Page 1
Biotechnology Explorer
™
Ligation and Transformation
Module
Instruction Manual
Catalog #166-5015EDU
explorer.bio-rad.com
For technical support call your local Bio-Rad office or in the U.S. call 1-800-424-6723.
PCR Fragment
This kit is shipped on blue ice. Open immediately upon arrival and store
reagents bags at –20°C.
Duplication of any part of this document is permitted for classroom use only.
Please visit explorer.bio-rad.com to access our selection of language translations
for Biotechnology Explorer kit curricula.
PCR Fragment
Page 2

Page 3

Table of Contents
Introduction ..................................................................................1
Kit Inventory Checklist ................................................................6
Safety Issues ................................................................................9
Background ................................................................................10
Quick Guide ................................................................................26
Instructor’s Advance Preparation ............................................32
Student Ligation Protocol..........................................................36
Student Transformation Protocol..............................................40
Appendix A Inoculating a Bacterial Colony for
Plasmid Miniprep ................................................46
Appendix B Restriction Digestion of Plasmid DNA
with Bgl II Enzyme ..............................................48
Page 4

Page 5

Introduction
Cloning is the production of multiple exact copies of a piece of DNA, usually
a gene, using molecular biology techniques. Cloning is frequently the first
step of a research project, producing enough DNA for further study.
Using the Ligation and Transformation module students can subclone virtually
any DNA fragment of interest that has been amplified using PCR. We
recommend that the DNA fragment be approximately 200–2,000 base pairs
(bp) in length for best results. Below is a typical workflow for cloning and
sequencing a gene. The steps that the Ligation and Transformation module
enable students to perform are in bold.
The Ligation and Transformation module is part of Bio-Rad’s Cloning and
Sequencing Explorer Series. The Cloning and Sequencing Explorer Series
is a sequence of individual modules that have been designed to work in
concert to give students the real world experience of a molecular biology
research workflow. The additional modules of the Cloning and Sequencing
Explorer Series can be purchased separately. Further information on the
separate modules is available in the Biotechnology Explorer™ catalog or
from explorer.bio-rad.com.
• Amplify gene of interest using PCR
1
• Purify PCR product
2
• Ligation of PCR product into pJet1.2 plasmid
• Transform ligated plasmid into bacteria
• Culture bacteria and grow minipreps
3
• Purify plasmid from minipreps
4
• Analyze plasmid by restriction digestion
• Electrophorese restriction digest reaction
5
• Sequence plasmid and analyze
6
1
GAPDH
PCR module (catalog #166-5010EDU) amplifies a fragment of the
GAPDH
gene
from a preparation of plant genomic DNA.
2
PCR Kleen™ Spin module (catalog #732-6300EDU) purifies 25 PCR products.
3
Microbial Culturing module (catalog #166-5020EDU) contains all required reagents for
culturing bacteria for transformation using the Ligation and Transformation module.
4
Aurum™ Plasmid Mini Purification module (catalog #732-6400EDU) contains reagents to
purify plasmid DNA from 100 minipreps.
5
Electrophoresis modules contain reagents to analyze plasmid restriction digests.
6
Sequencing and Bioinformatics module (catalog #166-5025EDU) is designed to allow
sequencing and bioinformatics analysis of plasmids generated using the Ligation and
Transformation module.
1
Page 6

Using the Ligation and Transformation module, students will clone a gene
of interest. Prior to starting this laboratory activity, students must have
already amplified a gene of interest using polymerase chain reaction
(PCR) and subsequently purified the PCR product to remove excess
primers, nucleotides, and DNA polymerase, which would otherwise interfere
with subsequent experiments. Students can then use the Ligation and
Transformation module to ligate the DNA fragment into the pJet1.2 blunted
vector, which encodes
amp
r
, an ampicillin-resistance gene. Following ligation,
students will perform transformation to introduce the plasmid into living
bacterial cells.
The pJet1.2 blunted vector enables positive selection of plasmids with the
desired insert due to the disruption of
eco47IR
, an otherwise lethal gene,
that allows growth of successful transformants. Bacteria are then plated
and incubated overnight at 37°C on the selective medium containing
ampicillin and isopropyl b-D-1-thiogalactopyranoside (IPTG), which is
added to increase expression of the
amp
r
gene. Since transformed cells
express an ampicillin-resistance gene, they will grow and divide, each
forming a colony on the plate that is the product of a single transformation
event.
The bacteria containing the cloned gene can be grown in liquid growth
medium and the plasmid containing the insert can be purified from the
bacteria. The pJet1.2 plasmid contains a Bgl II restriction enzyme recognition
site on either side of the insertion site. Using the Bgl II enzyme students
will analyze the cloned plasmid by restriction enzyme digestion and analyze
their digests by agarose gel electrophoresis to confirm the presence of an
insert and determine its size. The resulting fragment can then be compared
to the size of the PCR fragment ligated into the plasmid. Finally, the DNA
fragment can be then sequenced to determine the exact order of
nucleotides in the DNA molecule.
What Skills Do Students Need to Perform this Laboratory
Activity?
This laboratory activity assumes that students and instructors have basic
molecular biology and microbiology skills, such as proper pipeting
techniques, pouring and streaking agar plates and performing agarose gel
2
Page 7

electrophoresis. In addition, students must understand the principles of
PCR and be able to perform PCR reactions. Bio-Rad’s Biotechnology
Explorer program has a full range of kits to help teach basic skills in individual
laboratories.
What Is the Timeline for Completing the Ligation and
Transformation Protocol?
Before starting this activity, students must have already amplified a gene
of interest using PCR. In addition, the PCR product should be purified to
remove components of the amplification reaction that would otherwise
interfere with the ligation step.
The amount of time it takes to complete the ligation and transformation
protocols depends greatly on the level of your students and whether
additional/optional techniques and analyses are performed in addition to the
basic protocol. Steps using the Ligation and Transformation module are
highlighted in bold. Additionally, there are a few incubation steps that add
to the number of days it takes to complete the laboratory activity. A rough
guide is provided on pages 4 and 5.
3
Page 8
When Activity to Complete Duration
At least 1 day prior to Run a PCR reaction in thermal 3–4 h
starting the Ligation cycler to amplify a
and Transformation gene of interest
module Electrophorese the PCR 1 h
products (optional)
Purify PCR products 0.5 h
At least 3 days prior to Prepare LB and LB Amp 0.5 h
the transformation step IPTG agar plates
At least 2 days prior to Prepare LB broth 0.5 h
the transformation step Streak
E. coli
on a starter LB 5 min
agar plate
Grow
E. coli
starter plate at 37°C 16+ h
As late as possible the Inoculate starter culture 5 min
day before the Incubate starter culture at
transformation step 37°C in a shaking 8+ h
water bath or incubator
Day of ligation step Ligate PCR product 1 h
Note: Bolded steps use reagents from the Ligation and Transformation
module.
4
Page 9

When Activity to Complete Duration
Immediately following Transform
E. coli
with ligation 1 h
ligation or during the mixture and plate bacteria on
next laboratory activity LB Amp IPTG agar plates
Incubate transformed bacteria 16+ h
at 37°C
Next day after the Analyze results 0.5 h
transformation step Grow bacterial colony in LB Amp 16+ h
broth for miniprep
Day after growing Perform miniprep plasmid 1 h
bacterial culture for purification to isolate
miniprep plasmid carrying insert
Next laboratory activity Digest plasmid DNA with 1 h
Bgl II restriction enzyme
Analyze digest by agarose gel 1 h
electrophoresis
Prepare the DNA insert for 0.5 h
sequencing
Note: Bolded steps use reagents from the Ligation and Transformation
module.
5
Page 10

Kit Inventory Checklist
This section lists equipment and reagents necessary to perform the ligation
and transformation protocol in your classroom or teaching laboratory. Each
kit contains sufficient materials for 12 student workstations, 12 ligation
reactions, and 24 transformations. We recommend that students are
teamed up – two to four students per workstation. Please use the checklist
below to confirm inventory.
Kit Components Number/Kit (✔)
T4 DNA ligase, 10 µl 1
❒
Ligation reaction buffer (2x concentration), 100 µl 1 ❒
Proofreading polymerase, 10 µl 1 ❒
pJet1.2 blunted vector, 10 µl 1 ❒
Sterile water, 1 ml 1 ❒
Bgl II restriction enzyme, 50 µl 1 ❒
10x Bgl II reaction buffer, 1 ml 1 ❒
Isopropyl b-D-1-thiogalactopyranoside (IPTG), 1 M, 0.1 ml 1 ❒
Transformation reagent A, 1.25 ml 4 ❒
Transformation reagent B, 1.25 ml 4 ❒
C-growth medium, 30 ml 1 ❒
Microcentrifuge tubes, clear, 1.5 ml 30 ❒
Microcentrifuge tubes, multicolor, 2.0 ml 120 ❒
6
Page 11

