Page 1

iQ™5 Optical System Software
Instruction Manual
Compatible with the MyiQ™2,
™
MyiQ
, and iQ™5 real-time
PCR detection systems
Page 2

NOTICE TO PURCHASER
Bio-Rad’s real-time thermal cyclers are licensed real-time thermal cyclers under Applera’s United
States Patent 6,814,934 B1 for use in research, human
except the field of veterinary diagnostics. Purchase of this instrument conveys a limited nontransferable immunity from suit for the purchaser’s own internal research and development and
for use in applied fields under one or more of U.S. Patents 5,656,493, 5,333,675, 5,475,610
(claims 1, 44, 158, 160–163 and 167 only), and 6,703,236 (claims 1–7 only), or corresponding
claims in their non-U.S. counterparts, owned by Applera Corporation. No right is conveyed
expressly, by implication or by estoppel under any other patent claim, such as claims to
apparatus, reagents, kits, or methods such as 5' nuclease methods. Further information on
purchasing licenses may be obtained by contacting the Director of Licensing, Applied Biosystems,
850 Lincoln Centre Drive, Foster City, California 94404, USA.
Bio-Rad’s thermal cyclers and real-time thermal cyclers are covered by one or more of the
following U.S. patents or their foreign counterparts owned by Eppendorf AG: U.S. Patent Nos.
6,767,512 and 7,074,367.
SYBR is a trademark of Molecular Probes, Inc. Bio-Rad Laboratories, Inc. is licensed by Molecular
Probes, Inc. to sell reagents containing SYBR® Green I for use in real-time PCR, for research
purposes only.
Cy is a trademark of GE Healthcare. SYBR and Texas Red are trademarks of Molecular Probes,
Inc. HEX, TAMRA, TET, and VIC are trademarks of Applera Corp. Excel, Windows, and Windows
XP are trademarks of Microsoft Corporation. Phillips is a trademark of Phillips Screw Company
Corporation.
in vitro
diagnostics, and for all other fields
i
Page 3

Safety Information
Grounding
Always connect the MyiQTM2, MyiQTM, or iQTM5 optics module to a three-prong, grounded AC outlet
using the AC power cord provided with the system. Do not use an adaptor to a two-terminal
outlet. Always ensure that you set the module power switch to the off position when you connect
or disconnect power cords.
Handling
Handle all components of the real-time PCR detection system with care and with clean, dry hands
at all times. The optical system contains mirrors and lenses that may shatter if the unit is
dropped or struck with great force. If the unit is damaged such that internal components or wires
are exposed, contact your local Bio-Rad office immediately. Do not attempt to repair or power on
the instrument.
Servicing
The only user-serviceable parts of the optics module are the lamp and filters. Call your local
Bio-Rad office for all other optics module and thermal cycler related service. When you replace
the lamp or filters, open only the outer casing of the optics module. The camera lamp may get
extremely hot during system operation. Do not attempt to remove the lamp without powering off
the instrument and allowing the system to cool for at least 15 minutes. To prevent skin burns
and fire hazards, do not attempt to operate the real-time PCR detection system while the camera
case is open. Do not open casing of the optics module when the instrument is in use.
Operating Temperature
For normal operation, the maximum ambient temperature should not exceed 40°C. To ensure
adequate cooling of the system, maintain a clearance of at least 4 inches around the sides of the
MyiQ2, MyiQ, or iQ5 optics module. Do not block the fan vents near the lamp, as this may lead to
improper operation or cause physical damage to the detector. Do not operate the optics module
in extreme humidity (that is, greater than 90 percent) or where condensation can short internal
electrical circuits or fog optical elements.
Notice
The MyiQ2, MyiQ, and iQ5 instruments are designed and certified to meet EN-61010 safety
standards.
EN-61010 certified products are safe to use when operated in accordance with the instruction
manual. These instruments should not be modified in any way. Alteration of the instruments will:
• Void the manufacturer’s warranty
• Void the EN-61010 safety certification
• Create a potential safety hazard
Bio-Rad is not responsible for any injury or damage caused by the use of these instrument for
purposes other than those for which they are intended, or by modifications to the instruments
not performed by Bio-Rad or an authorized agent.
- ii -
Page 4

Table of Contents
NOTICE TO PURCHASER ..................................................................................................i
Safety Information .........................................................................................................ii
Section 1. Getting Started ..............................................................................................1
1.1 The MyiQ™2 Real-Time PCR Detection System ................................................................1
1.2 Setting Up the System Hardware ....................................................................................1
1.2.1 System Checklist......................................................................................................1
1.2.2 Installing the Optical Reaction Module on the iCycler Chassis......................................2
1.2.3 Installing the Support Bracket ..................................................................................3
1.2.4 Installing the Optics Module .....................................................................................3
1.2.5 Connecting Power and Communication Cables to the System .....................................3
1.3 Installing the iQ5 Software .............................................................................................4
1.3.1 Installing the Camera Drivers ………………………………………………………………………………4
1.4 Recommended Computer Settings...................................................................................4
1.5 Calibrating the Instrument ..............................................................................................6
1.6 Compatibility with Earlier Versions of the iQ5 Optical System Software ..............................6
Section 2. Quick Guides ..................................................................................................7
2.1 Protocol Quick Guide ......................................................................................................7
2.2 Plate Setup Quick Guide .................................................................................................8
2.3 Running a Real-Time Experiment Quick Guide .................................................................9
2.3.1 Beginning a Run ......................................................................................................9
2.3.2 Monitoring the Run ................................................................................................10
2.4 Data Analysis Quick Guides...........................................................................................10
2.4.1 PCR Quant Tab Quick Guide ...................................................................................10
2.4.2 End Point Quick Guide............................................................................................11
2.4.3 Allelic Discrimination Quick Guide – For Multiplex Data Only ........................................12
2.4.4 Gene Expression Quick Guide.....................................................................................13
2.4.5 Post-Run Plate Setup Editing Quick Guide ...................................................................16
Section 3. Introduction to the iQ5 Optical System Software (Version 2.1) .................18
Section 4. Workshop Module........................................................................................19
4.1 Setup Tab....................................................................................................................19
4.1.1 File Browser Section...............................................................................................20
4.1.2 Selected Protocol Area ...........................................................................................20
4.1.3 Selected Plate Setup Area ......................................................................................20
- iii -
Page 5

