Page 1

ProteinChip®
SEND ID Array
Instruction Manual
Catalog #C57-30081
For Technical Support, contact your
local Bio-Rad office, or in the U.S.,
call 1-800-4BIORAD (1-800-424-6723).
Page 2

Use
n
Peptide analysis
Introduction
Fundamental to most applications of laser desorption/ionization
mass spectrometry (LDI-MS) is the addition of matrix to the
analyte. Matrix signals interfere in the low molecular weight (MW)
range of the spectra, rendering matrix-assisted laser desorption/
ionization (MALDI)-MS and surface-enhanced laser desorption/
ionization (SELDI)-MS techniques problematic for peptide
analysis. Surface-enhanced neat desorption (SEND) technology
is unique in that the matrix is integral to the ProteinChip array
surface. The chemical noise in the spectra from the matrix is
significantly reduced when compared to addition of matrix
on-spot (particularly in the region 600 to 1,500 Da). This allows
the use of SELDI for lower MW species analysis with a reduced
amount of interfering peaks in the spectra.
The ProteinChip SEND ID array has C-18 as a functional group,
allowing the use of the array for on-chip cleanup, removing salt
and denaturant (such as urea) prior to analysis by SELDI.
The primary application of the ProteinChip SEND ID array is
peptide analysis. Successful mass determination of molecules
lower than 600 Da will be determined by how well these
molecules are ionized, desorbed, and detected by the mass
spectrometer. If laser intensity has to be increased above a
certain level to detect the molecule, the background peaks below
the 600 Da range may interfere with detection of analyte peaks.
1 © 2007 Bio-Rad Laboratories, Inc.
Page 3

Storage and Packaging
Store ProteinChip SEND ID arrays at room temperature. They
should be stored in the foil pouch in which they are supplied to
limit their exposure to light.
ProteinChip SEND ID arrays are packaged in a 12-array
ProteinChip cassette. A bioprocessor reservoir is included in the
package to protect the arrays during shipment (see Figure 1).
The spare ProteinChip cassette included to separate the
reservoirs from the arrays should be removed before use in
the ProteinChip cassette-compatible bioprocessor (catalog
#C50-30011). As the recommended protocols are rarely done
with more than a few microliters of sample, this reservoir is not
needed and should be discarded.
The arrays can be used in the cassette, or individual arrays can be
removed for processing. Take care to avoid touching the arrays.
A pair of ProteinChip array forceps (catalog #C20-10002) helps to
remove the arrays from the cassette (see Figure 2).
Fig. 1. ProteinChip cassette
and reservoir.
2 © 2007 Bio-Rad Laboratories, Inc.
Fig. 2. Removal of ProteinChip
arrays from the cassette using
array forceps.
Page 4