Required Accessories Number/Kit (✔)
PCR product (previously amplified and 1 per team
❒
purified by students)
Microbial Culturing module (catalog #166-5020EDU)* 1
❒
containing the following:
• LB broth capsules (each for making 50 ml of LB broth) 12
❒
• LB nutrient agar powder (to make 500 ml) 1 pouch ❒
• Ampicillin 2 vials ❒
•
E. coli
HB101 K-12, lyophilized bacteria 1 vial ❒
• Culture tubes, sterile, 15 ml 75 ❒
• Petri dishes, sterile 40 ❒
• Sterile inoculating loops 80 ❒
Variable speed microcentrifuge (catalog #166-0602EDU) 1 ❒
Shaking water bath or shaking incubator (37°C) 1 ❒
Water bath (catalog #166-0504EDU), heating block, 1 ❒
(catalog #166-0562EDU)or incubator (70°C)
Adjustable-volume micropipet
0.5–10 µl (catalog #166-0505EDU) 12
❒
20–200 µl (catalog #166-0507EDU) 12 ❒
100–1,000 µl (catalog #166-0508EDU) 12 ❒
Pipet Tips
0.5–10 µl (catalog #223-9354EDU) 1 box
❒
2–200 µl (catalog #223-9347EDU) 1 box ❒
100–1,000 µl (catalog #223-9350EDU) 1 box ❒
Ice bath 1 ❒
Parafilm sealing film 1 ❒
Marking pens 1 ❒
*Note: Standard microbiological reagents may be used in place of the
Microbial Culturing module (see Instructor’s Advanced Prep section for
requirements). Any
E. coli
strain commonly used for transformation (for
example, DH5a, DH10, JM107) may be used in place of
E. coli
HB101.
7
Page 12

Optional Accessories
Vortex mixer (catalog #166-0610EDU)
Vacuum source
Agarose electrophoresis equipment
GAPDH
PCR module (catalog #166-5010EDU)
PCR Kleen™ Spin Purification module (catalog #732-6300EDU)
pGLO™ Plasmid, 20 µg (catalog #166-0405EDU)
Aurum™ Plasmid Mini Purification Module (catalog #732-6400EDU)
Electrophoresis reagents:
Small Ethidium Bromide DNA Electrophoresis Reagent Pack (catalog
#166-0451EDU)
Small Fast Blast™ DNA Electrophoresis Reagent Pack (catalog
#166-0450EDU)
Sample Loading Dye, 5x, 1 ml (catalog #166-0401EDU)
EZ Load™ 500 bp Molecular Ruler (catalog #170-8354EDU)
Sequencing and Bioinformatics module (catalog #166-5025EDU)
Refills Available Separately
Ligation module reagent refill (catalog #166-5016EDU)
Includes T4 DNA ligase, 2x ligation reaction buffer, proofreading
polymerase, pJet1.2 blunted vector, sterile water
Bgl II reagent refill (catalog #166-5018EDU)
Includes Bgl II restriction enzyme and 10x Bgl II reaction buffer
Transformation module reagent refill (catalog #166-5017EDU)
Includes transformation reagent A, transformation reagent B, 1 M IPTG,
C-growth medium
8
Page 13

Storage Instructions
The kit is shipped on blue ice. Open immediately upon arrival and store
reagent bags immediately at –20°C.
Safety Issues
Eating, drinking, smoking, and applying cosmetics are not permitted in the
work area. Wearing protective eyewear and gloves is strongly recommended.
Transformation reagent B contains dimethyl sulfoxide (DMSO, CAS #67-68-5),
an organic solvent. Handle with care and follow standard laboratory
practices, including wearing eye protection, gloves, and a laboratory coat
to avoid contact with eyes, skin, and clothing. If the solution comes into
contact with gloves, change the gloves. DMSO passes directly through
latex gloves, readily penetrates skin, and may result in the absorption of
toxic materials and allergens dissolved in the solvent. After handling, wash
hands and any areas that came into contact with the solution thoroughly.
Refer to MSDS for complete safety information.
Ampicillin may cause allergic reactions or irritation to the eyes, respiratory
system, and skin. In case of contact with eyes, rinse immediately with
plenty of water and seek medical advice. Wear suitable protective clothing.
Ampicillin is a member of the penicillin family of antibiotics. Those with
allergies to penicillin or any other member of the penicillin family of
antibiotics should avoid contact with ampicillin.
The
E. coli
HB101 K-12 strain is not pathogenic. However, handling of
E. coli
HB101 K-12 requires the use of standard microbiological practices.
These practices include, but are not limited to, the following: (1) work surfaces
are decontaminated once a day after any spill of viable material; (2) all contaminated liquid or solid wastes are decontaminated before disposal; (3)
persons should wash their hands: (a) after they handle materials involving
organisms containing recombinant DNA molecules, and (b) before exiting
the laboratory; (4) all procedures should be performed carefully to minimize
the creation of aerosols and; (5) mechanical pipetting devices should be
used—mouth pipetting is prohibited.
9
Page 14

Background
Cloning
Cloning is the production of multiple exact copies of a piece of DNA,
usually a gene, using molecular biology techniques. Cloning is frequently
the first step used in a research project, producing enough DNA for further
study. Once a gene or part of a gene has been amplified using PCR, the
next step is to insert the DNA into a plasmid or cloning vector so that the
DNA fragment can be propagated.
Plasmids as Cloning Vectors
Many cloning vectors are derived from bacterial plasmids. Plasmids are
circular extrachromosomal DNA molecules, usually around 2,000–100,000
base pairs (bp) long, although most plasmids used in cloning are
2,000–10,000 bp. Bacteria may naturally contain many copies of a single
plasmid, or single copies of others. Plasmids are able to replicate
independently of the host DNA and most plasmids carry at least one gene.
Frequently these genes code for a factor or function that helps the bacteria
survive. For example, resistance to the antibiotic ampicillin is conveyed by
a plasmid carrying an ampicillin-resistance gene. Plasmids are capable of
being transferred from one bacterium to another. These characteristics
have resulted both in wonderful new uses for plasmids (such as their use in
cloning, making many of the techniques of molecular biology possible) and
in the emergence of dangerous pathogenic organisms (namely bacteria
resistant to multiple antibiotics).
Plasmids thus already have many of the characteristics needed for use as
cloning vectors, and other useful features have been added through genetic
engineering. A wide variety of vectors are available commercially for various
applications. A plasmid designed to clone a gene is different from a plasmid
designed to express a cDNA (complementary DNA) in a mammalian cell
line, which is different again from one designed to add a tag to a protein for
easy purification. The primary characteristics of any good vector include:
10
Page 15

• Self-replication — Plasmids have an origin of replication so they can
reproduce independently within the host cell; since the origin of
replication engineered into most cloning vectors is bacterial, the plasmid
can be replicated by enzymes already present in the host bacteria
• Size — Most bacterial vectors are small, between 2,000–10,000 bp
long (2–10 kilobases or kb), making them easy to manipulate
• Copy number — Each plasmid is found at specific levels in its host
bacterial strain. A high copy number plasmid might have hundreds of
copies in each bacterium, while a low copy number plasmid might
have only one or two copies per cell. Cloning vectors derived from
specific plasmids have the same copy number range as the original
plasmid. Most commonly used cloning vectors are high copy number
• Multiple cloning site (MCS) — Vectors have been engineered to contain
an MCS, a series of restriction sites, to simplify insertion of foreign
DNA into the plasmid. An MCS may have 20 or more different enzyme
sites, each site unique both in the MCS and in the plasmid. This
means that for each restriction site included in the MCS, the
corresponding restriction enzyme will cut the plasmid only at its single
site in the MCS
• Selectable markers — Plasmids carry one or more resistance genes
for antibiotics, so if the transformation is successful (that is, if the
plasmid enters and replicates in the host cell), the host cell will grow in
the presence of the antibiotic. Commonly used selectable markers are
genes for resistance to ampicillin (
amp
r
), tetracycline (
tet
r
), kanamycin
(
kan
r
), streptomycin (
sm
r
), and chloramphenicol (
cm
r
)
11
Page 16

• Screening — When bacteria are being transformed with a ligation reaction,
not all of the religated vectors will necessarily contain the DNA fragment
of interest. To produce visible indicators that cells contain an insert,
vectors frequently contain reporter genes, which distinguish them from
cells that do not have inserts. Two common reporter genes are
beta-galactosidase (b-gal) and green fluorescent protein (GFP)
Some newer plasmid vectors use positive selection, in which the
inserted DNA interrupts a gene that would otherwise be lethal to the
bacteria. If foreign DNA is not successfully inserted into the MCS, the
lethal gene is expressed and transformed cells die. If the foreign DNA
is successfully inserted, the lethal gene is not expressed and the
transformed bacteria survive and divide. Positive selection eliminates
the need for reporter genes, as only cells transformed with vector
containing an insert will survive
• Control mechanism — Most vectors have some control mechanism for
transcription of the antibiotic resistance or other engineered gene. One
of the best-known control mechanisms is the lac operon (an operon is
a group of genes). When lactose (a sugar) is absent in the cell, the lac
repressor protein binds to the lac operon, preventing transcription of the
gene. When lactose is present in the cell, it binds to the lac repressor
protein, causing the repressor protein to detach from the operon. With
the repressor protein no longer bound to the operon, RNA polymerase
can bind and the genes can be transcribed. In this system, lactose acts
as an inducer. (A closely related compound, (IPTG), is often used in the
laboratory as an artificial inducer.) Genes from the lac operon have
been engineered into many cloning vectors
• Size of insert — Plasmid vectors have limitations on the size of inserts
that they can accept, usually less than the size of the vector. Other
vectors have been developed for use if the target DNA is larger, for
example, lambda phage (inserts up to 25 kb), cosmids (inserts up to
45 kb), bacterial artificial chromosomes (BACs; inserts from 100–300 kb),
yeast artificial chromosomes (YACs; inserts from 100–3,000 kb), and
bacteriophage P1 (inserts up to 125 kb)
12
Page 17