4.1.4 Selected Data File Area ..........................................................................................21
4.2 Plate Setup ..................................................................................................................21
4.2.1 Plate Setup Editor Window .....................................................................................21
4.2.2 Well Definition Icons ..............................................................................................22
4.2.3 Editing or Creating a New Plate Setup .....................................................................23
4.2.4 Fluorophore Selection ............................................................................................24
4.2.5 Whole Plate Loading ..............................................................................................26
4.2.6 Specifying Replicates..............................................................................................26
4.2.7 Sample Volume, Seal Type, and Vessel Type ...........................................................27
4.2.8 Defining a Dilution Series .......................................................................................28
4.2.9 Plate Setup Editor Spreadsheets .............................................................................29
4.2.10 Plate Summary ....................................................................................................30
4.3 Protocol.......................................................................................................................31
4.3.1 Selecting a Protocol ...............................................................................................31
4.3.2 Editing or Creating a Protocol .................................................................................32
4.3.3 Add Protocol Options .............................................................................................33
4.3.4 Gradient ................................................................................................................35
4.3.5 Melt Curve/Peak ....................................................................................................36
4.3.6 Sample Protocol Files .............................................................................................37
4.4 Run Set .......................................................................................................................37
4.4.1 Selecting a Run Set................................................................................................38
4.4.2 Creating a Run Set.................................................................................................38
4.5 Data File......................................................................................................................38
4.5.1 Selecting a Data File ..............................................................................................39
4.5.2 Opening a Data File ...............................................................................................39
4.5.3 Opening a Pure Dye Calibration file .........................................................................39
4.5.4 Applying Alternate Pure Dye Calibrations (Alternate RME) ........................................39
4.5.5 Applying Alternate Background or Well Factors ........................................................40
Section 5. Run-Time Central Module............................................................................41
5.1 Initiate Run Window: Run Selected ...............................................................................41
5.2 Well Factors.................................................................................................................42
5.2.1 Dynamic Well Factors .............................................................................................42
5.2.2 Persistent Well Factors ...........................................................................................42
5.3 Real-Time PCR Experiments Using DNA-Binding Dyes.....................................................43
5.3.1 Spiking Real-Time PCR Experiments Using DNA binding Dyes With Fluorescein..........43
5.4 Initiate Run Window: Run End Point Selected ................................................................43
- iv -
Page 6

5.5 Show Plate Window .....................................................................................................44
5.6 Monitor Run Window ....................................................................................................45
5.7 Run-Time Protocol Editing ............................................................................................45
Section 6. Data Analysis Module ..................................................................................47
6.1 PCR Quant Tab ............................................................................................................48
6.1.1 Customizing the PCR Quant Display ........................................................................49
6.2 Amplification Chart.......................................................................................................49
6.2.1 Fluorophore Selector Buttons..................................................................................49
6.2.2 Select Analysis Mode ..............................................................................................50
6.2.3 Log View Button ....................................................................................................50
6.2.4 Selecting and Viewing Traces .................................................................................51
6.3 Standard Curve Chart ...................................................................................................53
6.3.1 Standard Curve Chart Menu....................................................................................54
6.4 Results Section ............................................................................................................55
6.4.1 Plate Spreadsheet ..................................................................................................55
6.4.2 Standard Curve/CT Results Spreadsheet ..................................................................56
6.4.3 Amplification Data (RFU) Spreadsheet.....................................................................57
6.5 Amplification Plot Context Menu....................................................................................57
6.5.1 Data Analysis Options ............................................................................................57
6.5.2 Amplification Plot Data Display Options ...................................................................61
6.5.3 Data Export Options...............................................................................................64
6.5.4 Exporting Results to Microsoft Excel ........................................................................64
6.5.5 Printing Results......................................................................................................65
6.6 Melt Curve and Melt Peak Charts...................................................................................65
6.6.1 Melt Curve and Melt Peak Chart Menu .....................................................................67
6.6.2 Delete Selected Peaks ............................................................................................67
6.6.3 Edit Melt Peak Begin/End Temperature ...................................................................67
6.6.4 Melt Curve/Peak Control Area .................................................................................68
6.6.5 Melt Curve/Peak Spreadsheet .................................................................................68
6.7 Allelic Discrimination Module (For Multiplex Data Only) ...................................................69
6.7.1 Allelic Discrimination Plot........................................................................................69
6.7.2 Allelic Discrimination Plot Menu...............................................................................71
6.7.3 Allelic Data Spreadsheet.........................................................................................71
6.7.4 Automatic/Manual Call ...........................................................................................72
6.7.5 Manual Calls ..........................................................................................................73
6.7.6 Display Mode .........................................................................................................73
- v -
Page 7