Technical Considerations
n
If your sample has salt or urea contamination, proceed directly
to Protocol 2. Otherwise, perform Protocol 1 first. If there is
insufficient signal after reading the arrays, a cleanup step is
recommended: Add 5 µl of 0.1% trifluoroacetic acid (TFA) to
the spot; do not agitate; remove the 0.1% TFA droplet after
30 sec; and add 5 µl 25% acetonitrile (ACN) and 0.1% TFA to
the spot. Allow to dry, and read the arrays again
n
With ProteinChip SEND ID arrays, it is essential that you mix
your sample with a solution of 50% ACN and 0.2% TFA at a
1:1 (v/v) ratio before adding to the spot
n
Ideally, the final concentration of ACN after dilution is 25%;
a final concentration of greater than 40% ACN is not compatible
with the ProteinChip SEND ID array
Factors That Can Result in Weak Signal
n
We do not recommend washing on-spot (by pipetting the
sample up and down) because this can reduce signal
n
Weak signal can be improved by adding ACN to your sample
as described in Protocol 1
n
A predominant peak at mass-to-charge ratio (m/z) 211
indicates sodium contamination, and you should perform
sample cleanup as described in Protocol 2, step 5. This
contamination can suppress sample peaks and result in
weak signal
3 © 2007 Bio-Rad Laboratories, Inc.
Page 5
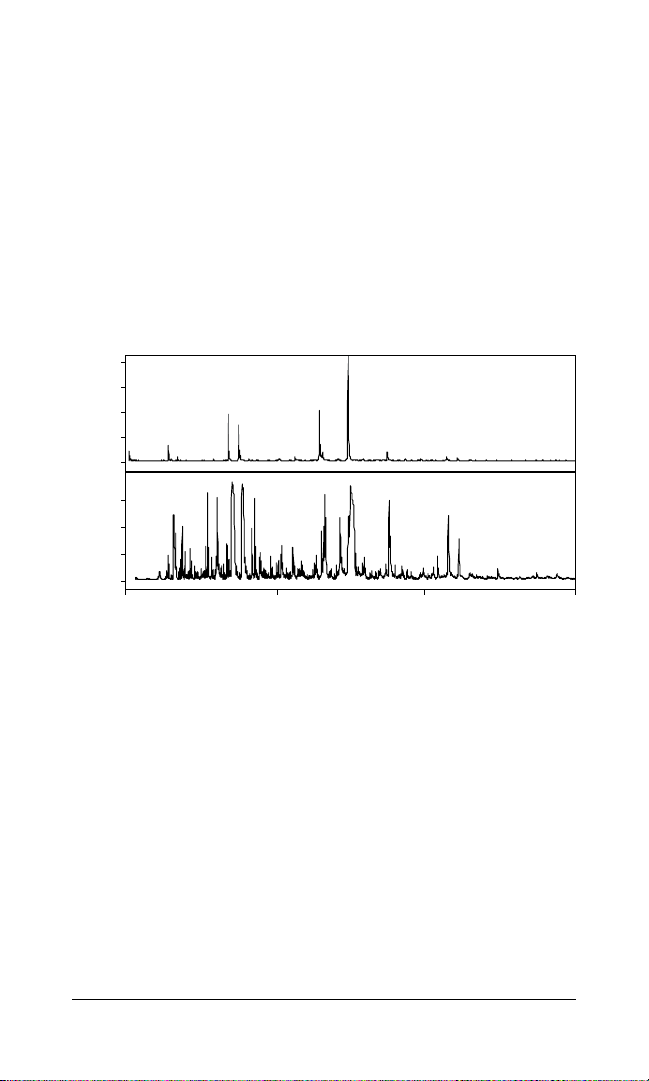
Chemical Noise
Because ProteinChip alpha-cyano-4-hydroxycinnamic acid
(CHCA) matrix is integral to the array surface, chemical noise
from matrix peaks are significantly reduced with the ProteinChip
SEND ID array when compared to standard MALDI or SELDI
analysis. Some chemical noise will be seen; the amount seen is
affected by two main factors (Figure 3):
n
Increased laser intensity will increase chemical noise
n
The higher the concentration of sample, the lower the intensity
of chemical noise peaks
A
20
15
10
Intensity
5
0
B
75
50
Intensity
25
0
0 250 500 750
171.0+H
189.0+H
Fig. 3. Comparison of chemical noise peaks due to variation in sample
concentration and laser intensity. A, minimal chemical noise is seen when
concentration of sample is high and laser intensity is low (1 pmol of sample applied
directly to the array); B, increased chemical noise is seen below 600 Da when sample
concentration is decreased to 50 fmol and increased laser intensity is necessary.
m/z, mass-to-charge ratio.
378.0+H
m/z
4 © 2007 Bio-Rad Laboratories, Inc.
Page 6