DNA Ligation
Ligation is the process of joining two pieces of linear DNA into a single
piece through the use of an enzyme called DNA ligase. DNA ligase catalyzes
the formation of a phosphodiester bond between the 3'-hydroxyl on one
piece of DNA and the 5'-phosphate on a second piece of DNA.
The most commonly used DNA ligase is T4 DNA ligase (named because it
originated in a bacteriophage named T4). There are several ways that the
efficiency of DNA ligation can be optimized. First, like any enzyme, there
are conditions that are optimal for ligase activity:
• T4 DNA ligase requires ATP and magnesium ions for activity
• The concentration of vector and insert DNA in solution must be high for
efficient ligation
• The molar ratio of insert to vector DNA should be approximately equal,
although the optimal ratio may not be 1:1
13
Page 18
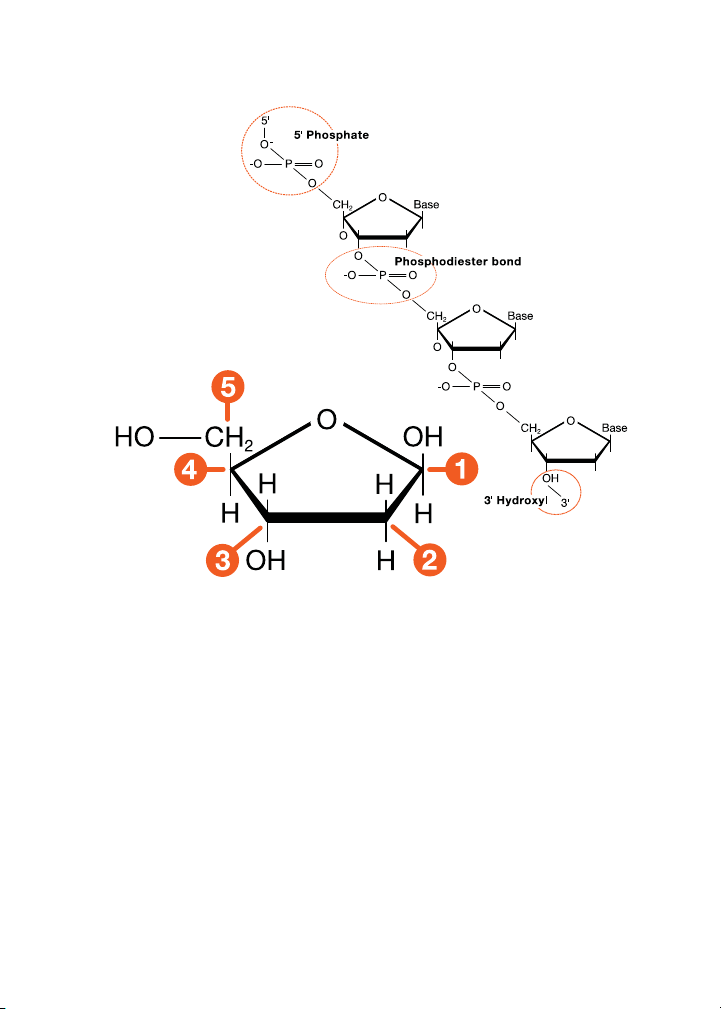
Chemical structure of deoxyribose sugar and deoxyribose nucleic acid (DNA).
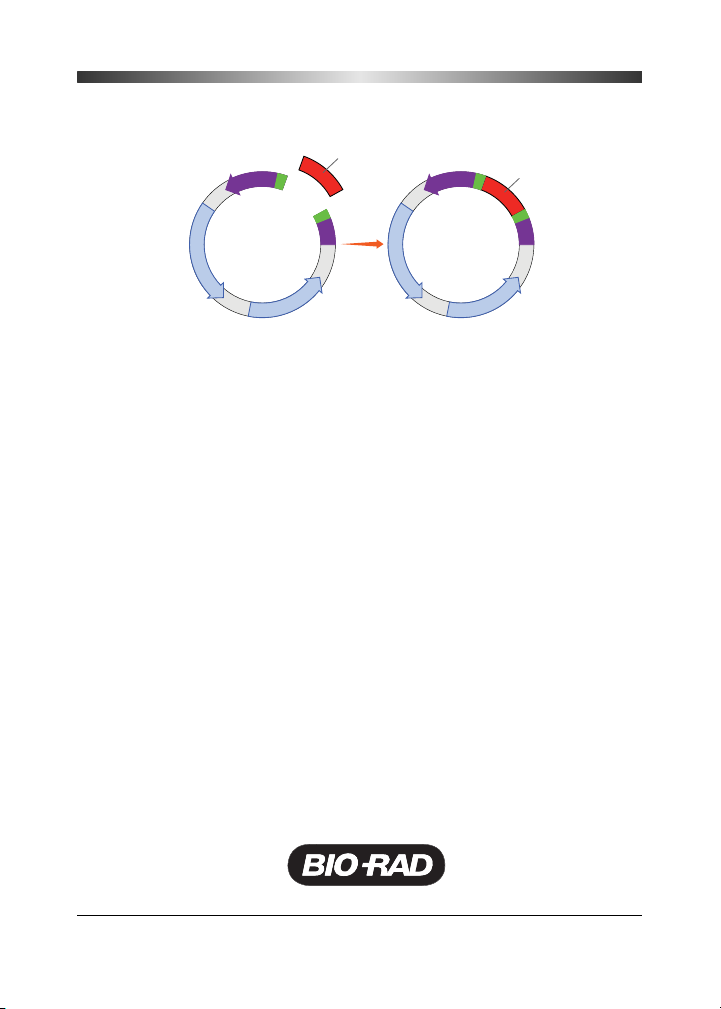
Ligation is used to join vector DNA and insert DNA. There are two ways in
which DNA can be ligated into a cloning vector, one using DNA with
so-called sticky ends and the other using DNA with blunt ends. Unlike DNA
with blunt ends, DNA with sticky ends has one or more unpaired bases at
its ends that do not have complementary bases on the other strand of the
double helix. When a DNA fragment is generated by
Ta q
DNA polymerase
by a process like PCR, it typically has sticky ends with a single adenosine
(A). When a DNA fragment is generated by cutting a piece of DNA with a
restriction enzyme (an enzyme that cuts both strands of double-stranded
DNA), it may have either sticky ends or blunt ends, depending on the
restriction enzyme.
14
Page 19

DNA ligation with sticky ends — To prepare a cloning vector for ligation
with insert DNA, it is cut with a restriction enzyme within the MCS, opening
it to receive the inserted DNA. If the insert has sticky ends, that is, overhangs
on the end of the DNA strands, then the vector should be cut with the
same enzyme, producing sticky ends that will be complementary to the
ends of the insert DNA. For example, if the insert DNA has been prepared
by cutting it at both ends with Bgl II, then the vector would also be cut with
Bgl II. Having complementary sticky ends improves the efficiency of ligation,
whereas mismatches in the sequences reduce efficiency.
Because the sticky ends on the vector and the insert are complementary,
when they come into contact during the ligation reaction they will base-pair
with each other using hydrogen bonds. (The base-paired sticky ends of the
insert and vector are not stably associated, and they can dissociate prior
to ligation.) While the insert and vector are associated, T4 DNA ligase
forms a phosphodiester bond, covalently linking the two pieces of DNA.
Actually, there are two ligations. The first ligation is intermolecular, between
one end of the vector and one end of the insert, resulting in a linear DNA
molecule. The second ligation is intramolecular, circularizing the molecule.
Sticky end digestion.
15
Page 20

One advantage to sticky-end ligation is that it makes directional
cloning possible. If it is desirable to have the insert in one orientation
only (for instance, in the A Æ B direction in the vector, but not in the
B Æ A direction), then the insert and vector can both be digested with two
different restriction enzymes so that their ends are asymmetric. This is important
if the DNA insert is a cDNA encoding a protein to be expressed in the
transformed cell. When this is done, only the complementary ends will ligate
and the insert will have a single orientation in the ligation products.
Blunt end digestion.
16
Page 21

DNA ligation with blunt ends — Blunt-end ligation, in which both the
inserted DNA and the vector have blunt ends, has an advantage compared
to sticky-end ligation in that all DNA ends are compatible with all other
ends. In other words, it is not necessary to cut the vector and insert with
the same restriction enzymes to get complementary overhangs as for
sticky-end ligation. Vectors used for blunt-end ligation have a blunt-ended
ligation site in the MCS. They still have an MCS, as the restriction enzyme
sites are very useful for subsequent manipulation of the inserted DNA.
LIgation of PCR fragment into vector.
In PCR,
Ta q
DNA polymerase adds a single nucleotide to the 3'-end of the
PCR product, usually an A. Since this A overhang would prevent blunt-end
ligation, it must be removed prior to ligation. Treating the PCR product with
a proofreading DNA polymerase removes the 3'-A, leaving blunt ends
ready for ligation. (Not all thermally stable DNA polymerases used in PCR
leave an A overhang; some polymerases like
Pfu
DNA polymerase have
proofreading ability.)
Features of the pJet1.2 blunted vector
The pJet1.2 blunted vector has several features that make it a good choice
for this laboratory activity:
• It is a vector designed for blunt-end cloning and is already linearized
with blunt ends
17
PCR Fragment
PCR Fragment
Page 22

• Its MCS has restriction enzyme sites that can be used for later
manipulation of the DNA
• It is a high copy number plasmid
• It contains the b-lactamase gene,
amp
r
, which confers resistance to
ampicillin
• It contains the
eco47IR
gene, which allows positive selection of
transformants. This gene codes for the
Eco47I
restriction enzyme, and
when the enzyme is expressed, it is toxic to
E. coli
. When the
eco47IR
gene is disrupted by the insertion of DNA into the cloning site, the gene
will no longer be expressed and the transformed cells will grow and
divide on selective media
• It is 2,974 bp long and its sequence is readily available — the accession
number for pJet1.2 is EF694056
pJet1.2 blunted vector.
18
Page 23