6.7.7 Vertical Threshold ..................................................................................................74
6.7.8 Horizontal Threshold ..............................................................................................74
6.7.9 Normalize Data ......................................................................................................74
6.7.10 Restore Default....................................................................................................75
6.8 End Point Analysis........................................................................................................75
6.8.1 The End Point Analysis Settings ………………………………………………………………………….75
6.8.2 End Point Analysis Spreadsheet ..............................................................................77
6.8.3 Recalculate Button .................................................................................................78
6.9 Gene Expression Analysis. ............................................................................................78
6.9.1 Normalized Gene Expression (ddC
6.9.2 Relative Quantity (dC
6.9.3 Specifying Gene and Condition Labels for Gene Expression Analysis ..........................82
6.9.4 Setting Analysis Parameters for Gene Expression Analysis Using the Settings Tab......85
6.9.4.1 Gene List: Assigning Reference Genes and Target-Specific Reaction Efficiency…85
6.9.4.2 Assigning and Naming Control Samples Using the Condition List ……………………87
6.9.4.3 Data Set List …………………………………………………………………………………………….88
6.9.5 Applying an Analysis Method ..................................................................................89
6.9.6 Data Table for Gene Expression Analysis .................................................................89
)....................................................................................81
T, ∆CT
∆∆CT) ...............................................................80
T,
6.9.7 Graphing Options for Expression Data .....................................................................92
6.9.8 Normalized Expression Calculations.........................................................................96
6.9.9 Relative Quantity Calculations.................................................................................97
6.9.10 Gene Expression - Frequently Asked Questions ......................................................98
6.10 Gene Study: Multi File Gene Expression Analysis ..........................................................99
6.10.1 The Gene Study File .............................................................................................99
6.10.2 Creating a Gene Study File .................................................................................101
6.10.3 Gene Expression Study Manager .........................................................................102
6.10.4 Editing a Gene Study..........................................................................................102
6.10.5 Excluding/Including Samples Using the Gene Expression Plate Interface ............... 103
6.10.6 Inter-Run Calibration..........................................................................................103
6.11 Post-Run Plate Editing ..............................................................................................105
6.12 The Reports Tool .....................................................................................................107
6.12.1 Report Viewer Options .......................................................................................108
6.12.2 Parts of the Data Report.....................................................................................108
6.12.3 Data Analysis Report Types ................................................................................108
Section 7. Calibrating the Instrument........................................................................111
7.1 Calibration Overview ..................................................................................................111
- vi -
Page 8

7.2 Components Required for Calibration ..........................................................................111
7.3 Preparing Calibration Plates ........................................................................................112
7.4 Performing Calibrations ..............................................................................................112
7.4.1 Performing Mask Alignment ..................................................................................113
7.4.2 Performing Background Calibration .......................................................................114
7.4.3 Generating Persistent Well Factors........................................................................115
7.4.4 Performing Pure Dye Calibration ...........................................................................116
7.4.5 Editing and Creating Pure Dye Plate Setups ...........................................................118
7.5 Viewing Calibration Files............................................................................................. 119
7.6 Troubleshooting Optics with the Mask Image Window .................................................. 120
7.6.1 Filter Position.......................................................................................................120
7.6.2 Camera Status ..................................................................................................... 120
Section 8. User Profiles ..............................................................................................122
8.1 User Preferences........................................................................................................122
8.1.1 File Paths Preferences ..........................................................................................123
8.1.2 Plate Setup Preferences ....................................................................................... 124
8.1.3 Protocol Preferences ............................................................................................ 125
8.1.4 File Selection at Application Startup Preferences ....................................................125
8.1.5 PCR Quant Screen Preferences .............................................................................126
8.1.6 Allelic Discrimination Module Preferences ..............................................................127
8.1.7 End Point Module Preferences ..............................................................................127
8.1.8 Gene Expression Module Preferences ....................................................................128
8.2 User Administration....................................................................................................128
8.2.1 Adding New Users................................................................................................129
8.2.2 Deleting Existing Users.........................................................................................130
8.3 Logging on to the iQ5 Software ..................................................................................130
8.3.1 Switching Users ................................................................................................... 131
8.3.2 Changing Password..............................................................................................131
8.3.3 Defining Roles .....................................................................................................132
Section 9. Instrument Maintenance ...........................................................................133
9.1 Cleaning the Real-Time PCR Detection System.............................................................133
9.2 Filter Description and Installation Instructions..............................................................133
9.2.1 Recommended Fluorophores and Filter Specifications.............................................133
9.2.2 Accessing the Filters ............................................................................................ 134
9.2.3 Cleaning the Filters ..............................................................................................134
9.3 Replacing the Lamp ................................................................................................... 135
- vii -
Page 9

Appendix A: Warranty ................................................................................................136
Appendix B: Troubleshooting Error Messages ...........................................................137
Appendix C: Product Ordering Information ...............................................................142
- viii -
Page 10

Section 1 Getting Started
Section 1. Getting Started
This section contains information on the following topics:
o The MyiQ2 Real-Time Detection System (page 1)
o Setting up the system hardware (page 1)
o Installing the iQ5 software (page 4)
o Recommended computer settings (page 4)
o Compatibility with earlier versions of the software (page 6)
1.1 The MyiQ™2 Real-Time PCR Detection System
The MyiQ2 Real-Time PCR Detection System is the latest addition to a family of real-time PCR
detection systems that are comprised of an optical detection module and iCycler® thermal cycler.
The two-color MyiQ™2 features a broad-spectrum light source (tungsten-halogen lamp) and
paired filter optical design to ensure optimal excitation and emission. This results in excellent
sensitivity and discrimination between multiple fluorophores. A CCD detector captures a
simultaneous image of all 96 wells of the block resulting in a comprehensive data set illustrating
the kinetic behavior of the data during each cycle.
The iQ™5 Optical System software (version 2.1) is used to control and collect and analyze data
from the MyiQ2 Real-Time PCR Detection System. The software is also capable of controlling, and
collecting and analyzing data from the single color MyiQ™ and five color iQ™5 Real-Time PCR
detection systems. This manual provides instructions on use of the software applicable to each of
these three systems. The online Help manual is available at all times by pressing the F1 key.
1.2 Setting Up the System Hardware
The MyiQ2, MyiQ, or iQ5 system should be installed on a clean, dry, and level surface. Identify an
appropriate work area for the installation process prior to unpacking any system components.
The entire installation process should take approximately 15 minutes to complete. Handle the
optics module and iCycler thermal cycler with care and with clean, dry hands during unpacking
and assembly.
1.2.1 System Checklist
MyiQ2 Optics Module (catalog #170-9758)
• Optics module
• Power cord
• iQ5 optical system software installation disk (170-9753)
• Amplification tech notes CD (version 1.0)
iCycler Chassis (catalog #170-8701)
• Power cord
• iCycler chassis
• iCycler thermal cycler instruction manual
- 1 -
Page 11

