is open. Do not open casing of the optics module when the instrument is in use.
Operating Temperature
For normal operation, the maximum ambient temperature should not exceed 40°C. To ensure adequate cooling of the system, maintain a clearance of at least 4 inches around the sides of the MyiQ2, MyiQ, or iQ5 optics module. Do not block the fan vents near the lamp, as this may lead to improper operation or cause physical damage to the detector. Do not operate the optics module in extreme humidity (that is, greater than 90 percent) or where condensation can short internal electrical circuits or fog optical elements.
Notice
The MyiQ2, MyiQ, and iQ5 instruments are designed and certified to meet EN-61010 safety standards.
EN-61010 certified products are safe to use when operated in accordance with the instruction manual. These instruments should not be modified in any way. Alteration of the instruments will:
•Void the manufacturer’s warranty
•Void the EN-61010 safety certification
•Create a potential safety hazard
Bio-Rad is not responsible for any injury or damage caused by the use of these instrument for purposes other than those for which they are intended, or by modifications to the instruments not performed by Bio-Rad or an authorized agent.
- ii -
Table of Contents |
|
NOTICE TO PURCHASER.................................................................................................. |
i |
Safety Information......................................................................................................... |
ii |
Section 1. Getting Started.............................................................................................. |
1 |
1.1 The MyiQ™2 Real-Time PCR Detection System ................................................................ |
1 |
1.2 Setting Up the System Hardware .................................................................................... |
1 |
1.2.1 System Checklist...................................................................................................... |
1 |
1.2.2 Installing the Optical Reaction Module on the iCycler Chassis...................................... |
2 |
1.2.3 Installing the Support Bracket .................................................................................. |
3 |
1.2.4 Installing the Optics Module ..................................................................................... |
3 |
1.2.5 Connecting Power and Communication Cables to the System ..................................... |
3 |
1.3 Installing the iQ5 Software ............................................................................................. |
4 |
1.3.1 Installing the Camera Drivers ………………………………………………………………………………4 |
|
1.4 Recommended Computer Settings................................................................................... |
4 |
1.5 Calibrating the Instrument.............................................................................................. |
6 |
1.6 Compatibility with Earlier Versions of the iQ5 Optical System Software .............................. |
6 |
Section 2. Quick Guides.................................................................................................. |
7 |
2.1 Protocol Quick Guide...................................................................................................... |
7 |
2.2 Plate Setup Quick Guide ................................................................................................. |
8 |
2.3 Running a Real-Time Experiment Quick Guide ................................................................. |
9 |
2.3.1 Beginning a Run ...................................................................................................... |
9 |
2.3.2 Monitoring the Run ................................................................................................ |
10 |
2.4 Data Analysis Quick Guides........................................................................................... |
10 |
2.4.1 PCR Quant Tab Quick Guide ................................................................................... |
10 |
2.4.2 End Point Quick Guide............................................................................................ |
11 |
2.4.3 Allelic Discrimination Quick Guide – For Multiplex Data Only ........................................ |
12 |
2.4.4 Gene Expression Quick Guide..................................................................................... |
13 |
2.4.5 Post-Run Plate Setup Editing Quick Guide................................................................... |
16 |
Section 3. Introduction to the iQ5 Optical System Software (Version 2.1)................. |
18 |
Section 4. Workshop Module........................................................................................ |
19 |
4.1 Setup Tab.................................................................................................................... |
19 |
4.1.1 File Browser Section............................................................................................... |
20 |
4.1.2 Selected Protocol Area ........................................................................................... |
20 |
4.1.3 Selected Plate Setup Area ...................................................................................... |
20 |
- iii - |
|
4.1.4 Selected Data File Area .......................................................................................... |
21 |
4.2 Plate Setup.................................................................................................................. |
21 |
4.2.1 Plate Setup Editor Window ..................................................................................... |
21 |
4.2.2 Well Definition Icons.............................................................................................. |
22 |
4.2.3 Editing or Creating a New Plate Setup..................................................................... |
23 |
4.2.4 Fluorophore Selection ............................................................................................ |
24 |
4.2.5 Whole Plate Loading .............................................................................................. |
26 |
4.2.6 Specifying Replicates.............................................................................................. |
26 |
4.2.7 Sample Volume, Seal Type, and Vessel Type ........................................................... |
27 |
4.2.8 Defining a Dilution Series ....................................................................................... |
28 |
4.2.9 Plate Setup Editor Spreadsheets ............................................................................. |
29 |
4.2.10 Plate Summary .................................................................................................... |
30 |
4.3 Protocol....................................................................................................................... |
31 |
4.3.1 Selecting a Protocol ............................................................................................... |
31 |
4.3.2 Editing or Creating a Protocol ................................................................................. |
32 |
4.3.3 Add Protocol Options ............................................................................................. |
33 |
4.3.4 Gradient................................................................................................................ |
35 |
4.3.5 Melt Curve/Peak .................................................................................................... |
36 |
4.3.6 Sample Protocol Files............................................................................................. |
37 |
4.4 Run Set....................................................................................................................... |
37 |
4.4.1 Selecting a Run Set................................................................................................ |
38 |
4.4.2 Creating a Run Set................................................................................................. |
38 |
4.5 Data File...................................................................................................................... |
38 |
4.5.1 Selecting a Data File .............................................................................................. |
39 |
4.5.2 Opening a Data File ............................................................................................... |
39 |
4.5.3 Opening a Pure Dye Calibration file......................................................................... |
39 |
4.5.4 Applying Alternate Pure Dye Calibrations (Alternate RME) ........................................ |
39 |
4.5.5 Applying Alternate Background or Well Factors........................................................ |
40 |
Section 5. Run-Time Central Module............................................................................ |
41 |
5.1 Initiate Run Window: Run Selected............................................................................... |
41 |
5.2 Well Factors................................................................................................................. |
42 |
5.2.1 Dynamic Well Factors............................................................................................. |
42 |
5.2.2 Persistent Well Factors........................................................................................... |
42 |
5.3 Real-Time PCR Experiments Using DNA-Binding Dyes..................................................... |
43 |
5.3.1 Spiking Real-Time PCR Experiments Using DNA binding Dyes With Fluorescein.......... |
43 |
5.4 Initiate Run Window: Run End Point Selected................................................................ |
43 |
- iv -
5.5 |
Show Plate Window ..................................................................................................... |
44 |
5.6 |
Monitor Run Window.................................................................................................... |
45 |
5.7 |
Run-Time Protocol Editing ............................................................................................ |
45 |
Section 6. Data Analysis Module .................................................................................. |
47 |
|
6.1 |
PCR Quant Tab............................................................................................................ |
48 |
6.1.1 Customizing the PCR Quant Display ........................................................................ |
49 |
|
6.2 |
Amplification Chart....................................................................................................... |
49 |
6.2.1 Fluorophore Selector Buttons.................................................................................. |
49 |
|
6.2.2 Select Analysis Mode.............................................................................................. |
50 |
|
6.2.3 Log View Button .................................................................................................... |
50 |
|
6.2.4 Selecting and Viewing Traces ................................................................................. |
51 |
|
6.3 |
Standard Curve Chart................................................................................................... |
53 |
6.3.1 Standard Curve Chart Menu.................................................................................... |
54 |
|
6.4 |
Results Section ............................................................................................................ |
55 |
6.4.1 Plate Spreadsheet.................................................................................................. |
55 |
|
6.4.2 Standard Curve/CT Results Spreadsheet .................................................................. |
56 |
|
6.4.3 Amplification Data (RFU) Spreadsheet..................................................................... |
57 |
|
6.5 |