Products of Ligation
Ligation is a very inefficient process; from millions of vectors and inserts,
only 1–100 are expected to ligate together as desired and lead to growth
of colonies from the desired transformants. There are several possible
products from a ligation with a vector such as the pJet1.2 blunted vector:
• Self-ligation of the vector — A self-ligated vector, without any DNA
inserted in the MCS, should have an intact lethal gene. The product of
the lethal gene should kill any bacteria transformed by these vector-only
constructs. (Note: The eco47IR gene is not 100% lethal, and so some
bacteria may grow, resulting in tiny satellite colonies, which may be
observed in no-insert ligation controls)
• Ligation of a vector with primer-dimers or other short DNA fragments
from the PCR reaction — Even if these fragments are a small proportion
of the total fragments available for ligation, small fragments ligate more
easily than larger fragments
• Ligation of a vector with multiple inserts — Since the inserts all have
the same blunt ends, they can ligate to each other and also ligate to
the vector, giving a product with multiple inserts in a row. The number
of these products can be reduced by controlling the molar ratio of insert
to vector. If the ratio is high (that is, if there are many more insert
molecules than vector), then it is more probable that inserts will ligate
to each other rather than ligate to the vector. So the molar ratio can be
an important factor in setting up ligation reactions
• Self-ligation of insert — It is possible for the insert molecules to
self-ligate, forming closed circles. Since these molecules do not have
any of the vector genes, they will not be able to replicate, so they are
not of consequence
• Ligation of one insert into a vector — This is the desired result, and in
ligation to a vector such as pJet1.2 blunted vector, there is no
directionality in the cloning. In other words, the insert can ligate into the
vector in either direction, because all of the ends are identical. So the
products should be a 50:50 mix, with half of the inserts in one
orientation and the other half in the reverse orientation
19
Page 24

Possible ligation products.
Transformation
Once a gene or part of a gene has been amplified using PCR and ligated
into a plasmid, the next step in cloning is transformation, introducing the
plasmid into living bacterial cells so that it can be replicated. Heat shock
transformation and electroporation are the two methods of bacterial
transformation commonly used in the laboratory. Both methods require
competent cells, bacterial cells that can take up DNA. Not all cells are
naturally competent. For example some species, such as
Bacillus subtilis
,
can be easily transformed, but for other species, such as
Escherichia coli
,
only a small number of cells in a culture may be able to take up DNA.
Competent cells may be prepared in the laboratory or purchased
commercially.
20
No interruption of
lethal gene so
transformed
bacteria will die
Can be minimized
by controlling
molar ratio of
inserts
Product cannot
replicate
Desired product –
insert can be in
either orientation
Self-ligation
of vector
Multiple
inserts
Self-ligation
of inserts
Ligation
of vector
and insert
PCR Fragment
PCR Fragment
PCR Fragment
PCR Fragment
PCR Fragment
PCR Fragment
Page 25

• Heat shock is the most easily accomplished transformation method, as
it does not require any equipment other than a water bath. Plasmid
DNA and heat-shock competent cells in calcium chloride are mixed
together and incubated on ice for several minutes. Although the
mechanism is not fully understood, calcium chloride causes DNA to
bind to the bacterial cell wall. The cells are then subjected to a brief
heat shock resulting in the uptake of DNA into the bacteria. Traditionally
bacteria are heat shocked by incubation at 42°C for 50 sec. However in
this laboratory the bacteria are heat shocked by spreading the ice cold
bacteria directly onto warm agar plates at 37°C. Cells intended for heat
shock transformation must be in the exponential growth phase to be
highly competent
• Electroporation is also commonly used for transformation, and its
mechanism of enabling DNA uptake is somewhat better understood
than heat-shock transformation. When bacterial cells are subjected to a
brief electrical shock, small pores open in their cell wall, allowing DNA
to enter the cells. For electroporation, electrocompetent bacteria and
plasmid DNA are mixed and placed in a special type of cuvette, a square
test tube with metal electrodes on two sides (see figure on page 22).
The cuvette is placed in an instrument called an electroporator that
delivers an electrical charge of specific strength and duration to the
cells. The electricity travels through the cells between the two electrodes,
which is why electrocompetent cells must be prepared in a solution of
very low ionic strength. For electroporation to be successful, the cells
themselves must carry the current across the gap between the electrodes.
If there are many ions (like Na
+
) in the solution, the ions will carry the
current instead of the cells, causing the cells to overheat and die
21
Page 26
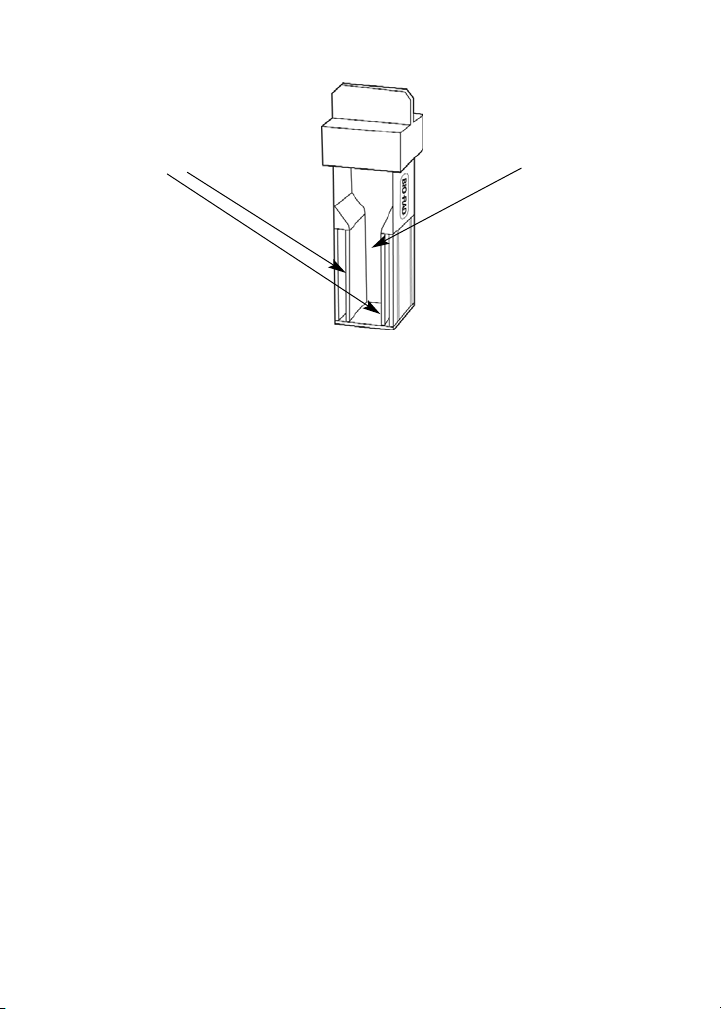
Bio-Rad electroporation cuvette.
There are ways to increase the number of competent cells in a bacterial
culture. To prepare competent cells for heat shock transformation, the
bacteria must be washed to remove the growth medium, then resuspended
in an ice cold calcium chloride solution. For electroporation, the cells must be
washed repeatedly in chilled buffer, and resuspended in a chilled sterile
solution that has very low ionic strength. In both cases, the cells must be
in solution and at a high concentration for transformation to be successful
and the cells must be kept cold at all times prior to transformation. The
cells are extremely fragile at this stage and the cold temperature keeps
them inert. If they are warmed up in transformation solution, they will start
to die. The transformation protocol also requires the cells to be cold; even
though the exact process of transformation is still not fully understood, it is
presumed that the cold hardens the cell membranes of the bacteria and
the heat shock rapidly melts or breaks the membranes, which allows the
DNA inside the cells.
Since bacteria have defense mechanisms that use restriction enzymes to
degrade foreign DNA, only mutant strains that no longer have restriction
activity can be used for transformation. Normal bacteria would degrade the
plasmid DNA as soon as it enters the cell. Mutant strains for transformation
are widely available, for example, the
E. coli
HB101 K-12 strain recommended
for use in this activity.
What Happens After Transformation
After either transformation method, the cells are usually incubated in
nutrient medium for up to 1 hour to allow them to recover from the stress
22
Chamber for cells
Metal electrodes
Page 27

of the transformation (either the heat shock or the electrical pulse) and
begin to express the genes on the plasmid (such as an antibiotic resistance
gene), although this step may be omitted. The cells are plated on a selective
medium for growth, usually agar plates containing nutrient medium and the
antibiotic for which resistance is carried by the plasmid. For example, if the
plasmid contains the
amp
r
gene, providing resistance to ampicillin, the
agar plates should also contain ampicillin. This means that only bacteria
that have been successfully transformed and now carry the plasmid will be
able to survive and divide on the ampicillin-containing plates. The plasmid
will replicate in the bacterial cells (using the host cell’s replication machinery)
and, as the bacteria divide, the plasmids will be passed on to their offspring.
The plasmid that is used in this laboratory activity, pJet1.2 blunted vector,
contains the
amp
r
gene. IPTG is added to the selective medium to artificially
increase expression of the
amp
r
gene, which is under the control of the
lac
operon (normally regulated by lactose); this approach increases transformation
efficiency with this plasmid.
Even though the efficiency of bacterial transformation can be optimized by
using competent cells and by determining the best experimental conditions
for transformation and selection, transformation is still an inefficient
process, with only a small percentage of available DNA being taken into a
small percentage of the competent bacteria. After the bacteria are plated
on the selective medium, the antibiotic will prevent the untransformed cells
from growing. Transformed cells, however, will grow and divide, each
forming a colony on the plate that is the product of a single transformation
event. In other words, all the cells in each colony are clones, hence the
origin of the term cloning.
23
Page 28