Section 1 Getting Started
iQ5 Optical Accessory Kit (catalog #170-9752)
• Optical reaction block
• Modified sliding rear cover for iCycler thermal cycler
• Serial cable
• USB cable
• Filter extraction tool
• Optical tape applicator tools (3)
• Optics support bracket
• Support bracket screws
• Hex driver
• Hex screws
• Spare part: halogen lamp
MyiQ2 Calibrator Dye Solution Kit (catalog #170-8791)
• 1x calibration dye solutions (4 dyes, 3 tubes of each dye)
External Well Factor Solution (catalog #170-8794)
• External well factor solution (5 tubes)
Sample Consumables
• PCR plates and sealers
• Reagents
Contact your local Bio-Rad office if any system components are missing or damaged.
Accessories
The following accessories are required to complete the installation:
• Scissors
• #2 Phillips screwdriver
• Calibrated micropipet(s)
• Aerosol barrier pipet tips
• Optical-quality sealing film or tube caps
• Optical-quality PCR plates or tubes
1.2.2 Installing the Optical Reaction Module on the iCycler Chassis
1. Remove the existing rear cover from the iCycler chassis by sliding the cover towards the
front of the iCycler base.
2. Install the modified sliding rear cover provided with the optical reaction block, ensuring
that the notch is oriented towards the rear of the iCycler thermal cycler.
3. Push the sliding rear cover on top of the chassis as far back as possible.
4. Rotate the green latches on the optical reaction block up towards the open lid.
5. Lift the optical reaction block by the handle and install it onto the chassis. Lower the
front portion of the reaction block so that it engages with the chassis before the rear
portion. The rear of the block lid should fit over the front lip of the sliding rear cover.
6. Secure the optical reaction block in place by rotating the green latches downward.
7. Close the optical lid.
- 2 -
Page 12

Section 1 Getting Started
1.2.3 Installing the Support Bracket
A support bracket with roller is provided for the MyiQ2, MyiQ, or iQ5 optics module. It is mounted
to the rear of the iCycler thermal cycler.
Align the optics module support bracket with the two holes on the rear of the iCycler thermal
cycler.
Using a #2 Phillips screwdriver, adjust the height of the bracket with two of the appropriate
screws provided with the system accessories. Both of the screws should be approximately in the
center of the slots on the bracket.
1.2.4 Installing the Optics Module
1. Remove the plastic sheath and protective label from the optics module and place the
optical module on a flat surface, taking care not to touch the inside of the nose portion of
the module.
2. Remove the protective label from the optical lid.
3. Slide the optics module onto the U-bracket.
4. Secure the optics module to the U-bracket using the two long, thin hex screws and the
hex driver provided.
5. After the optics module has been installed, confirm that the optics module and lid
assembly can be opened and closed readily.
If the lid is difficult to open, lower the support bracket slightly before tightening the bracket
mounting screws. If the lid is difficult to close, try raising the support bracket slightly before
tightening the rear screws.
1.2.5 Connecting Power and Communication Cables to the System
Before connecting any communication or power cables to the system, confirm that the power
switch for the iCycler thermal cycler and the optics module are in the OFF position.
1. Close the optical reaction block by sliding the lid forward and pressing down on the lid
handle. On the right side of the optics module are three connectors.
2. Using the cables provided, establish power and communication with the computer as
follows:
• Recessed 3-pin power connector — Connect the supplied power cord between the
optics module and a grounded power outlet. This connection provides power only to
the optical module; a separate power cord must be connected to the iCycler thermal
cycler
• Serial connector — A serial connector is located at the rear of the iCycler thermal
cycler. Connect the serial cable to the rear of the iCycler chassis and to the serial port
on the side of the optics module. This connection enables communication between the
iQ5 software and the iCycler thermal cycler
• USB port connector — Connect the supplied USB cable between the optics module and
a USB 2.0 high-speed enabled port on the computer. Data are transferred to the
computer via this cable. This single connection directs the operation of both the optics
module and the iCycler thermal cycler by the iQ5 Optical System software
• At the right rear corner of the optical reaction block is a single connector the Positive
docking connector — This self-aligning connector is secured into place when the optics
- 3 -
Page 13