Amplification Plot Context Menu.................................................................................... |
57 |
6.5.1 Data Analysis Options ............................................................................................ |
57 |
|
6.5.2 Amplification Plot Data Display Options ................................................................... |
61 |
|
6.5.3 Data Export Options............................................................................................... |
64 |
|
6.5.4 Exporting Results to Microsoft Excel........................................................................ |
64 |
|
6.5.5 Printing Results...................................................................................................... |
65 |
|
6.6 |
Melt Curve and Melt Peak Charts................................................................................... |
65 |
6.6.1 Melt Curve and Melt Peak Chart Menu..................................................................... |
67 |
|
6.6.2 Delete Selected Peaks............................................................................................ |
67 |
|
6.6.3 Edit Melt Peak Begin/End Temperature ................................................................... |
67 |
|
6.6.4 Melt Curve/Peak Control Area................................................................................. |
68 |
|
6.6.5 Melt Curve/Peak Spreadsheet................................................................................. |
68 |
|
6.7 |
Allelic Discrimination Module (For Multiplex Data Only)................................................... |
69 |
6.7.1 Allelic Discrimination Plot........................................................................................ |
69 |
|
6.7.2 Allelic Discrimination Plot Menu............................................................................... |
71 |
|
6.7.3 Allelic Data Spreadsheet......................................................................................... |
71 |
|
6.7.4 Automatic/Manual Call ........................................................................................... |
72 |
|
6.7.5 Manual Calls.......................................................................................................... |
73 |
|
6.7.6 Display Mode......................................................................................................... |
73 |
|
|
- v - |
|
6.7.7 Vertical Threshold.................................................................................................. |
74 |
6.7.8 Horizontal Threshold.............................................................................................. |
74 |
6.7.9 Normalize Data...................................................................................................... |
74 |
6.7.10 Restore Default.................................................................................................... |
75 |
6.8 End Point Analysis........................................................................................................ |
75 |
6.8.1 The End Point Analysis Settings ………………………………………………………………………….75 |
|
6.8.2 End Point Analysis Spreadsheet .............................................................................. |
77 |
6.8.3 Recalculate Button................................................................................................. |
78 |
6.9 Gene Expression Analysis. ............................................................................................ |
78 |
6.9.1 Normalized Gene Expression (ddCT, ΔΔCT) ............................................................... |
80 |
6.9.2 Relative Quantity (dCT, ΔCT).................................................................................... |
81 |
6.9.3 Specifying Gene and Condition Labels for Gene Expression Analysis.......................... |
82 |
6.9.4 Setting Analysis Parameters for Gene Expression Analysis Using the Settings Tab...... |
85 |
6.9.4.1Gene List: Assigning Reference Genes and Target-Specific Reaction Efficiency…85
6.9.4.2Assigning and Naming Control Samples Using the Condition List ……………………87
6.9.4.3Data Set List …………………………………………………………………………………………….88
6.9.5 Applying an Analysis Method .................................................................................. |
89 |
6.9.6 Data Table for Gene Expression Analysis................................................................. |
89 |
6.9.7 Graphing Options for Expression Data..................................................................... |
92 |
6.9.8 Normalized Expression Calculations......................................................................... |
96 |
6.9.9 Relative Quantity Calculations................................................................................. |
97 |
6.9.10 Gene Expression - Frequently Asked Questions...................................................... |
98 |
6.10 Gene Study: Multi File Gene Expression Analysis .......................................................... |
99 |
6.10.1 The Gene Study File............................................................................................. |
99 |
6.10.2 Creating a Gene Study File ................................................................................. |
101 |
6.10.3 Gene Expression Study Manager......................................................................... |
102 |
6.10.4 Editing a Gene Study.......................................................................................... |
102 |
6.10.5 Excluding/Including Samples Using the Gene Expression Plate Interface ............... |
103 |
6.10.6 Inter-Run Calibration.......................................................................................... |
103 |
6.11 Post-Run Plate Editing.............................................................................................. |
105 |
6.12 The Reports Tool ..................................................................................................... |
107 |
6.12.1 Report Viewer Options ....................................................................................... |
108 |
6.12.2 Parts of the Data Report..................................................................................... |
108 |
6.12.3 Data Analysis Report Types ................................................................................ |
108 |
Section 7. Calibrating the Instrument........................................................................ |
111 |
7.1 Calibration Overview .................................................................................................. |
111 |
- vi -
7.2 |
Components Required for Calibration .......................................................................... |
111 |
7.3 |
Preparing Calibration Plates........................................................................................ |
112 |
7.4 |
Performing Calibrations .............................................................................................. |
112 |
7.4.1 Performing Mask Alignment.................................................................................. |
113 |
|
7.4.2 Performing Background Calibration ....................................................................... |
114 |
|
7.4.3 Generating Persistent Well Factors........................................................................ |
115 |
|
7.4.4 Performing Pure Dye Calibration........................................................................... |
116 |
|
7.4.5 Editing and Creating Pure Dye Plate Setups........................................................... |
118 |
|
7.5 |
Viewing Calibration Files............................................................................................. |
119 |
7.6 |
Troubleshooting Optics with the Mask Image Window.................................................. |
120 |
7.6.1 Filter Position....................................................................................................... |
120 |
|
7.6.2 Camera Status..................................................................................................... |
120 |
|
Section 8. User Profiles .............................................................................................. |
122 |
|
8.1 |
User Preferences........................................................................................................ |
122 |
8.1.1 File Paths Preferences.......................................................................................... |
123 |
|
8.1.2 Plate Setup Preferences ....................................................................................... |
124 |
|
8.1.3 Protocol Preferences ............................................................................................ |
125 |
|
8.1.4 File Selection at Application Startup Preferences.................................................... |
125 |
|
8.1.5 PCR Quant Screen Preferences ............................................................................. |
126 |
|
8.1.6 Allelic Discrimination Module Preferences .............................................................. |
127 |
|
8.1.7 End Point Module Preferences .............................................................................. |
127 |
|
8.1.8 Gene Expression Module Preferences.................................................................... |
128 |
|
8.2 |
User Administration.................................................................................................... |
128 |
8.2.1 Adding New Users................................................................................................ |
129 |
|
8.2.2 Deleting Existing Users......................................................................................... |
130 |
|
8.3 |
Logging on to the iQ5 Software .................................................................................. |
130 |
8.3.1 Switching Users................................................................................................... |
131 |
|
8.3.2 Changing Password.............................................................................................. |
131 |
|
8.3.3 Defining Roles ..................................................................................................... |
132 |
|
Section 9. Instrument Maintenance........................................................................... |
133 |
|
9.1 |
Cleaning the Real-Time PCR Detection System............................................................. |
133 |
9.2 |
Filter Description and Installation Instructions.............................................................. |
133 |
9.2.1 Recommended Fluorophores and Filter Specifications............................................. |
133 |
|
9.2.2 Accessing the Filters ............................................................................................ |
134 |
|
9.2.3 Cleaning the Filters.............................................................................................. |
134 |
|
9.3 |
Replacing the Lamp ................................................................................................... |
135 |
|
- vii - |
|
Appendix A: Warranty ................................................................................................ |
136 |
Appendix B: Troubleshooting Error Messages ........................................................... |
137 |
Appendix C: Product Ordering Information ............................................................... |
142 |
- viii -
Section 1 Getting Started
Section 1. Getting Started
This section contains information on the following topics:
oThe MyiQ2 Real-Time Detection System (page 1)
oSetting up the system hardware (page 1)
oInstalling the iQ5 software (page 4)
oRecommended computer settings (page 4)
oCompatibility with earlier versions of the software (page 6)
1.1The MyiQ™2 Real-Time PCR Detection System
The MyiQ2 Real-Time PCR Detection System is the latest addition to a family of real-time PCR detection systems that are comprised of an optical detection module and iCycler® thermal cycler.