Process of bacterial transformation. Competent
E. coli
are transformed with plasmid DNA.
Only a few bacteria take up the plasmid DNA. Bacteria are then plated on selective media and
incubated overnight. Only bacteria that contain the plasmid will grow and form colonies. Bacteria
colonies are then picked and grown for use in plasmid minipreps.
Minipreps of Plasmid DNA
Once a plasmid has been introduced into competent bacterial cells and
the cells have grown into colonies on a medium selective for cells containing
the plasmid, the next step is to prepare a miniprep of the plasmid DNA in
preparation for sequencing or further experiments. The three steps are: 1)
growing cells in liquid culture; 2) purifying the plasmid DNA from the culture;
and 3) performing a restriction digest on the purified DNA to determine
whether the DNA insert in the plasmid is the expected size. To start the
liquid culture, cells from an isolated bacterial colony are placed in selective
medium (nutrient broth with an antibiotic to which the plasmid provides
resistance). This placing of the cells into the medium is called inoculation.
It is important to choose a single isolated colony from the plate, so that the
liquid culture will contain cells that all have the same plasmid. If cells from
more than one colony are used for inoculation, the miniprep may contain
multiple plasmids and the mixed DNA will not be useful for sequencing or
further experiments.
Restriction digestion of plasmid DNA
Before proceeding with further experiments using the purified plasmid
DNA, it is important to verify that the isolated plasmid contains the insert
of interest using restriction digestion followed by gel electrophoresis.
Plasmid DNA
24
Competent E. coli
Transformation
Transformed cell
Transfer to
selective media
Incubate at
37°C overnight
Pick colonies
Page 29

Looking back at earlier steps in the experiment, a gene or portion of a
gene was ligated into the plasmid vector. From previous work, the size of
this insert should be known. By digesting a small portion of the miniprep
DNA with a specific restriction enzyme such as Bgl II (see below), the
insert should be cut out of the vector. Running the products of the restriction
digestion on an agarose gel should give two DNA bands, one the size of
the vector and the other the size of the inserted DNA.
If there are more than two bands in any digest, it may mean that the insert
contains a Bgl II site. Does the size of the excised bands add up to the
size of the original PCR fragment? Alternatively, two similarly sized fragments
may indicate that a mixed culture was used to start the miniprep, instead
of an isolated colony.
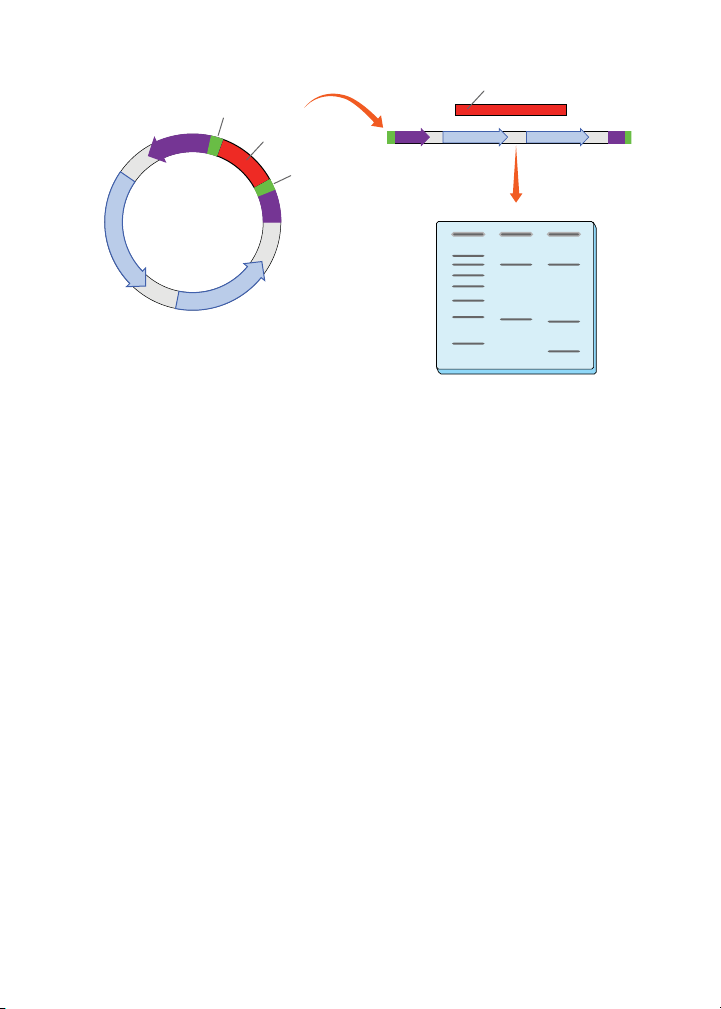
Restriction enzyme digestion analysis of plasmid DNA. Circular plasmid DNA purified from
bacterial minipreps is isolated and digested with Bgl II, a restriction enzyme producing at least
two linearlized fragments — vector DNA and PCR fragment (lane 2). These fragments can be
visualized using agarose gel electrophoresis. If the PCR fragment contains a Bgl II restriction
site, three DNA bands may be observed (lane 3). A 500 bp molecular weight ruler is shown in
lane 1.
25
BgI II
PCR
Fragment
BgI II
Bgl II
Digest
PCR Fragment
12
3
Page 30

Ligation – Quick Guide
1. Label a microcentrifuge tube with
your initials, plant name, and
"ligation."
2. Briefly spin down the stock tubes
of 2x reaction buffer and proof
reading polymerase to collect the
contents at the bottom of the
tube.
3. Set up blunting reaction with the
following reagents.
Reagent Amount
2x ligation reaction buffer 5.0 µl
Purified PCR product 1.0 µl
Sterile water 2.5 µl
Proofreading polymerase 0.5 µl
Total 9 µl
4. Close the cap and mix well.
Centrifuge briefly to collect the
contents at the bottom of the
tube.
5. Incubate the tube at 70°C for
5 min.
26
70°C water bath
Page 31

6. Cool tube on ice for 2 min.
7. Once cool, centrifuge briefly to
bring contents to the bottom of
the tube and keep tube at room
temperature.
8. Spin down stock tubes of pJet1.2
vector and the T4 DNA ligase to
collect the contents at the bottom
of the tube.
9. Set up a ligation reaction with the
following reagents.
Reagent Amount
Blunting reaction (already in 9.0 µl
microcentrifuge tube)
T4 DNA ligase 0.5 µl
pJet1.2 blunted vector 0.5 µl
Total 10 µl
10. Close the cap and mix well.
Centrifuge briefly to collect the
contents at the bottom of the
tube.
11. Incubate the tube at room
temperature for 5–10 min.
12. Store the ligation at –20°C or if
proceeding directly to
transformation step, store on ice.
27
Ice
Page 32

Transformation – Quick
Guide
Preparation for Competent Cells
1. Label an LB Amp IPTG agar
plate with your initials and place
at 37°C.
2. If not already done, pipet 1.5 ml
C-growth medium into a 15 ml
culture tube. Label with your
initials and warm to 37°C for at
least 10 min. Also ensure your
starter culture is shaking at 37°C.
3. Pipet 150 µl of fresh starter
culture (inoculated yesterday)
into the pre-warmed C-growth
medium and place in shaking
water bath at 37°C.
4. Label a 1.5 ml microcentrifuge
tube with your initials and
"competent cells."
5. Prepare transformation (TF) buffer by
combining 250 µl of transformation
reagent A and 250 µl of
transformation reagent B into a
tube labeled transformation
buffer. Keep on ice.
6. After a 20–40 min incubation,
transfer the actively growing
culture in the C-growth medium
to your competent cells
microcentrifuge tube.
28
37°C shaking
water bath
Starter
culture
C-growth
medium
Ice
B
A
37°C incubator
Page 33

7. Centrifuge at top speed for one
minute and immediately put tube
on ice.
8. Use a 1000 µl pipet or a vacuum
source to remove culture
supernatant avoiding the pellet.
Keep the tube on ice.
9. Resuspend the bacterial pellet in
300 µl of ice cold transformation
buffer by very gently pipetting up
and down in the solution above
the pellet – do not touch the pellet.
10. Incubate the resuspended bacteria
on ice for 5 min.
11. Centrifuge the bacteria at top
speed for 1 min. Ensure the
bacteria are on ice immediately
prior to and immediately following
centrifugation.
12. Using a 1000 µl pipet or a vacuum
source, remove the supernatant
avoiding the bacterial pellet.
13. Very gently resuspend the
bacterial pellet in 120 µl of ice
cold transformation buffer. Keep
tube on ice.
14. Incubate resuspended bacteria
on ice for 5 min. The cells are
now competent for transformation.
29
Ice
TF buffer
TF buffer
Ice
Ice
Page 34

Experimental Procedure for
Transformation
15. Label one microcentrifuge tube
with your initials and
"transformation."
16. Pipet 5 µl of your ligation reaction
into your transformation
microcentrifuge tube. Keep on
ice.
17. Using a fresh tip, very gently
pipet the competent cells up and
down two times then pipet 50 µl
of competent cells into your
transformation tube. Very gently
pipet up and down two times to
mix and return to ice.
18. Incubate the transformation on
ice for 10 min.
30
Ice
Ice
Ligation
LigationCompetent
cells
Page 35