Section 1 Getting Started
module is installed on the iCycler chassis. This connection senses when the lid handle
is lifted.
1.3 Installing the iQ5 Software
Locate the software installation disk for iQ5 Optical System software version 2.1 provided with
the MyiQ2 system. This installation disk is compatible with computers running the Windows XP
and Windows Vista 32-bit operating system.
1. Insert the iQ5 Optical System software installation CD in a CD-ROM drive.
2. If the installation program does not begin automatically, click Run… in the Start menu
and then type X:\iQ5\Setup, where X is the drive letter of the CD-ROM drive. For
example, if the CD-ROM is the E drive, type E:\iQ5\Setup.
3. Follow all screen prompts to finalize the installation. Certain configurations of Windows
operating systems initialize new folders by assigning Read and Execute permission for
the members of the Users group. If you have this type of operating system and this is a
first-time installation, the administrator must change the protection for the Program
Files/Bio-Rad folder or the Program Files/Bio-Rad/iQ5 folder so that you can save
protocol, plate setup, and data files.
4. If you still cannot write to the folders beneath the Program Files/Bio-Rad/iQ5 folder after
changing the protection on either of these folders, check the properties of each folder.
Specifically, in the Properties window Securities tab, ensure that the checkbox that allows
inheritable permissions to propagate to that folder has been selected.
5. Confirm that the iQ5 system software is working properly by double-clicking on the
shortcut icon located on the Windows desktop, or by selecting the iQ5 program icon from
the Bio-Rad folder in the Windows Start menu.
1.3.1 Installing the Camera Drivers
Before using the real-time PCR detection system for the first time the camera drivers must be
installed.
1. Power on the iCycler thermal cycler and the optics module.
2. Windows will display a “Found New Hardware Wizard” dialog box. To install the camera
drivers, select the option to Install from a list or specific location. Click Next to
continue.
3. In the new window that appears, select Search for the best driver in these
locations, and then click the Include this location in the search checkbox. Use the
browse button to navigate to the “iQ5\Drivers” folder. The default location of the drivers
is: C:\Program Files\Bio-Rad\iQ5\Drivers. Click Next to continue.
4. Next Windows will display a Hardware Installation dialog box regarding Windows Logo
Testing, a service offered by Microsoft. To complete driver installation for the iQ5 or
MyiQ systems, click Continue Anyway.
1.4 Recommended Computer Settings
For the MyiQ2, MyiQ and iQ5 systems to communicate properly with the iQ5 Optical System
software, version 2.1, the computer settings should be set as described below.
- 4 -
Page 14

Section 1 Getting Started
Computer Power Management Settings on Windows Vista
1. From the Start menu, choose Control Panel. Switch the Control Panel view to Classic
View.
2. Choose Power options, and then choose Create a Power Plan.
3. Choose the High Performance plan, then enter the Plan name iQ5 software, and press
Next.
4. Change the Turn Off The Display and Put the Computer to Sleep options to “Never”.
5. Click Save Changes.
6. Select your new iQ5 software power plan by clicking on the radio button, open Change
Advanced Power Settings to set the following conditions.
a. Adjust settings to turn off hard disk after 180 minutes.
b. For Sleep Settings:
i. Set Sleep After setting to “Never”
ii. Set Hybrid Sleep setting to “On”
iii. Set Hibernate After setting to “Never”
c. For USB Settings, set USB Selective Suspend setting to “Disabled”.
d. Set Search and Indexing, Power Savings Mode setting to “Balanced”,
e. Set Display, Turn Display Off After setting to “Never”.
When completed, click Apply and then Done.
Computer Power Management Settings on Windows XP
1. From the Start menu, choose Settings, then Control Panel.
2. Choose Power Options, the Power Options Properties window will open.
3. On the Power Schemes tab, set all power scheme settings to Never.
4. On the Hibernate tab, deselect the Enable Hibernation checkbox.
5. Return to the Control Panel window, and select Display; the Display Properties window
will open.
6. On the Screen Saver tab, select None.
7. Return to the Control Panel window, and select System, the System Properties window
will open.
8. On the Hardware tab, click on the Device Manager button to open the Device Manager
file tree. Click on the “+” button to expand the Universal Serial Bus Controllers list.
9. Scroll down and highlight the first listed USB Root Hub. Right-click and select the
Properties option to open the USB Root Hub Properties window.
10. Select the Power Management tab, and deselect the Allow the Computer to Turn Off This
Device to Save Power checkbox.
11. Repeat steps 9–10 for each individual USB root hub listed.
- 5 -
Page 15

Section 1 Getting Started
1.5
Calibrating the Instrument
Before using the real-time PCR detection system for the first time, mask alignment, background
calibration, persistent well factor collection, and pure dye calibration (for MyiQ2 and iQ5 systems
only) must be performed. See section 7, Calibrating the Instrument, for detailed instructions on
calibration.
1.6 Compatibility with Earlier Versions of the iQ5 Optical
System Software
Data Files
Protocol, plate setup, and data files generated or saved with version 3.0 or 3.1 of the iQ RealTime Detection System software and earlier versions of the iQ5 software are recognized by
version 2.1 of the iQ5 software. These .tmo, .pts, and .opd files can be opened from within iQ5
software for analysis.
To open or import files generated by older versions of the iQ Real-Tme Detection System
software (version 2.3b and earlier), you must open the file in version 3.0 or 3.1 of the iQ
software first and then save the file again.
Calibration Files
Earlier versions of the iQ5 Optical System software (earlier than version 2.1) have volumedependent calibration files for both persistent well factors and pure dye calibration collection. In
version 2.1, there is no volume dependency for these calibration files. If a computer is upgraded
from an earlier version of the iQ5 Optical System software to version 2.1, existing calibration files
will be upgraded to remove calibration volume dependency.
For example, Persistent_Plates_Film_25ul_IQ2Emulator_IQ2Emulator.xml will be upgraded
and renamed as Persistent_Plates_Film_IQ2Emulator_IQ2Emulator.xml.
In all cases, the original calibration files with the volume dependency will be backed up to the
backup folder after upgrading.
- 6 -
Page 16

Section 2 Quick Guides
Section 2. Quick Guides
This section contains quick guides for the following topics:
o Protocol Quick Guide (page 7)
o Plate Setup Quick Guide (page 8)
o Running a Real-Time Experiment Quick Guide (page 9)
o Data Analysis Quick Guides
o PCR Quant Tab Quick Guide (page 10)
o End Point Quick Guide (page 11)
o Allelic Discrimination Quick Guide (page 12)
o Gene Expression Quick Guide (page 13)
o Post-Run Plate Editing Quick Guide (page 16)
The fundamental steps of running a real-time PCR experiment on the MyiQ2, MyiQ, or iQ5 RealTime Detection System are the following:
1. Select, Edit, or Create a Protocol.
2. Select, Edit, or Create a Plate Setup.
3. Click Run
4. Select the appropriate Well Factor option and begin the run
5. Analyze the data
The quick guides below outline key procedures from each step. These quick guides are also
located in the online help of the iQ5 Optical System software.
For more complete information regarding the procedures and features of the iQ5 Optical System
software, refer to the relevant sections of this user manual.
2.1 Protocol Quick Guide
Within the Workshop module click the Protocol button.
Selecting a Protocol
1. Navigate to your desired protocol file using the file tree browser.
2. Click the protocol name once. The selected protocol appears in the Selected Protocol
pane.
NOTE: The iQ5 software has a number of sample protocol files which may be used.
- 7 -
Page 17