The two-color MyiQ™2 features a broad-spectrum light source (tungsten-halogen lamp) and paired filter optical design to ensure optimal excitation and emission. This results in excellent sensitivity and discrimination between multiple fluorophores. A CCD detector captures a simultaneous image of all 96 wells of the block resulting in a comprehensive data set illustrating the kinetic behavior of the data during each cycle.
The iQ™5 Optical System software (version 2.1) is used to control and collect and analyze data from the MyiQ2 Real-Time PCR Detection System. The software is also capable of controlling, and collecting and analyzing data from the single color MyiQ™ and five color iQ™5 Real-Time PCR detection systems. This manual provides instructions on use of the software applicable to each of these three systems. The online Help manual is available at all times by pressing the F1 key.
1.2 Setting Up the System Hardware
The MyiQ2, MyiQ, or iQ5 system should be installed on a clean, dry, and level surface. Identify an appropriate work area for the installation process prior to unpacking any system components. The entire installation process should take approximately 15 minutes to complete. Handle the optics module and iCycler thermal cycler with care and with clean, dry hands during unpacking and assembly.
1.2.1 System Checklist
MyiQ2 Optics Module (catalog #170-9758)
•Optics module
•Power cord
•iQ5 optical system software installation disk (170-9753)
•Amplification tech notes CD (version 1.0)
iCycler Chassis (catalog #170-8701)
•Power cord
•iCycler chassis
•iCycler thermal cycler instruction manual
- 1 -
Section 1 Getting Started
iQ5 Optical Accessory Kit (catalog #170-9752)
•Optical reaction block
•Modified sliding rear cover for iCycler thermal cycler
•Serial cable
•USB cable
•Filter extraction tool
•Optical tape applicator tools (3)
•Optics support bracket
•Support bracket screws
•Hex driver
•Hex screws
•Spare part: halogen lamp
MyiQ2 Calibrator Dye Solution Kit (catalog #170-8791)
•1x calibration dye solutions (4 dyes, 3 tubes of each dye)
External Well Factor Solution (catalog #170-8794)
•External well factor solution (5 tubes)
Sample Consumables
•PCR plates and sealers
•Reagents
Contact your local Bio-Rad office if any system components are missing or damaged.
Accessories
The following accessories are required to complete the installation:
•Scissors
•#2 Phillips screwdriver
•Calibrated micropipet(s)
•Aerosol barrier pipet tips
•Optical-quality sealing film or tube caps
•Optical-quality PCR plates or tubes
1.2.2 Installing the Optical Reaction Module on the iCycler Chassis
1.Remove the existing rear cover from the iCycler chassis by sliding the cover towards the front of the iCycler base.
2.Install the modified sliding rear cover provided with the optical reaction block, ensuring that the notch is oriented towards the rear of the iCycler thermal cycler.
3.Push the sliding rear cover on top of the chassis as far back as possible.
4.Rotate the green latches on the optical reaction block up towards the open lid.
5.Lift the optical reaction block by the handle and install it onto the chassis. Lower the front portion of the reaction block so that it engages with the chassis before the rear portion. The rear of the block lid should fit over the front lip of the sliding rear cover.
6.Secure the optical reaction block in place by rotating the green latches downward.
7.Close the optical lid.
-2 -
Section 1 Getting Started
1.2.3 Installing the Support Bracket
A support bracket with roller is provided for the MyiQ2, MyiQ, or iQ5 optics module. It is mounted to the rear of the iCycler thermal cycler.
Align the optics module support bracket with the two holes on the rear of the iCycler thermal cycler.
Using a #2 Phillips screwdriver, adjust the height of the bracket with two of the appropriate screws provided with the system accessories. Both of the screws should be approximately in the center of the slots on the bracket.
1.2.4 Installing the Optics Module
1.Remove the plastic sheath and protective label from the optics module and place the optical module on a flat surface, taking care not to touch the inside of the nose portion of the module.
2.Remove the protective label from the optical lid.
3.Slide the optics module onto the U-bracket.
4.Secure the optics module to the U-bracket using the two long, thin hex screws and the hex driver provided.
5.After the optics module has been installed, confirm that the optics module and lid assembly can be opened and closed readily.
If the lid is difficult to open, lower the support bracket slightly before tightening the bracket mounting screws. If the lid is difficult to close, try raising the support bracket slightly before tightening the rear screws.
1.2.5 Connecting Power and Communication Cables to the System
Before connecting any communication or power cables to the system, confirm that the power switch for the iCycler thermal cycler and the optics module are in the OFF position.
1.Close the optical reaction block by sliding the lid forward and pressing down on the lid handle. On the right side of the optics module are three connectors.
2.Using the cables provided, establish power and communication with the computer as follows:
•Recessed 3-pin power connector — Connect the supplied power cord between the optics module and a grounded power outlet. This connection provides power only to the optical module; a separate power cord must be connected to the iCycler thermal cycler
•Serial connector — A serial connector is located at the rear of the iCycler thermal cycler. Connect the serial cable to the rear of the iCycler chassis and to the serial port on the side of the optics module. This connection enables communication between the iQ5 software and the iCycler thermal cycler
•USB port connector — Connect the supplied USB cable between the optics module and a USB 2.0 high-speed enabled port on the computer. Data are transferred to the computer via this cable. This single connection directs the operation of both the optics module and the iCycler thermal cycler by the iQ5 Optical System software
•At the right rear corner of the optical reaction block is a single connector the Positive docking connector — This self-aligning connector is secured into place when the optics
-3 -
Section 1 Getting Started
module is installed on the iCycler chassis. This connection senses when the lid handle is lifted.