19. Retrieve the warm LP Amp IPTG
agar plate from the 37°C
incubator and pipet the entire
volume of the transformation onto
the labeled agar plate. Use an
inoculation loop to very gently
spread the bacteria around the
plate — do not spread for more
than 10 seconds.
20. Immediately place agar plate,
upside down at 37°C and
incubate overnight.
21. The next day, analyze results or
wrap plate in Parafilm and store
at 4°C until required.
31
37°C incubator
Page 36

Instructor’s Advance Preparation
In the first part of this activity, students will insert (ligate) the purified PCR
products into the pJet1.2 blunted vector. They will then transform competent
bacteria with the ligation reaction mixture.
Note: Students can either proceed directly to transformation activity or
store the ligation reaction at 4°C or –20°C until the next laboratory session.
Note: In order to complete the laboratory more efficiently, preparation for
the transformation step can be initiated prior to performing the ligation
reaction, allowing immediate transformation of competent cells with the
products of the ligation reaction. Refer to Tasks to Perform Prior to the
Transformation Laboratory on page 34 for details.
Ligation Reaction
The pJet1.2 blunted vector is supplied ready to use, already opened, with
blunt ends ready for ligation to PCR products. The pJet1.2 plasmid selects
successful ligations through the disruption of an otherwise lethal gene,
eco47IR
, which enables positive selection of the recombinants. Before
ligation, a 3'-A overhang must be removed from the PCR products by
treating the PCR product with a proofreading DNA polymerase, leaving
them with blunt ends ready to be ligated to the pJet1.2 blunted vector. This
DNA polymerase is active at 70°C but not at lower temperatures, so it is
not necessary to inactivate this enzyme after use. Once blunted, the PCR
product is combined with the plasmid and T4 DNA ligase under conditions
optimal for ligation. The ligation reaction is fast, complete in 5–10 min. Only
a minimal increase in the number of transformants is gained by extending
the ligation time beyond 10 min. Following ligation of a gene into a plasmid,
students will perform transformation to introduce the plasmid into competent
E. coli
cells.
Note: Prior to starting this laboratory activity, students must have already
performed a polymerase chain reaction (PCR) to amplify the DNA fragment
they wish to clone and subsequently purify the PCR product to remove
excess primers, nucleotides, and DNA polymerase, which would otherwise
interfere with subsequent experiments.
32
Page 37

Note: Components needed to carry out PCR reactions are not included in
the Ligation and Transformation module. A kit that utilizes size exclusion
chromatography, such as Bio-Rad’s PCR Kleen™ Spin Purification module
(catalog #732-6300EDU), can be used for purifying the PCR products.
Once the purified PCR product is available, students may decide to
electrophorese the PCR products to assess the quality and quantity of the
sample. Electrophoresis of the purified sample is an optional activity and
could be skipped.
Tasks to Perform Prior to the Ligation Laboratory
1. Thaw the 2x ligation reaction buffer, proofreading polymerase, T4 DNA
ligase, pJet1.2 blunted vector, purified PCR fragment, and sterile water
and store on ice. Just before use, mix and centrifuge the reagents to
collect contents at the bottom of the tubes. Do not aliquot the ligation
reagents for students; the volumes required are too small. Have these
reagents available at the common workstation for students. Also ensure
students are familiar with methods to pipet small volumes.
Transformation Laboratory
At this transformation step, students will transform bacteria with the ligation reaction. Following transformation, pJet1.2 enables positive selection
of plasmids with the desired insert due to the disruption of an otherwise
lethal gene,
eco47IR
, which allows growth of successful transformants.
Bacterial transformation with ligation reactions is a very inefficient process
(much more inefficient than transformation with plasmid DNA), so students
should be encouraged to take special care during this protocol. There are
many steps that if performed improperly can lead to reduced transformation
efficiency or even no colonies. It is recommended that students perform a
control transformation using 1 µl of 50–200 ng/µl of an ampicillin resistant
control plasmid if available. (pGLO Plasmid (20 µg) (catalog #166-0405EDU)
can be purchased separately and used as a control.) Each student team
would require an additional LB Amp IPTG agar plate for their control
transformation. Note: Each preparation of competent cells derives sufficient
cells for two transformations.
The transformation protocol involves creating competent cells and
immediately performing the transformation. Once made, the competent
cells must be used immediately or discarded according to local regulations.
They cannot be stored for later use. This transformation method permits
33
Page 38

relatively high transformation efficiency (106transformants per µg DNA)
without a requirement for a refrigerated centrifuge, commercial competent
cells, or a –70°C freezer to store the competent cells.
Note: Bio-Rad offers both chemically competent and electrocompetent
cells for purchase should your teaching goals include electroporation or
more traditional chemical transformation techniques. The commercially
available cells require storage at –70°C and have transformation efficiencies
of 10
9
/µg DNA.
Note: The Ligation and Transformation module requires microbial culturing
reagents, such as LB broth, LB agar, ampicillin, petri dishes, cell culture
tubes, and inoculation loops, which are not included in this module. You
may choose to purchase the Microbial Culturing module (catalog #166-5020EDU)
for these components.
Tasks to Perform Prior to the Transformation Laboratory
1. Prepare solid and liquid growth media at least 5 days prior to the
transformation step. The Microbial Culturing module contains all
reagents required for this activity, or alternatively the reagents may be
prepared using standard protocols. (Note: Complete instructions for
reagent preparation and streaking plates are available in the Microbial
Culturing module instruction manual.) Each student team requires:
1 LB agar plate for preparing starter colonies, 1–2 LB Amp IPTG agar
plate with final concentrations of 50 µg/ml ampicillin and 0.2 mM IPTG
for plating transformations (if performing a positive control transformation
2 plates will be required), 5 ml of LB broth for growing starter culture
and 20 ml of LB Amp broth with final concentration of 50 µg/ml ampicillin
for growing 4 minipreps. All reagents may be stored at 4°C for up to
1 month.
2. Innoculate growth media and culture cells
a. Streak an
E. coli
HB101 starter plate: At least 2 days prior to the
transformation, streak an
E. coli
culture appropriate for transformation on an LB agar plate using standard microbial techniques to
allow formation of single colonies. If using the Microbial Culturing
module, rehydrate the
E. coli
HB101 vial with 250 µl of sterile water
and use 10 µl of the rehydrated bacteria to streak plates. Incubate
plate at 37°C overnight. Once colonies have grown, wrap plate in
Parafilm and store at 4°C for up to 2 weeks.
34
Page 39

b. Prepare starter culture: As late as possible the day before the
transformation, inoculate a 2–5 ml LB culture with a starter colony
from the
E. coli
starter plate. Incubate cultures in a shaking water
bath or incubator overnight at 37°C and at least 200 rpm.
Note: It is important to use a fresh starter culture (<24 hours since
inoculation) for the transformation, or transformation efficiency will be
reduced.
Note: If a shaking water bath or incubator is not available, incubate starter
cultures at 37°C 1 day prior to the transformation, manually shaking the
vial containing the culture as frequently as possible to oxygenate the
culture (a reduction in transformation efficiency may be observed).
3. Place 1 LB Amp IPTG agar plate per student team in 37°C incubator
just before the transformation lab. Note: If performing a positive control
transformation, a second plate per student team will be needed.
4. Pipet 1.5 ml of C-growth medium to one 15 ml culture tube per team
and incubate at 37°C for at least 10 min prior to the transformation
laboratory.
35
Page 40

Student Ligation Protocol
Note: Before use, the appropriate reagents must be defrosted, thoroughly
mixed, and centrifuged to collect contents at the bottom of the tubes. Refer
to Tasks to Perform Prior to the Ligation Laboratory on page 33 for
details.
Listed below are materials and reagents required at the workstations prior
to starting the ligation activity.
Instructor’s (Common) Station Quantity (✔)
Ice bucket containing stock tubes of:
o 2x ligation reaction buffer 100 µl
❒
o Proofreading polymerase 10 µl ❒
o T4 DNA ligase 10 µl ❒
o pJet1.2 blunted vector 10 µl ❒
o Sterile water 1 ml ❒
Water bath, heating block, or incubator at 70°C 1 ❒
Microcentrifuge (refrigerated, if available) 1 ❒
Materials Required at Each Student Station Quantity (✔)
Purified PCR product 1–2 µl
❒
1.5 ml microcentrifuge tube 1 ❒
10 µl adjustable-volume micropipet and tips 1 ❒
Ice bucket 1 ❒
Marking pen 1 ❒
36
Page 41

Setting Up the Blunting Reaction
This reaction removes the 3' nucleotide overhang left by the
Ta q
DNA
polymerase that would prevent blunt end ligation
.
1. Label a microcentrifuge tube with your initials, the name of your amplified
gene, and "ligation."
2. Pulse spin the stock tubes containing the ligation reaction buffer and
proofreading polymerase in a microcentrifuge for 10 sec to force contents
to bottom of tubes prior to use.
Note: When pipetting very small volumes, take special care. When pulling
up reagents, make sure only the soft stop of the pipet is used even though
it may feel like a very small movement. Also, look at the end of the pipet tip
and make sure that the correct volume of reagent is in the tip. After adding
the reagent to the tube, be sure that the pipet tip is empty. Never reuse a
pipet tip.
3. Set up a blunting reaction with the following reagents:
Reagent Amount
2x ligation reaction buffer 5.0 µl
Purified PCR product 1.0 µl
Sterile water 2.5 µl
Proofreading polymerase 0.5 µl
Total 9 µl
Note: The amount of PCR product added to the reaction may be
increased to 2 µl if the PCR product is not very intense when analyzed on
an agarose gel — for example if it is not as intense as the 1 kb band in the
molecular weight marker. If the amount of PCR product is increased,
remember to decrease the volume of sterile water to 1.5 µl to compensate
and keep the total volume of 9 µl.
37
Page 42