Section 2 Quick Guides
Editing or Creating a Protocol
Edit the selected protocol by clicking Edit in the Selected Protocol window, or create a protocol
from a protocol template by clicking Create New in the Selected Protocol window.
NOTE: Clicking Edit or Create New in the Selected Protocol pane opens the Protocol Editor.
NOTE: You can only exit the Protocol Editor by clicking Save & Exit Protocol Editing or
Cancel & Exit Protocol Editing.
1. Edit the protocol by performing one or more of the following five tasks:
• Edit the Dwell Time and Setpoint temperature: Click in the Dwell Time or
Setpoint cell, and then enter the Dwell Time or Setpoint temperature. To enter 10
seconds, type 0 followed by a colon (:), then type 10 (that is, as the time appears in
the spreadsheet). Alternatively, 10 seconds can be entered as 0.10.
• Set Data Acquisition Step: Click in the Data Acquisition column, and then click
Real-Time at the step you want to collect real-time data. Click Melt Curve if data
from a melt curve is required.
NOTE: Ensure that every thermal protocol has at least one Data Acquisition step.
• Insert a cycle: Insert a cycle by clicking in the Insert column within the cycle row.
Cycles have a blue background. The iQ5 software inserts the new cycle below the
current cycle
• Insert a step: Insert a step by clicking in the Insert column within a step row.
Steps have a white background. The iQ5 software inserts the new step below the
current cycle. You can use the Options cell to customize whether the step or cycle is
inserted before or after the current cycle/step, as well as how many steps the iQ5
software will insert when you insert a cycle
• Delete a cycle: Delete a cycle by clicking in the Delete column within a cycle row.
Cycles are indicated with a blue background.
• Delete a step: Delete a step by clicking in the Delete column within a step row.
Steps are indicated with a white background
2. Save the protocol by clicking Save & Exit Protocol Editing. Enter the name of the
protocol in the Save As dialog box, and then click Save.
2.2 Plate Setup Quick Guide
Within the Workshop module:
1. Open the Plate Setup Editor Window using one of the following methods:
• Click Create New in the plate setup display pane to enter the Plate Setup Editor
or
• Click Plate and select the desired plate setup file from the file tree directory. Double-
click the file name to go directly to the Plate Setup Editor
or
• Click Plate and select the desired plate setup file from the file tree directory. Click
the file name to open the plate setup in the bottom right section of the Workshop
- 8 -
Page 18

Section 2 Quick Guides
window. Click Edit to open the plate setup in the Plate Setup Editor
or
• Click Data File and select the desired data setup file from the file tree directory.
Click the file name to open the plate setup associated with the data file in the bottom
right section of the Workshop window. Click Edit to open the plate setup in the Plate
Setup Editor
2. Enter or edit any notes about the plate setup in the Notes box.
3. Enter or edit the sample volume, seal type, and vessel type.
4. Enter or edit a name for the experiment.
5. Click Select/Add Fluorophores and select or edit the fluorophores to be used on the
plate.
6. For most experiments, leave the Whole Plate Loading box checked. With Whole Plate
Loading, changes made to any fluorophore within a well are extended to the other
fluorophores within the well and the replicate group. If you are editing a plate, the Whole
Plate Loading checkbox may be unavailable because it is not appropriate based on the
current definition of the plate.
7. Click a sample type icon.
8. Select the type of replicate loading desired.
9. Click a fluorophore.
10. Click or drag across the plate to define wells with the selected fluorophore and sample
type.
11. Continue defining the remaining wells that will contain the first fluorophore by changing
to any other sample type icons required.
To calculate standard concentrations automatically, click Dilution Series and enter the
upper or lower concentrations, and units of the standards, set the dilution factor, and
click Apply Dilution Series.
12. Repeat steps 7–11 for any additional dye layers/fluorophores as required. Remember
that if the Whole Plate Loading box is checked, changes made in standard concentrations
will be applied to all dye layers for that well and extended to all replicates in the group.
NOTE: To delete a previously defined well, click the Delete All icon, and then click the
well. To delete the selected fluorophore from a previously defined well, click the Delete
Fluorophore icon, and then click the well.
13. Click Save & Exit Plate Editing.
2.3 Running a Real-Time Experiment Quick Guide
After you click Run in the Workshop module, the iQ5 software opens the Initiate Run tab within
the Run-Time Central module.
2.3.1 Beginning a Run
To begin a run:
1. Insert the experimental plate into the iCycler reaction module.
- 9 -
Page 19

Section 2 Quick Guides
2. From the Workshop Setup window, select your desired Plate (.pts) and Protocol (.tmo)
file individually, or your desired Run Set (.run).
3. Click Run.
4. Check that the desired Protocol and Plate Setup are displayed in the bottom half of the
Initiate Run screen.
5. Select the type of well factors to use by selecting either:
• Use Persistent Well Factors
or
• Collect Well Factors from Experimental Plate
6. Click Begin Run.
7. Name the file with a unique name in the Save Optical Data File dialog box.
8. Click OK.
2.3.2 Monitoring the Run
When the real-time PCR detection system begins the run, the iQ5 software opens the Monitor
Run window. You can see the progress of the run in this window.
At the end of the run, the Run Status dialog box appears. You can choose between displaying the
data in the Data Analysis module or returning to the Workshop module.
Click Yes to proceed to the Data Analysis module.
2.4 Data Analysis Quick Guides
When the iQ5 software opens, the Data Analysis module is grayed out and inactive. To analyze a
data file, click the Data File tab of the Workshop module, select the data file, and then click
Analyze.
The Data Analysis module consists of six tabs:
• PCR Quant
• Melt Curve/Peak
• End Point
• Allelic Disc
• Gene Expr
• Edit Plate
2.4.1 PCR Quant Tab Quick Guide
Use the PCR Quant tab to set the analysis conditions for the data file including setting the PCR
baseline, setting the threshold and determining which wells to exclude or include in the
experiment.
To analyze a data file:
1. Click the Data File tab in the Workshop module.
- 10 -
Page 20