1.3 Installing the iQ5 Software
Locate the software installation disk for iQ5 Optical System software version 2.1 provided with the MyiQ2 system. This installation disk is compatible with computers running the Windows XP and Windows Vista 32-bit operating system.
1.Insert the iQ5 Optical System software installation CD in a CD-ROM drive.
2.If the installation program does not begin automatically, click Run… in the Start menu and then type X:\iQ5\Setup, where X is the drive letter of the CD-ROM drive. For example, if the CD-ROM is the E drive, type E:\iQ5\Setup.
3.Follow all screen prompts to finalize the installation. Certain configurations of Windows operating systems initialize new folders by assigning Read and Execute permission for the members of the Users group. If you have this type of operating system and this is a first-time installation, the administrator must change the protection for the Program Files/Bio-Rad folder or the Program Files/Bio-Rad/iQ5 folder so that you can save protocol, plate setup, and data files.
4.If you still cannot write to the folders beneath the Program Files/Bio-Rad/iQ5 folder after changing the protection on either of these folders, check the properties of each folder. Specifically, in the Properties window Securities tab, ensure that the checkbox that allows inheritable permissions to propagate to that folder has been selected.
5.Confirm that the iQ5 system software is working properly by double-clicking on the shortcut icon located on the Windows desktop, or by selecting the iQ5 program icon from the Bio-Rad folder in the Windows Start menu.
1.3.1 Installing the Camera Drivers
Before using the real-time PCR detection system for the first time the camera drivers must be installed.
1.Power on the iCycler thermal cycler and the optics module.
2.Windows will display a “Found New Hardware Wizard” dialog box. To install the camera drivers, select the option to Install from a list or specific location. Click Next to continue.
3.In the new window that appears, select Search for the best driver in these locations, and then click the Include this location in the search checkbox. Use the browse button to navigate to the “iQ5\Drivers” folder. The default location of the drivers is: C:\Program Files\Bio-Rad\iQ5\Drivers. Click Next to continue.
4.Next Windows will display a Hardware Installation dialog box regarding Windows Logo Testing, a service offered by Microsoft. To complete driver installation for the iQ5 or MyiQ systems, click Continue Anyway.
1.4Recommended Computer Settings
For the MyiQ2, MyiQ and iQ5 systems to communicate properly with the iQ5 Optical System software, version 2.1, the computer settings should be set as described below.
- 4 -
Section 1 Getting Started
Computer Power Management Settings on Windows Vista
1.From the Start menu, choose Control Panel. Switch the Control Panel view to Classic View.
2.Choose Power options, and then choose Create a Power Plan.
3.Choose the High Performance plan, then enter the Plan name iQ5 software, and press Next.
4.Change the Turn Off The Display and Put the Computer to Sleep options to “Never”.
5.Click Save Changes.
6.Select your new iQ5 software power plan by clicking on the radio button, open Change Advanced Power Settings to set the following conditions.
a.Adjust settings to turn off hard disk after 180 minutes.
b.For Sleep Settings:
i.Set Sleep After setting to “Never”
ii.Set Hybrid Sleep setting to “On”
iii.Set Hibernate After setting to “Never”
c.For USB Settings, set USB Selective Suspend setting to “Disabled”.
d.Set Search and Indexing, Power Savings Mode setting to “Balanced”,
e.Set Display, Turn Display Off After setting to “Never”.
When completed, click Apply and then Done.
Computer Power Management Settings on Windows XP
1.From the Start menu, choose Settings, then Control Panel.
2.Choose Power Options, the Power Options Properties window will open.
3.On the Power Schemes tab, set all power scheme settings to Never.
4.On the Hibernate tab, deselect the Enable Hibernation checkbox.
5.Return to the Control Panel window, and select Display; the Display Properties window will open.
6.On the Screen Saver tab, select None.
7.Return to the Control Panel window, and select System, the System Properties window will open.
8.On the Hardware tab, click on the Device Manager button to open the Device Manager file tree. Click on the “+” button to expand the Universal Serial Bus Controllers list.
9.Scroll down and highlight the first listed USB Root Hub. Right-click and select the Properties option to open the USB Root Hub Properties window.
10.Select the Power Management tab, and deselect the Allow the Computer to Turn Off This Device to Save Power checkbox.
11.Repeat steps 9–10 for each individual USB root hub listed.
-5 -
Section 1 Getting Started
1.5 Calibrating the Instrument
Before using the real-time PCR detection system for the first time, mask alignment, background calibration, persistent well factor collection, and pure dye calibration (for MyiQ2 and iQ5 systems only) must be performed. See section 7, Calibrating the Instrument, for detailed instructions on calibration.
1.6Compatibility with Earlier Versions of the iQ5 Optical System Software
Data Files
Protocol, plate setup, and data files generated or saved with version 3.0 or 3.1 of the iQ RealTime Detection System software and earlier versions of the iQ5 software are recognized by version 2.1 of the iQ5 software. These .tmo, .pts, and .opd files can be opened from within iQ5 software for analysis.
To open or import files generated by older versions of the iQ Real-Tme Detection System software (version 2.3b and earlier), you must open the file in version 3.0 or 3.1 of the iQ software first and then save the file again.
Calibration Files
Earlier versions of the iQ5 Optical System software (earlier than version 2.1) have volumedependent calibration files for both persistent well factors and pure dye calibration collection. In version 2.1, there is no volume dependency for these calibration files. If a computer is upgraded from an earlier version of the iQ5 Optical System software to version 2.1, existing calibration files will be upgraded to remove calibration volume dependency.
For example, Persistent_Plates_Film_25ul_IQ2Emulator_IQ2Emulator.xml will be upgraded and renamed as Persistent_Plates_Film_IQ2Emulator_IQ2Emulator.xml.
In all cases, the original calibration files with the volume dependency will be backed up to the backup folder after upgrading.
- 6 -
Section 2 Quick Guides
Section 2. Quick Guides
This section contains quick guides for the following topics:
oProtocol Quick Guide (page 7)
oPlate Setup Quick Guide (page 8)
oRunning a Real-Time Experiment Quick Guide (page 9)
oData Analysis Quick Guides
oPCR Quant Tab Quick Guide (page 10)
oEnd Point Quick Guide (page 11)
oAllelic Discrimination Quick Guide (page 12)
oGene Expression Quick Guide (page 13)
oPost-Run Plate Editing Quick Guide (page 16)
The fundamental steps of running a real-time PCR experiment on the MyiQ2, MyiQ, or iQ5 RealTime Detection System are the following:
1.Select, Edit, or Create a Protocol.