Protocol 1
The following method can be used to apply a sample directly to
the ProteinChip SEND ID array for peptide analysis.
1. Mix your sample 1:1 (v/v) with 50% ACN (v/v) and 0.2% TFA
in deionized (DI) water.
2. Add 1–2 µl of sample to each spot on the ProteinChip
SEND ID array.
3. Allow the spots to air-dry.
4. Read the arrays in the ProteinChip SELDI reader.
Protocol 2
The following method can be used to clean up a sample on
the ProteinChip array to remove salt and urea contamination
(see Figure 4). The C-18 chemistry on the surface selectively retains
peptides through their hydrophobic regions while impurities are
washed away.
1. Mix sample with 0.2% TFA at a 1:1 (v/v) ratio.
2. Apply 5 µl of 0.1% TFA to the ProteinChip SEND ID array.
Remove within 30 sec; do not agitate. Repeat.
3. Spot 2 µl of sample onto the array.
4. Incubate the samples for 10 min in a humidity chamber
at room temperature. Remove the samples.
5. Add 5 µl of 0.1% TFA to each spot. Remove within
30 sec; do not agitate.
6. Add 2 µl of 25% ACN (v/v) and 0.1% TFA in DI water to
each spot.
7. Allow the array to air-dry.
8. Read the array in the ProteinChip SELDI reader.
5 © 2007 Bio-Rad Laboratories, Inc.
Page 7
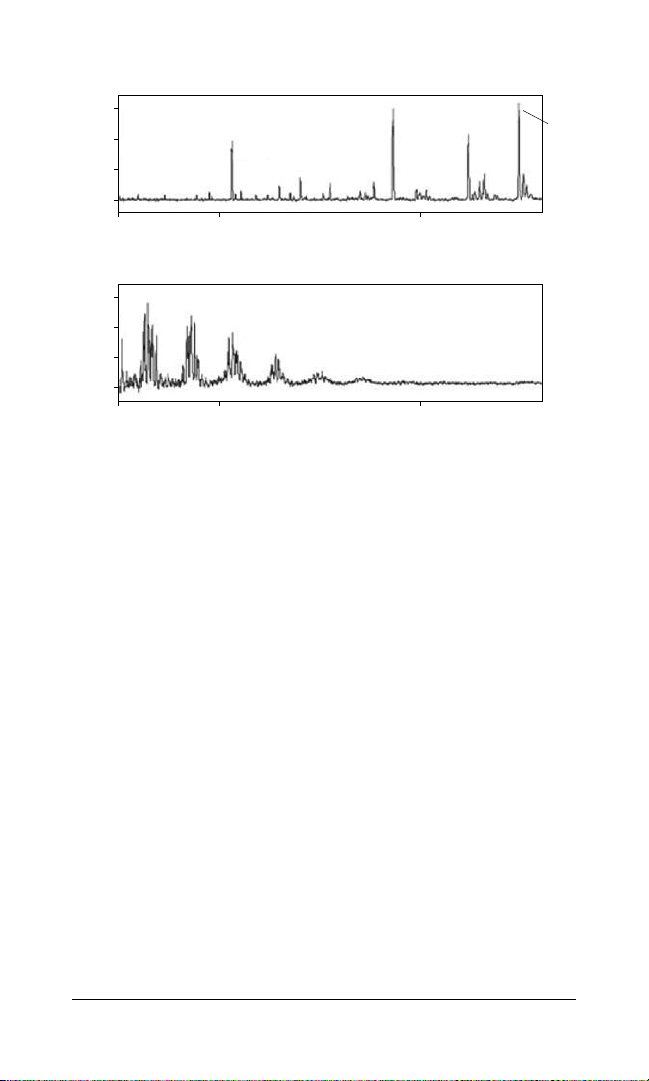
ProteinChip SEND ID array
30
20
10
Relative intensity
0
1,000 2,000
1,060.46
1,859.01
2,482.0
2,230.97
ProteinChip NP20 array with CHCA matrix
60
40
20
Relative intensity
0
1,000 2,000
m/z
Fig. 4. Clearly visible peptide peaks with the ProteinChip SEND ID array.
Upper panel, shown after on-chip cleanup of a bovine serum albumin (BSA) digest
contaminated with 2 M urea; bottom panel, only matrix peaks are visible when the
same sample is applied to a ProteinChip NP20 array with no cleanup and CHCA
added separately.
Ordering Information
Catalog # Description
C57-30081
C20-10002
ProteinChip SEND ID Arrays, A–H format, 12
ProteinChip Array Forceps, 1 pair
6 © 2007 Bio-Rad Laboratories, Inc.
Page 8

The SELDI process is covered by U.S. patents 5,719,060, 6,225,047, 6,579,719,
Life Science
Group
07-0780 1207 Sig 1207
10008249 US/EG Rev D
Bio-Rad
Laboratories, Inc.
Web site www.bio-rad.com USA 800 4BIORAD
Australia 61 02 9914 2800 Austria 01 877 89 01
Belgium 09 385 55 11 Brazil 55 21 3237 9400
Canada 905 364 3435 China 86 21 6426 0808
Czech Republic 420 241 430 532 Denmark 44 52 10 00
Finland 09 804 22 00 France 01 47 95 69 65
Germany 089 318 84 0 Greece 30 210 777 4396
Hong Kong 852 2789 3300 Hungary 36 1 455 8800
India 91 124 4029300 Israel 03 963 6050 Italy 39 02 216091
Japan 03 6361 7000 Korea 82 2 3473 4460
Mexico 52 555 488 7670 The Netherlands 0318 540666
New Zealand 0508 805 500 Norway 23 38 41 30
Poland 48 22 331 99 99 Portugal 351 21 472 7700
Russia 7 495 721 14 04 Singapore 65 6415 3188
South Africa 27 861 246 723 Spain 34 91 590 5200
Sweden 08 55512700 Switzerland 061 717 95 55
Taiwan 886 2 2578 7189 United Kingdom 0208328 2000
6,818,411, and other issued patents and pending applications in the U.S. and
other jurisdictions.
 Loading...
Loading...