Section 2 Quick Guides
2. Select a data file from the file tree browser, and then click Analyze. The file opens in the
PCR Quant tab within the Data Analysis module.
3. Select or deselect wells to be included in the analysis by selecting Analyze Wells. Select
or deselect wells to be displayed by selecting Display Wells.
4. The iQ5 software automatically chooses the data analysis conditions including baselines
and thresholds. If the data file is being opened for the first time, an automated analysis
of baselines and threshold will be conducted, including every defined well. If the file was
previously saved after an analysis, the last set of analysis conditions are applied again.
5. Make any manual adjustments to the threshold by clicking and dragging the green
threshold line.
6. Make any manual adjustments to the baseline by right-clicking on the amplification traces
plot to access the baseline threshold parameters popup window and editing User Defined
options.
NOTE: You can revert to software auto calculated threshold and baseline by rightclicking on the amplification traces plot to access the baseline threshold parameters
popup window and selecting Auto Calculated.
2.4.2 End Point Quick Guide
You can implement End Point analysis in two ways:
• Click Run End Point to initiate the collection of End Point data from a sample plate
• Click the End Point tab in the Data Analysis module for an existing data file
Initiating an End Point Run
1. Insert the experimental plate into the iCycler reaction module.
2. From the Workshop Setup Window Select or Create the Plate Setup from the Plate Setup
tab.
3. Click Run End Point.
4. Check that the desired Protocol and Plate Setup are displayed in the bottom half of the
Initiate Run screen.
5. In the Run-Time Central/Initiate Run tab, specify the setpoint for data collection, and
then click Begin Run.
NOTE: You must use Persistent Well Factors for every End Point Run.
6. Name the file with a unique name in the Save Optical Data File dialog box, and then click
Save.
End Point Analysis of an Newly Completed End Point Run
1. Once the real-time PCR detection system completes the run, the End Point tab is
displayed.
2. Make selections for the following parameters:
• Method — Use Negatives to differentiate samples that do not amplify the target
sequence from those that do amplify the target sequence.
• End Point Tolerance and Tolerance Parameter.
- 11 -
Page 21

Section 2 Quick Guides
3. Select the wells to analyze by clicking Analyze Wells.
4. Define the positive and/or negative controls in the Define Controls column within the End
Point Analysis table.
5. Click Recalculate. The End Point Analysis table displays a positive, negative, or blank label
for each unknown under the Unknowns Call column.
6. Click Reports to obtain customized reports of the End Point Analysis.
End Point Analysis of an Existing Data File
1. Within the Workshop module click Data and select your desired data file using the file
tree browser.
2. Click Analyze.
3. Click the End Point tab.
4. Make selections for the following parameters:
• Method
• End Point Tolerance and Tolerance Parameter
5. Select the wells to analyze by checking Analyze Wells. Click on Analyze Selected Wells,
and close the Select Wells for Analysis floating window when finished.
6. Define the positive and/or negative controls in the Define Controls column of the end
point analysis table.
7. Click Recalculate. The End Point Analysis table displays a positive, negative, or blank
label for each unknown under the Unknowns Call column.
8. Click Reports to obtain customized reports of the End Point Analysis.
2.4.3 Allelic Discrimination Quick Guide – For Multiplex Data Only
The Allelic Discrimination feature of the Data Analysis module is available post-run and offers
flexibility for analyzing allelic discrimination data from multiplex PCR experiments. You can display
samples on a scatter plot based on threshold cycle or relative fluorescence units (RFU) values at
any PCR cycle. You can have the iQ5 software automatically make allele calls, or you may
manually make the allele calls.
To analyze an allelic discrimination file:
1. Within the Workshop module, click Data, and select your desired data file using the file
tree browser
2. Click Analyze.
3. The file opens in the PCR Quant tab of the Data Analysis module. The iQ5 software
automatically chooses the Data Analysis conditions. To manually adjust these conditions,
refer to the PCR Quant Tab.
4. You can select or deselect wells that will be included in the analysis by selecting the
Analyze Wells checkbox.
5. Click the Allelic Disc tab in the Data Analysis module.
6. Select the Fluorophores that represent Allele1 and Allele2 in the Assign Fluorophores
area.
- 12 -
Page 22