2.Select, Edit, or Create a Plate Setup.
3.Click Run
4.Select the appropriate Well Factor option and begin the run
5.Analyze the data
The quick guides below outline key procedures from each step. These quick guides are also located in the online help of the iQ5 Optical System software.
For more complete information regarding the procedures and features of the iQ5 Optical System software, refer to the relevant sections of this user manual.
2.1 Protocol Quick Guide
Within the Workshop module click the Protocol button.
Selecting a Protocol
1.Navigate to your desired protocol file using the file tree browser.
2.Click the protocol name once. The selected protocol appears in the Selected Protocol pane.
NOTE: The iQ5 software has a number of sample protocol files which may be used.
- 7 -
Section 2 Quick Guides
Editing or Creating a Protocol
Edit the selected protocol by clicking Edit in the Selected Protocol window, or create a protocol from a protocol template by clicking Create New in the Selected Protocol window.
NOTE: Clicking Edit or Create New in the Selected Protocol pane opens the Protocol Editor.
NOTE: You can only exit the Protocol Editor by clicking Save & Exit Protocol Editing or
Cancel & Exit Protocol Editing.
1.Edit the protocol by performing one or more of the following five tasks:
•Edit the Dwell Time and Setpoint temperature: Click in the Dwell Time or Setpoint cell, and then enter the Dwell Time or Setpoint temperature. To enter 10 seconds, type 0 followed by a colon (:), then type 10 (that is, as the time appears in the spreadsheet). Alternatively, 10 seconds can be entered as 0.10.
•Set Data Acquisition Step: Click in the Data Acquisition column, and then click Real-Time at the step you want to collect real-time data. Click Melt Curve if data from a melt curve is required.
NOTE: Ensure that every thermal protocol has at least one Data Acquisition step.
•Insert a cycle: Insert a cycle by clicking in the Insert column within the cycle row. Cycles have a blue background. The iQ5 software inserts the new cycle below the current cycle
•Insert a step: Insert a step by clicking in the Insert column within a step row. Steps have a white background. The iQ5 software inserts the new step below the current cycle. You can use the Options cell to customize whether the step or cycle is inserted before or after the current cycle/step, as well as how many steps the iQ5 software will insert when you insert a cycle
•Delete a cycle: Delete a cycle by clicking in the Delete column within a cycle row. Cycles are indicated with a blue background.
•Delete a step: Delete a step by clicking in the Delete column within a step row. Steps are indicated with a white background
2.Save the protocol by clicking Save & Exit Protocol Editing. Enter the name of the protocol in the Save As dialog box, and then click Save.
2.2 Plate Setup Quick Guide
Within the Workshop module:
1.Open the Plate Setup Editor Window using one of the following methods:
•Click Create New in the plate setup display pane to enter the Plate Setup Editor or
•Click Plate and select the desired plate setup file from the file tree directory. Doubleclick the file name to go directly to the Plate Setup Editor
or
•Click Plate and select the desired plate setup file from the file tree directory. Click the file name to open the plate setup in the bottom right section of the Workshop
-8 -
Section 2 Quick Guides
window. Click Edit to open the plate setup in the Plate Setup Editor
or
•Click Data File and select the desired data setup file from the file tree directory. Click the file name to open the plate setup associated with the data file in the bottom right section of the Workshop window. Click Edit to open the plate setup in the Plate Setup Editor
2.Enter or edit any notes about the plate setup in the Notes box.
3.Enter or edit the sample volume, seal type, and vessel type.
4.Enter or edit a name for the experiment.
5.Click Select/Add Fluorophores and select or edit the fluorophores to be used on the plate.
6.For most experiments, leave the Whole Plate Loading box checked. With Whole Plate Loading, changes made to any fluorophore within a well are extended to the other fluorophores within the well and the replicate group. If you are editing a plate, the Whole Plate Loading checkbox may be unavailable because it is not appropriate based on the current definition of the plate.
7.Click a sample type icon.
8.Select the type of replicate loading desired.
9.Click a fluorophore.
10.Click or drag across the plate to define wells with the selected fluorophore and sample type.
11.Continue defining the remaining wells that will contain the first fluorophore by changing to any other sample type icons required.
To calculate standard concentrations automatically, click Dilution Series and enter the upper or lower concentrations, and units of the standards, set the dilution factor, and click Apply Dilution Series.
12.Repeat steps 7–11 for any additional dye layers/fluorophores as required. Remember that if the Whole Plate Loading box is checked, changes made in standard concentrations will be applied to all dye layers for that well and extended to all replicates in the group.
NOTE: To delete a previously defined well, click the Delete All icon, and then click the well. To delete the selected fluorophore from a previously defined well, click the Delete Fluorophore icon, and then click the well.
13.Click Save & Exit Plate Editing.
2.3Running a Real-Time Experiment Quick Guide
After you click Run in the Workshop module, the iQ5 software opens the Initiate Run tab within the Run-Time Central module.
2.3.1 Beginning a Run
To begin a run:
1. Insert the experimental plate into the iCycler reaction module.
- 9 -
Section 2 Quick Guides
2.From the Workshop Setup window, select your desired Plate (.pts) and Protocol (.tmo) file individually, or your desired Run Set (.run).
3.Click Run.
4.Check that the desired Protocol and Plate Setup are displayed in the bottom half of the Initiate Run screen.
5.Select the type of well factors to use by selecting either:
•Use Persistent Well Factors or
•Collect Well Factors from Experimental Plate
6.Click Begin Run.
7.Name the file with a unique name in the Save Optical Data File dialog box.
8.Click OK.
2.3.2 Monitoring the Run
When the real-time PCR detection system begins the run, the iQ5 software opens the Monitor Run window. You can see the progress of the run in this window.
At the end of the run, the Run Status dialog box appears. You can choose between displaying the data in the Data Analysis module or returning to the Workshop module.
Click Yes to proceed to the Data Analysis module.
2.4 Data Analysis Quick Guides
When the iQ5 software opens, the Data Analysis module is grayed out and inactive. To analyze a data file, click the Data File tab of the Workshop module, select the data file, and then click Analyze.
The Data Analysis module consists of six tabs:
•PCR Quant
•Melt Curve/Peak
•End Point
•Allelic Disc
•Gene Expr
•Edit Plate
2.4.1 PCR Quant Tab Quick Guide
Use the PCR Quant tab to set the analysis conditions for the data file including setting the PCR baseline, setting the threshold and determining which wells to exclude or include in the experiment.
To analyze a data file:
1. Click the Data File tab in the Workshop module.
- 10 -
Section 2 Quick Guides
2.Select a data file from the file tree browser, and then click Analyze. The file opens in the PCR Quant tab within the Data Analysis module.
3.Select or deselect wells to be included in the analysis by selecting Analyze Wells. Select or deselect wells to be displayed by selecting Display Wells.