4. Close the cap and mix well. Centrifuge in a microcentrifuge for 10 sec
to collect the contents at the bottom of the tube.
This step is essential due to the very small volume used in this reaction
.
5. Place the tube at 70°C for 5 min.
6. Place tube on ice to cool for 2 min.
This recondenses water vapor to maintain reaction volume.
7. Once cool, centrifuge the tube briefly to collect the contents at the bottom
of the tube. Place tube at room temperature.
Setting Up the Ligation Reaction
This reaction inserts the PCR product into the pJet1.2 plasmid vector
.
8. Briefly centrifuge the stock tubes containing the pJet1.2 blunted vector
and the T4 DNA ligase in a microcentrifuge to force the contents to
bottom of tubes prior to using.
9. Setup a ligation reaction with the following reagents:
Reagent Amount
Blunting reaction (already in 9.0 µl
microcentrifuge tube)
T4 DNA ligase 0.5 µl
pJet1.2 blunted vector 0.5 µl
Total 10 µl
10. Close the cap and mix well. Centrifuge briefly in a microcentrifuge to
collect the contents at the bottom of the tube.
38
Page 43

11. Incubate tube at room temperature for 5–10 min.
12. Store the ligation reaction at –20°C. If you are proceeding directly to
transformation, pipet 5 µl of the ligation reaction into a microcentrifuge
tube labeled with your initials, the name of your gene, and
"transformation," and store it on ice until needed for the transformation.
39
Page 44

Student Transformation Protocol
Listed are materials and reagents required at the workstations prior to
beginning the transformation activity.
Instructor’s (Common) Workstation Quantity (✔)
Water bath, heating block, or incubator (37°C) 1 ❒
Microcentrifuge (refrigerated, if available) 1 ❒
Each student team will require the following items to transform bacteria
with one PCR product.
Materials Required at Each Student Station Quantity/Team (✔)
Ligation reaction from ligation laboratory (on ice) 5 µl ❒
(Optional) Control plasmid 50–200 ng/µl (on ice) 1 µl ❒
1.5 ml microcentrifuge tubes 4 ❒
15 ml culture tube containing 1.5 ml of 1 ❒
C-growth medium
Transformation reagent A (on ice) 250 µl
❒
Transformation reagent B (on ice) 250 µl ❒
Sterile inoculating loops 2 ❒
Fresh LB Amp IPTG agar plate prewarmed to 37°C 1–2 ❒
Fresh starter culture plate of
E. coli
HB101 1 ❒
200 µl adjustable-volume micropipet and tips 1 ❒
1,000 µl adjustable-volume micropipet and tips 1 ❒
Marking pens 1 ❒
Parafilm sealing film 1 strip ❒
(Optional) Vortex mixer 1 ❒
(Optional) Vacuum source 1 ❒
40
Page 45

Detailed Protocol for Transformation
Preparation of Competent Cells
1. Approximately 20–40 min prior to starting the transformation, prepare
competent cells by pipeting 150 µl of fresh starter culture (inoculated
one day prior) into the prewarmed C-growth medium and placing in a
shaking 37°C water bath or incubator for 20–40 min.
Note: If a shaking waterbath is not available, manually shake the culture
tubes every 5 min during the 20–40 min growth phase to oxygenate the
culture.
Note: Your instructor may have already completed this step to save time.
2. Label 1–2 LB Amp IPTG agar plates with your initials. Also label one
agar plate for your ligation (pJet + your insert name) and if you are
performing a positive control transformation label the second agar plate
for the positive control plasmid.
Place agar plates at 37°C.
3. If not already done, pipet 1.5 ml of C-growth medium to the 15 ml culture
tube. Label tube with your initials and warm it at 37°C for at least 10
min.
Also ensure that the
E. coli
starter culture is at 37°C.
4. Label a microcentrifuge tube with your initials and "competent cells".
5. Prepare the transformation buffer by combining 250 µl of transformation
reagent A and 250 µl of transformation reagent B into a microcentrifuge
tube labeled "TF buffer" and mix thoroughly with a vortex mixer (if
available). Keep on ice until use. (Note: This mixture must be used on
the day of preparation.)
41
Page 46

6. After bacteria have grown in C-growth medium for 20–40 min at 37°C
with shaking, transfer the culture to your competent cells tube by
decanting or pipetting it. It is better not to put the actively growing cell
culture on ice at this step.
7. Centrifuge the culture in a microcentrifuge at top speed for 1 min. Make
sure that the microcentrifuge is balanced and accommodate tubes of
classmates to ensure economic use of the microcentrifuge. Immediately
put the pelleted culture on ice.
Note: After this step, it is very important to keep the bacteria on ice as
much as possible during this procedure. Transformation efficiency will be
severely compromised if the cells warm up.
Note: It is very important to treat the bacteria extremely gently during this
procedure — the bacteria are very fragile and your transformation efficiency
will be compromised unless you are very gentle.
8. Locate the pellet of bacteria at the bottom of the tube. Remove the
culture supernatant, avoiding the pellet, using a 1,000 µl pipet or a
vacuum source. Keep the cells on ice.
42
Page 47

9. Resuspend the bacterial pellet in 300 µl of ice-cold transformation
buffer by gently pipetting up and down in the solution above the pellet
with a 1,000 µl pipet, and gradually wear away the pellet from the bottom
of the tube. Make sure that the bacteria are fully resuspended, with no
clumps. Avoid removing the cells from the ice bucket for more than a
few seconds.
10. Incubate the resuspended bacteria on ice for 5 min.
11. Centrifuge the bacteria in a microcentrifuge for 1 min. Note: Ensure
that the bacteria are on ice immediately prior to and immediately
following centrifugation. If the centrifuge is not close to the laboratory
bench, take the entire ice bucket to the microcentrifuge so that the
bacteria are only out of the ice bucket for 1 min. Use a refrigerated
microcentrifuge, if available.
12. Remove the supernatant from the pellet using a 1,000 µl pipet or
vacuum source.
13. Resuspend the bacterial pellet in 120 µl of ice-cold transformation
buffer by gently pipetting up and down with a 200 µl pipet. Be sure that
bacteria are fully resuspended with no clumps. Avoid removing the
cells from the ice bucket for more than a few seconds.
14. Incubate the resuspended bacteria on ice for 5 min.
The cells are now competent for transformation
.
Experimental Procedure for Transformation
15. Label one microcentrifuge tube with your initials, insert name, and
"TF" (referred to below as the "gene TF" tube).
43
Page 48

44
Note: If you are performing the ligation and transformation steps on the
same day, use the microcentrifuge containing 5 µl of the ligation mixture
that was prepared and labeled at the end of the ligation step.
16. If not already done, pipet 5 µl of the ligation reaction from the previous
stage into the "gene TF" microcentrifuge tube. Store any remaining ligation reaction at 4°C or –20°C.
17. (Optional) If performing a positive control transformation, pipet 1 µl of
the positive control plasmid into an appropriately labeled microcentrifuge
tube and keep on ice.
18. Using a fresh tip, pipet 50 µl of competent bacteria directly into the
ice-cold "gene TF" tube containing 5 µl of your ligation and gently pipet
up and down two times to mix.
19. (Optional) If performing a positive control transformation, pipet 50 µl of
competent bacteria directly into the ice-cold positive control plasmid
tube and gently pipet up and down two times to mix.
20. Incubate the transformations for 10 min on ice.
21. Retrieve LB Amp IPTG agar plates from the 37°C incubator.
22. Pipet the entire volume of each transformation onto the corresponding
labeled LB Amp IPTG agar plate, and using an inoculation loop or a
sterile spreader, very gently spread the bacteria around the plate —
remember that the bacteria are still very fragile! Once the plate is
covered, stop spreading. Do not spread for more than 10 sec.
It is vital that the LB Amp IPTG agar plate be warm at this step to ensure
sufficiently high transformation efficiency. Spreading the plate until it is dry
will also reduce transformation efficiency.
Ice
Page 49

23. Immediately place LB Amp IPTG agar plates upside down in the 37°C
incubator and incubate them overnight.
24. The next day, analyze the results, or wrap the plates in Parafilm and
place them at 4°C until required for inoculation of miniprep cultures
(see Appendix A).
Analysis of Results of Ligation and Transformation
Count the number of bacterial colonies that grew on the LB Amp IPTG
agar plates.
Note: Occasionally, satellite colonies may grow using this ligation method.
Count the large individual colonies, not the tiny colonies surrounding larger
colonies.
Transformation Number of Colonies
Ligation of gene of interest
Control plasmid
Note: If the number of colonies is very high and uncountable, enter "TNC"
for "too numerous" to count in the results table.
45
Page 50