4.The iQ5 software automatically chooses the data analysis conditions including baselines and thresholds. If the data file is being opened for the first time, an automated analysis of baselines and threshold will be conducted, including every defined well. If the file was previously saved after an analysis, the last set of analysis conditions are applied again.
5.Make any manual adjustments to the threshold by clicking and dragging the green threshold line.
6.Make any manual adjustments to the baseline by right-clicking on the amplification traces plot to access the baseline threshold parameters popup window and editing User Defined options.
NOTE: You can revert to software auto calculated threshold and baseline by rightclicking on the amplification traces plot to access the baseline threshold parameters popup window and selecting Auto Calculated.
2.4.2 End Point Quick Guide
You can implement End Point analysis in two ways:
•Click Run End Point to initiate the collection of End Point data from a sample plate
•Click the End Point tab in the Data Analysis module for an existing data file
Initiating an End Point Run
1.Insert the experimental plate into the iCycler reaction module.
2.From the Workshop Setup Window Select or Create the Plate Setup from the Plate Setup tab.
3.Click Run End Point.
4.Check that the desired Protocol and Plate Setup are displayed in the bottom half of the Initiate Run screen.
5.In the Run-Time Central/Initiate Run tab, specify the setpoint for data collection, and then click Begin Run.
NOTE: You must use Persistent Well Factors for every End Point Run.
6.Name the file with a unique name in the Save Optical Data File dialog box, and then click Save.
End Point Analysis of an Newly Completed End Point Run
1.Once the real-time PCR detection system completes the run, the End Point tab is displayed.
2.Make selections for the following parameters:
•Method — Use Negatives to differentiate samples that do not amplify the target sequence from those that do amplify the target sequence.
•End Point Tolerance and Tolerance Parameter.
-11 -
Section 2 Quick Guides
3.Select the wells to analyze by clicking Analyze Wells.
4.Define the positive and/or negative controls in the Define Controls column within the End Point Analysis table.
5.Click Recalculate. The End Point Analysis table displays a positive, negative, or blank label for each unknown under the Unknowns Call column.
6.Click Reports to obtain customized reports of the End Point Analysis.
End Point Analysis of an Existing Data File
1.Within the Workshop module click Data and select your desired data file using the file tree browser.
2.Click Analyze.
3.Click the End Point tab.
4.Make selections for the following parameters:
•Method
•End Point Tolerance and Tolerance Parameter
5.Select the wells to analyze by checking Analyze Wells. Click on Analyze Selected Wells, and close the Select Wells for Analysis floating window when finished.
6.Define the positive and/or negative controls in the Define Controls column of the end point analysis table.
7.Click Recalculate. The End Point Analysis table displays a positive, negative, or blank label for each unknown under the Unknowns Call column.
8.Click Reports to obtain customized reports of the End Point Analysis.
2.4.3 Allelic Discrimination Quick Guide – For Multiplex Data Only
The Allelic Discrimination feature of the Data Analysis module is available post-run and offers flexibility for analyzing allelic discrimination data from multiplex PCR experiments. You can display samples on a scatter plot based on threshold cycle or relative fluorescence units (RFU) values at any PCR cycle. You can have the iQ5 software automatically make allele calls, or you may manually make the allele calls.
To analyze an allelic discrimination file:
1.Within the Workshop module, click Data, and select your desired data file using the file tree browser
2.Click Analyze.
3.The file opens in the PCR Quant tab of the Data Analysis module. The iQ5 software automatically chooses the Data Analysis conditions. To manually adjust these conditions, refer to the PCR Quant Tab.
4.You can select or deselect wells that will be included in the analysis by selecting the Analyze Wells checkbox.
5.Click the Allelic Disc tab in the Data Analysis module.
6.Select the Fluorophores that represent Allele1 and Allele2 in the Assign Fluorophores area.
-12 -
Section 2 Quick Guides
7.Choose between the Threshold Cycle and RFU display modes in the Display Mode area to display the allelic discrimination data.
•Threshold Cycle displays the distribution of samples on the scatter plot based on the
threshold cycle (CT). Samples that do not cross threshold will be assigned the CT value of the last cycle run in the experiment.
•RFU displays the distribution of samples on the scatter plot based on the RFU generated by each sample at the last PCR cycle number. Click the drop-down list next to the Select Cycle box to generate the scatter plot based on an RFU from a different cycle of the PCR experiment.
8.Click Automatic Call or Manual Call. Automatic Call is the default parameter.
In Automatic Call: threshold bars are positioned automatically in one of two ways:
•Based on distribution of the control wells, when at least three wells have been assigned to Control 1 and three to Control 2
•At 90 percent of the CT range or 10 percent of the RFU range on each axis if no controls are named
Manual Call is the alternative analysis mode. Adjustments may be made either in the scatter plot or in the data spreadsheet:
•Scatter plot: Make a selection in the Allele Call box. Then click and drag the cursor over the corresponding sample(s) in the scatter plot. The iQ5 software reassigns the samples to the allele call selected by the radio button, and updates the data spreadsheet accordingly
•Data spreadsheet: Click in the Call cell of the spreadsheet. A menu appears that lets you select an allele call for that sample. Once you select an allele call, the scatter plot reflects the change
NOTE: You may click and drag threshold bars directly on the plot to adjust Automatic Call assignment.
9.Click Reports in the menu to obtain customized reports for the allelic discrimination data.
2.4.4 Gene Expression Quick Guide
The iQ5 software can present expression data normalized to one or more reference genes, or, for data normalized before PCR, as a relative quantity.
Calculating Relative Quantity (dCT, ΔCT)
To calculate Relative Quantity (dCT):
In the PCR Quant screen (with a .opd data file open), assess Threshold and Baseline information for the data file and make changes if necessary.
1.Click the Gene Expr tab.
2.Make any changes to Gene and Condition (for example, Sample and Treatment) assignments in the Gene Expression Plate interface.
•Expand the Gene Expression Plate Interface view by clicking on the “+” button to make well identification and selection easier. Highlight the wells in the gene expression plate interface you wish to edit
-13 -
Section 2 Quick Guides
•Change the gene assignments in these wells by typing your desired name into the gene pull-down men, then click enter to apply the name to the selected wells
•Change the condition assignments in these wells by typing your desired name into the condition pull-down menu, then click enter to apply the name to the selected wells
NOTE: The Gene and Condition Names have a character entry limit of 15 characters
•Minimize the Gene Expression Plate interface by clicking on the “-” button to return to the standard view of the Gene Expr tab window
3.Click Relative Quantity (dCt).
4.Click Recalculate to see your results.
5.Relative Quantity results are graphed. The Data Table spreadsheet is accessed by clicking Data Table. The Data Table spreadsheet lists the Condition and Gene name, calculated expression values and CT values. Right-click to print or export this data to Excel.
NOTE: Click the Settings tab, then select Gene List to enter a specific user-defined gene reaction efficiency.
NOTE: Click the Settings tab, then select Condition List to select a particular sample as a control sample.
If you need to compare these data to results obtained in other .opd files, you will need to enable this file for Multi-file Gene Expression analysis, also called a Gene Study. To enable your file for Gene Study:
1.Click the Enable for Gene Study button.
2.Go to the File menu to save your file.
Calculating Normalized Expression (ddCT, ΔΔCT)
To calculate Normalized Expression (ddCT):
1.In the PCR Quant screen (with a .opd data file open), assess Threshold and Baseline information for the data file and make changes if necessary.
2.Click the Gene Expr tab.
3.Make any changes to Gene and Condition (for example, Sample and Treatment) assignments in the Gene Expression Plate interface.
•Expand the Gene Expression Plate Interface view by clicking on the “+” button to make well identification and selection easier. Highlight the wells in the gene expression plate interface you wish to edit
•Change the gene assignments in these wells by typing your desired name into the gene pull-down men, then click enter to apply the name to the selected wells
•Change the condition assignments in these wells by typing your desired name into the condition pull-down menu, then click enter to apply the name to the selected wells
NOTE: The Gene and Condition Names have a character entry limit of 15 characters
-14 -
Section 2 Quick Guides
•Minimize the Gene Expression Plate interface by clicking on the “–” button to return to the standard view of the Gene Expr tab window
4.Click the Settings tab, then select Gene List to set your desired reference gene(s).
5.Click Normalized Expression (ddCt).
6.Click Recalculate to see your results.
7.Normalized expression results are graphed. The Data Table spreadsheet is accessed by clicking Data Table. The Data Table spreadsheet lists the Condition and Gene name,
calculated expression values and CT values. Right-click to print or export this data to Excel.
NOTE: Use the Settings tab, then select Gene List to enter a specific user-defined gene reaction efficiency. Use the Settings tab, then select Condition List to select a particular sample as a control sample.
If you need to compare these data to results obtained in other .opd files, you will need to enable this file for Multi-file Gene Expression analysis, also called a Gene Study. To enable your file for Gene Study:
1.Click the Enable for Gene Study button.
2.Go to the File menu to save your file.
Gene Study (Multiple File Gene Expression) Quick Guide
To create a Gene Study (for analysis of multiple .opd files):
1.For each .opd file you wish to combine to a Gene Study.
a.In the PCR Quant screen (with a .opd data file open), assess Threshold and Baseline information for the data file and make changes if necessary.
b.Click the Gene Expr tab.
c.Make sure all files to be included in the Gene Study have the Enable for Gene Study button active.
d.Save each individual file.
2.From the menu toolbar select File/New/Gene Study.
3.In the Gene Expression Study Manager select Add .opd’s.
4.Select the files that you wish to add to the Gene Study. More than one file can be selected.
5.Once the files are added to the Gene Expression Study Manger, select OK.
6.Make any changes to Gene and Condition (for example, Sample and Treatment) assignments in the Gene Expression Plate interface.
•Expand the Gene Expression Plate interface view by clicking on the “+” button to make well identification and selection easier. Highlight the wells in the gene expression plate interface you wish to edit
•Change the gene assignments in these wells by typing your desired name into the gene pull-down men, then click enter to apply the name to the selected wells
-15 -
Section 2 Quick Guides
•Change the condition assignments in these wells by typing your desired name into the condition pull-down menu, then click enter to apply the name to the selected wells
NOTE: The Gene and Condition Names have a character entry limit of 15 characters.
•Minimize the Gene Expression Plate interface by clicking on the “–” button to return to the standard view of the Gene Expr tab window
7.Select either Normalized expression (ddCt) or Relative quantity (dCt).
NOTE: For Normalized Expression analysis you must first assign reference gene(s). Click the Settings tab, then select Gene List spreadsheet to set your desired reference gene(s).
8.Click Recalculate to see your results.
9.Normalized Expression results are graphed. The Data Table spreadsheet is accessed by clicking Data Table. The Data Table spreadsheet lists the Condition and Gene name,
calculated expression values and CT values. Right-click on the spreadsheet to print or export this data to Excel.
NOTE: Click the Settings tab, then select Gene List to enter a specific user-defined gene reaction efficiency.
NOTE: Use the Settings tab, then select Condition List to select a particular sample as a control sample.
2.4.5 Post-Run Plate Setup Editing Quick Guide
For post-run editing of the Plate Setup saved within a data file:
1.From the Workshop module:
•Click Data File above the directory of the home workshop. Navigate the directory until the desired data file is found. Double-click the file name to bring the file directly into the Data Analysis module
or
•Click Data File above the directory of the home workshop. Navigate the directory until the desired data file is found. Click the file name once to open the plate setup associated with the data file in the bottom right section of the Workshop window. Click Analyze to bring the data to the Data Analysis module
2.At the top of the Data Analysis window, click Edit Plate.
3.A modified version of the Plate Setup Editor will open. In this modified window, you cannot add fluorophores to or remove fluorophores from the fluorophore list. Nor may you add a previously defined fluorophore or remove a previously defined fluorophore from a well.
4.Edit any notes about the plate setup in the Notes box.
5.Edit the name of the experiment in the Experiment Name box.
6.For most experiments the Whole Plate Loading box will be checked. With Whole Plate Loading, changes made to any fluorophore within a well are extended to the other fluorophores within the well and within the replicate group. If you are editing a plate, the Whole Plate Loading checkbox may be unavailable because it is not appropriate based on the current definition of the plate.
7.Click a fluorophore.
-16 -
Section 2 Quick Guides
8.Click a sample type icon.
9.Select the type of replicate loading desired.
10.Click or drag across the plate to define wells with the selected fluorophore and sample type.
11.Continue defining the remaining wells that will contain the first fluorophore by changing to other sample type icons as appropriate.
To calculate standard concentrations automatically, click Dilution Series and enter the upper or lower concentrations and units of the standards, set the dilution factor, and click Apply Dilution Series.
12.Repeat steps 7–11 for any additional dye layers/fluorophores as required. Remember that if the Whole Plate Loading box is checked, changes made in standard concentrations will be applied to all dye layers for that well and extended to all replicates in the group.
13.Click Apply Plate Changes to make the changes. To see the effect on analysis, go to one of the other Data Analysis windows.
NOTE: To delete a previously defined well, click the Delete All icon, and then click the well. To delete the selected fluorophore from a previously defined well, click the Delete Fluorophore icon, and then click the well.
NOTE: Use the Next # checkbox to enter a particular number to assign to the next standard or sample you define.
NOTE: The original plate setup is retained with the data file and may be restored at any time by clicking Restore Original Plate.
- 17 -

Section 2 Introduction to the iQ5 Optical System Software
Section 3. Introduction to the iQ5 Optical System Software (Version 2.1)
The iQ5 software is divided into five sections, called modules. Icons representing each of the modules are always shown on the left side of the screen. The active or selected module has an orange background, whereas unselected modules have a gray background. Each module is subdivided into windows that perform a specific function for that module.