Appendix A
Inoculating a Bacterial Colony for Plasmid
Miniprep
Once the plasmid has been introduced into living bacterial cells and the
cells have grown and divided on selective medium, the next step in the
experiment is to prepare a miniprep of the plasmid DNA for sequencing or
further experiments. The three steps are: 1) growing cells in liquid culture;
2) purifying the plasmid DNA from the culture; and 3) performing a
restriction digest on the purified DNA to determine if the DNA insert in the
plasmid is the expected size. To start the liquid culture, cells from an isolated
bacterial colony are placed in selective medium (nutrient broth with
antibiotic). It is important to choose an isolated single colony from the
plate, so that the liquid culture will contain cells that all have the same
plasmid. If cells from more than one colony are used for inoculation, the
miniprep may contain multiple plasmids and the DNA may not be used for
sequencing or further experiments.
Before proceeding to the miniprep protocol, grow transformed bacterial
colonies in liquid cultures using the following instructions:
1. Prepare 25 ml of LB Amp broth (if not already prepared; refer to
instructions in the Microbial Culturing module, catalog #166-5020EDU).
2. Label four 15 ml culture tubes with your initials and “pJet”, the name of
your gene, and #1 through #4.
3. Using sterile technique, pipet 5 ml of LB Amp broth into each of the
four 15 ml culture tubes.
4. One day prior to the next laboratory session, use a sterile loop or a
sterile pipet tip to pick a single colony from the LB Amp IPTG agar plate
containing the plated bacteria transformed with your ligation reaction.
Inoculate an LB Amp culture tube with the colony. Repeat for a total of
four miniprep cultures from four individual colonies.
Note: Occasionally satellite colonies may grow using this ligation method.
Pick the large individual colonies, not the tiny colonies surrounding larger
colonies. Be sure that a single colony is picked, or you may isolate multiple
plasmids from your miniprep and these cannot be sequenced.
5. Place the miniprep cultures to grow in a shaking incubator or water
bath at 37°C overnight.
46
Page 51

Note: If no colonies grew on your team’s agar plate from the pJet1.2 +
gene ligation reaction, use colonies from another team’s successful
transformation. Relabel your 15 ml culture tubes accordingly.
6. Perform a miniprep using the Aurum™ Plasmid Mini Purification module
(catalog #732-6400EDU).
47
Page 52

Appendix B
Restriction Digestion of Plasmid DNA with
Bgl II Enzyme
Instructors Advanced Preparation
The Ligation and Transformation module contains Bgl II enzyme and reaction
buffer to enable analysis of plasmids derived using the module and
subsequently purified using a plasmid purification protocol. Electrophoresis
reagents, including sample loading dye and a molecular weight ruler are
also required for the analysis. The pJet1.2 plasmid vector is 2,974 bp in
length, thus after restriction digestion of plasmids and subsequent
electrophoresis a band of around 3 kb and a smaller band corresponding
to the size of the PCR product insert is expected.
To digest and analyze four plasmids, each student team requires:
Item Qty
Microcentrifuge tubes 4–8
Bgl II restriction enzyme 4 µl
10x Bgl II reaction buffer 8 µl
Sterile water 30 µl
1% agarose gel 1
Electrophoresis running buffer (sufficient to fill
electrophoresis chamber)
Sample loading dye, 5x 50 µl
Molecular weight ruler (10 µl of EZ load molecular ruler per gel is
recommended)
2–20 µl adjustable volume pipet and tips 1
Background
Once transformed bacteria are grown overnight in a liquid culture (see
Appendix A), a miniprep is performed to purify the plasmid DNA from the
bacteria. Before proceeding with further experiments using the purified
plasmid DNA, it is important to verify that the isolated plasmid contains the
insert of interest. During the ligation stage, a gene of interest was
ligated into the pJet1.2 blunted vector, which contains a Bgl II restriction
enzyme recognition site on either side of the insertion site. To determine
the size of the fragment inserted, the purified plasmid can be analyzed by
restriction enzyme digestion with Bgl II enzyme and subsequent agarose
gel electrophoresis.
48
Page 53

By digesting a small portion of the miniprep DNA with Bgl II enzyme (see
instructions below), the insert should be cut out of the vector. Running the
products of the restriction digestion on an agarose gel should give two DNA
bands, one the size of the vector (2,974 bp) and the other the size of the
inserted DNA (the original PCR product). If there are more than two bands
in any digest, it may mean that the insert contains a Bgl II site. Does the
size of the excised bands add up to the size of the original PCR fragment?
Alternatively, two similarly sized fragments may indicate that a mixed
culture was used to start the miniprep, instead of an isolated colony.
Preparation for Restriction Digestion Analysis
1. Label one microcentrifuge tube with your initials and "Bgl II master
mix."
2. The restriction digestion reactions will be performed on each miniprep
in a final volume of 20 µl with 10 µl of plasmid DNA.
3. Prepare a master mix for Bgl II restriction enzyme digestion according
to the following table, using stock reagents from the common workstation.
Use a fresh tip for each reagent.
49
Page 54

Reagent Volume for 1 Reaction Volume for 5 Reactions
10x Bgl II reaction buffer 2 µl 10 µl
Sterile water 7 µl 35 µl
Bgl II restriction enzyme 1 µl 5 µl
Total 10 µl 50 µl
Experimental Procedure for Restriction Digestion Analysis
1. Label a microcentrifuge tube for each plasmid miniprep.
2. Prepare digestion reactions by combining 10 µl of the Bgl II master mix
and 10 µl of each plasmid DNA in the appropriately labeled
microcentrifuge tubes.
3. Mix tube components and spin briefly in a microcentrifuge to collect the
contents at the bottom of the tube.
4. Incubate reactions at 37°C for 1 hr. If the reactions will not be analyzed
by agarose gel electrophoresis immediately, store them at –20°C until
analysis.
Preparation for Electrophoresis
1. Place a 1% agarose gel in the electrophoresis chamber and add
electrophoresis running buffer to just cover the gel.
2. Plan your gel electrophoresis experiment.
3. Add the appropriate amount of sample loading dye to the DNA samples
and pipet up and down to mix.
4. Load 20 µl of the restriction digestion reactions on the gel according to
your plan and load an appropriate amount of molecular weight ruler in
a separate lane.
5. (Optional) It is recommended that undigested DNA (at the same dilution
as your restriction digests) also be run next to your digested samples.
6. Electrophorese samples at 100 V for 30 min and analyze results.
50
Page 55
Restriction enzyme digestion analysis of plasmid DNA. Circular plasmid DNA purified from
bacterial minipreps is isolated and digested with Bgl II, a restriction enzyme producing at least
two linearlized fragments — vector DNA and PCR fragment (lane 2). These fragments can be
visualized using agarose gel electrophoresis. If the PCR fragment contains a Bgl II restriction
site, three DNA bands may be observed (lane 3). A 500 bp molecular weight ruler is shown in
lane 1.
BgI II
51
PCR
Fragment
BgI II
Bgl II
Digest
PCR Fragment
12
3
Page 56

Legal Notices
Notice regarding Bio-Rad thermal cyclers and real-time systems: Purchase of
this instrument conveys a limited, non-transferable immunity from suit for the
purchaser’s own internal research and development and for use in applied
fields other than Human In Vitro Diagnostics under one or more of U.S.
Patents Nos. 5,656,493, 5,333,675, 5,475,610 (claims 1, 44, 158, 160–163,
and 167 only), and 6,703,236 (claims 1–7 only), or corresponding claims in
their non-U.S. counterparts, owned by Applera Corporation. No right is conveyed expressly, by implication, or by estoppel under any other patent claim,
such as claims to apparatus, reagents, kits, or methods such as 5' nuclease
methods. Further information on purchasing licenses may be obtained by
contacting the Director of Licensing, Applied Biosystems, 850 Lincoln Centre
Drive, Foster City, California 94404, USA. Bio-Rad’s real-time thermal cyclers
are licensed real-time thermal cyclers under Applera’s United States Patent
No. 6,814,934 B1 for use in research and for all other fields except the fields
of human diagnostics and veterinary diagnostics. Purchase of iTaq
™
DNA
polymerase includes an immunity from suit under patents specified in the
product insert to use only the amount purchased for the purchaser’s own
internal research. No other patent rights (such as 5’ Nuclease
Process patent
rights) are conveyed expressly, by implication, or by estoppel. Further
information on purchasing licenses may be obtained by contacting
the Director
of Licensing, Applied Biosystems, 850 Lincoln Centre Drive, Foster City,
California 94404, USA.
Trademarks
Parafilm is a trademark of the American National Can Company.
52
Page 57

Page 58

Page 59

Page 60

1665019 Rev A
Bio-Rad
Laboratories, Inc.
Life Science
Group
Bulletin 0000 Rev A US/EG
Web site www.bio-rad.com USA 800 424 6723
Australia 61 2 9914 2800 Aus tria 01 877 89 01 Belgium 09 38 5 55 11
Brazil 55 31 3689 6600 Canada 905 3 64 3435 China 86 21 6169 8500
Czech Republic 420 241 430 532 Denmark 44 52 10 00
Finland 09 804 22 00
Greece 30 210 777 4396 Hong Kong 852 2789 3300
Hungary 36 1 459 6100 India 91 124 4029300 Israel 03 963 6050
Italy 39 02 216091 Japan 03 6361 7000 Korea 82 2 3473 4460
Malaysia 60 3 2117 5260 Mexico 52 555 488 7670
The Neth erlands 0318 540666 New Zealand 64 9 415 2280
Norway 23 38 41 30 Poland 48 22 331 99 99 Portugal 351 21 472 770 0
Russia 7 495 721 14 04 Singapore 65 6415 3170
South Africa 27 861 246 723 Spain 34 91 590 5200
Sweden 08 555 12700 Switzerland 061 717 95 55
Tai wan 886 2 2578 7189 Thailand 66 2 6518311
United K ingdom 020 8 328 2000
France 01 47 95 69 65 Germany 089 31 884 0
00-0000 0000 Sig 0211
 Loading...
Loading...