The five modules in the iQ5 software are the:
Workshop Module: This module is used to select a Plate and Protocol and Run an experiment. It is also where experimental files are selected and opened for Data Analysis. The Workshop module consists of a Setup and Plate Summary window. The Protocol, Plate, Run Set and Data File tabs can be used to select, open, edit or create files. In Setup the selected Protocol and Plate Setup can be run or the selected data file can be opened for analysis in Data Analysis.
Run-Time Central Module: This module is used to initiate and monitor experimental runs. It is accessed from the Workshop module, once the Protocol and Plate Setup have been chosen, by clicking Run or Run End Point.
Data Analysis Module: This module contains a suite of tools enabling you to conduct thorough and varied analyses of your experimental data. Within this module are screens for Quantitative, Melt Curve/Peak, End Point, Allelic Discrimination, and Gene Expression analyses. The Edit Plate screen permits post-run editing of the experimental plate setup, allowing you to correct erroneous sample type assignments. The Data Analysis module is opened automatically when you open a saved data file from the Workshop.
Calibration Module: In order to extract the best data from your real-time PCR experiment, the MyiQ2, MyiQ or iQ5 systems must be calibrated. These simple and easily-performed calibration routines are accessed in the Calibration module. There are calibration routines for Pure Dyes, Mask Alignment, Background, and Well Factors collection.
User Profile Module: This module can be used by the Administrator to add new users and to set users access/restrictions to various functions of the iQ5 software.
All users can use this module to set their personal preferences for the iQ5 software.
- 18 -
Section 4 Workshop Module
Section 4. Workshop Module
This section contains information on the following topics:
oSetup Tab (page 19)
oPlate Setup (page 21)
oProtocol (page 31)
oRun Set (pg 37)
oOpening a Data File (page 39)
oApplying Alternate Calibration Files (page 39)
The Workshop module consists of the Setup and Plate Summary tabs. For detailed information on the information contained in the Plate Summary tab, refer to section 4.2.10
4.1 Setup Tab
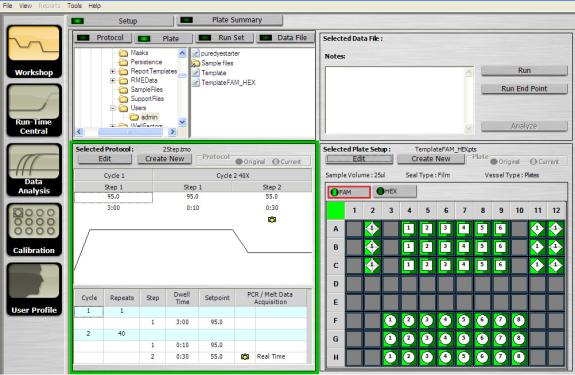
The Setup tab window (Figure 4.1) consists of four sections.
Fig. 4.1. The Setup tab.
- 19 -
Section 4 Workshop Module
4.1.1 File Browser Section
Located in the top left of the Setup window, the file browser area contains a folder tree on the left side and a file list on the right side.
The files displayed in the list depend upon the selected Setup window tab. With the Setup tab active, the displayed Setup window contains four additional tabs:
•Protocol: Used to select, edit, or create a protocol for running a real-time experiment. If the Protocol tab is selected, the iQ5 software displays protocol files, file extension
.tmo. The selected protocol appears in the Selected Protocol window
•Plate: Used to select, edit, or create a plate setup for running a real-time experiment. If the Plate tab is selected, the iQ5 software displays plate setup files, file extension .pts. The selected plate appears in the Selected Plate Setup window
•Run Set: Consists of a linked protocol and plate setup, useful when you repeat the same experiment on a regular basis. If the Run Set tab is selected, the iQ5 software displays run set files with the extension .run. The selected run set, which is a linked protocol and plate setup, is displayed in the Selected Protocol and Selected Plate Setup windows
•Data File: Primarily used to select and open a data file. You can also use this tab to run a real-time experiment using the same protocol and plate setup that were used to create the data file. The selected plate setup can be either the Original or Current (last saved) plate setup. If the Data File tab is selected, the iQ5 software displays data files created from previous experimental runs, which have the file extension .opd. The iQ5 software displays the selected data file name and any associated notes in the Selected Data File window. The Notes box is editable only after you open the data file. The iQ5 software displays the protocol and plate setup used in creating this data file in the Selected Protocol and Selected Plate Setup windows, respectively
4.1.2 Selected Protocol Area
Located in the bottom left of the Setup window, the Selected Protocol area displays the details of the protocol selected in the file browser area. The selected protocol file name appears at the top of this window. If a data file is selected, the iQ5 software displays the protocol used to create the data file and the data file name. The selected protocol is displayed both graphically and in spreadsheet format.
4.1.3 Selected Plate Setup Area
Located in the bottom right of the Setup window, the Selected Plate Setup window (Figure 4.2) displays the details of the plate setup selected in the file browser area. The selected plate setup name appears at the top of this window. If you have selected a data file, then the plate setup within the data file and the data file name are both selected in the iQ5 software.
When you select the Data File tab and a data file, the iQ5 software displays the Original and Current plate setup. The Original plate setup is the plate setup used to create the data file. The Current plate setup is the plate setup last saved with the data file. If you are looking at a data file that contains a plate setup that was edited and saved after the experimental data were collected, you can revert to the original plate setup by clicking Original in the Plate box. To go back to the present definition, click Current.
None of the information displayed on the plate in the Workshop home window can be edited from this screen.
- 20 -

Section 4 Workshop Module
Fig. 4.2. The Selected Plate Setup Area.
4.1.4 Selected Data File Area
Located in the top right of the Setup window, the Selected Data File area displays the selected data file or the run set file name that you selected in the file browser area. The Notes box displays the notes associated with either the run set or data file. Buttons in this area include:
•Run: Used to initiate a real-time PCR experimental run.
•Begin End Point: Used to initiate an End Point run.
•Analyze: Used to open a data file. You can also open the data file by double-clicking on the data file name in the file browser area.
4.2Plate Setup
The MyiQ2, MyiQ, and iQ5 Real-Time PCR Detection systems only display and analyze data from wells defined in plate setup as containing sample and at least one fluorophore. In the Plate Setup Editor window, you specify the type of sample and the fluorophores present in each well.
The Plate Setup Editor window is accessed from the main Workshop module window by clicking on either Edit or Create New in the Selected Plate Setup window (see Figure 4.1), or by doubleclicking on an existing plate setup file from the file browser folder tree.
4.2.1 Plate Setup Editor Window
The Plate Setup Editor window (Figure 4.3) is comprised of a 96-well plate layout, functions for specifying the sample type and fluorophores in each well, and a spreadsheet displaying the definition in each fluorophore for any individual well.
- 21 -
 Loading...
Loading...