Section 2 Quick Guides
7. Choose between the Threshold Cycle and RFU display modes in the Display Mode area to
display the allelic discrimination data.
• Threshold Cycle displays the distribution of samples on the scatter plot based on the
threshold cycle (CT). Samples that do not cross threshold will be assigned the CT value
of the last cycle run in the experiment.
• RFU displays the distribution of samples on the scatter plot based on the RFU
generated by each sample at the last PCR cycle number. Click the drop-down list next
to the Select Cycle box to generate the scatter plot based on an RFU from a different
cycle of the PCR experiment.
8. Click Automatic Call or Manual Call. Automatic Call is the default parameter.
In Automatic Call: threshold bars are positioned automatically in one of two ways:
• Based on distribution of the control wells, when at least three wells have been
assigned to Control 1 and three to Control 2
• At 90 percent of the CT range or 10 percent of the RFU range on each axis if no
controls are named
Manual Call is the alternative analysis mode. Adjustments may be made either in the
scatter plot or in the data spreadsheet:
• Scatter plot: Make a selection in the Allele Call box. Then click and drag the cursor
over the corresponding sample(s) in the scatter plot. The iQ5 software reassigns the
samples to the allele call selected by the radio button, and updates the data
spreadsheet accordingly
• Data spreadsheet: Click in the Call cell of the spreadsheet. A menu appears that lets
you select an allele call for that sample. Once you select an allele call, the scatter plot
reflects the change
NOTE: You may click and drag threshold bars directly on the plot to adjust Automatic
Call assignment.
9. Click Reports in the menu to obtain customized reports for the allelic discrimination
data.
2.4.4 Gene Expression Quick Guide
The iQ5 software can present expression data normalized to one or more reference genes, or, for
data normalized before PCR, as a relative quantity.
Calculating Relative Quantity (dCT, ∆CT)
To calculate Relative Quantity (dCT):
In the PCR Quant screen (with a .opd data file open), assess Threshold and Baseline information
for the data file and make changes if necessary.
1. Click the Gene Expr tab.
2. Make any changes to Gene and Condition (for example, Sample and Treatment)
assignments in the Gene Expression Plate interface.
• Expand the Gene Expression Plate Interface view by clicking on the “+” button to
make well identification and selection easier. Highlight the wells in the gene
expression plate interface you wish to edit
- 13 -
Page 23

Section 2 Quick Guides
• Change the gene assignments in these wells by typing your desired name into the
gene pull-down men, then click enter to apply the name to the selected wells
• Change the condition assignments in these wells by typing your desired name into
the condition pull-down menu, then click enter to apply the name to the selected
wells
NOTE: The Gene and Condition Names have a character entry limit of 15 characters
• Minimize the Gene Expression Plate interface by clicking on the “-” button to return
to the standard view of the Gene Expr tab window
3. Click Relative Quantity (dCt).
4. Click Recalculate to see your results.
5. Relative Quantity results are graphed. The Data Table spreadsheet is accessed by
clicking Data Table. The Data Table spreadsheet lists the Condition and Gene name,
calculated expression values and CT values. Right-click to print or export this data to
Excel.
NOTE: Click the Settings tab, then select Gene List to enter a specific user-defined gene
reaction efficiency.
NOTE: Click the Settings tab, then select Condition List to select a particular sample as a
control sample.
If you need to compare these data to results obtained in other .opd files, you will need to enable
this file for Multi-file Gene Expression analysis, also called a Gene Study. To enable your file for
Gene Study:
1. Click the Enable for Gene Study button.
2. Go to the File menu to save your file.
Calculating Normalized Expression (ddCT, ∆∆CT)
To calculate Normalized Expression (ddCT):
1. In the PCR Quant screen (with a .opd data file open), assess Threshold and Baseline
information for the data file and make changes if necessary.
2. Click the Gene Expr tab.
3. Make any changes to Gene and Condition (for example, Sample and Treatment)
assignments in the Gene Expression Plate interface.
• Expand the Gene Expression Plate Interface view by clicking on the “+” button to
make well identification and selection easier. Highlight the wells in the gene
expression plate interface you wish to edit
• Change the gene assignments in these wells by typing your desired name into the
gene pull-down men, then click enter to apply the name to the selected wells
• Change the condition assignments in these wells by typing your desired name into
the condition pull-down menu, then click enter to apply the name to the selected
wells
NOTE: The Gene and Condition Names have a character entry limit of 15 characters
- 14 -
Page 24

Section 2 Quick Guides
• Minimize the Gene Expression Plate interface by clicking on the “–” button to return
to the standard view of the Gene Expr tab window
4. Click the Settings tab, then select Gene List to set your desired reference gene(s).
5. Click Normalized Expression (ddCt).
6. Click Recalculate to see your results.
7. Normalized expression results are graphed. The Data Table spreadsheet is accessed by
clicking Data Table. The Data Table spreadsheet lists the Condition and Gene name,
calculated expression values and CT values. Right-click to print or export this data to
Excel.
NOTE: Use the Settings tab, then select Gene List to enter a specific user-defined
gene reaction efficiency. Use the Settings tab, then select Condition List to select a
particular sample as a control sample.
If you need to compare these data to results obtained in other .opd files, you will need to enable
this file for Multi-file Gene Expression analysis, also called a Gene Study. To enable your file for
Gene Study:
1. Click the Enable for Gene Study button.
2. Go to the File menu to save your file.
Gene Study (Multiple File Gene Expression) Quick Guide
To create a Gene Study (for analysis of multiple .opd files):
1. For each .opd file you wish to combine to a Gene Study.
a. In the PCR Quant screen (with a .opd data file open), assess Threshold and
Baseline information for the data file and make changes if necessary.
b. Click the Gene Expr tab.
c. Make sure all files to be included in the Gene Study have the Enable for Gene
Study button active.
d. Save each individual file.
2. From the menu toolbar select File/New/Gene Study.
3. In the Gene Expression Study Manager select Add .opd’s.
4. Select the files that you wish to add to the Gene Study. More than one file can be
selected.
5. Once the files are added to the Gene Expression Study Manger, select OK.
6. Make any changes to Gene and Condition (for example, Sample and Treatment)
assignments in the Gene Expression Plate interface.
• Expand the Gene Expression Plate interface view by clicking on the “+” button to
make well identification and selection easier. Highlight the wells in the gene
expression plate interface you wish to edit
• Change the gene assignments in these wells by typing your desired name into the
gene pull-down men, then click enter to apply the name to the selected wells
- 15 -
Page 25

Section 2 Quick Guides
• Change the condition assignments in these wells by typing your desired name into
the condition pull-down menu, then click enter to apply the name to the selected
wells
NOTE: The Gene and Condition Names have a character entry limit of 15 characters.
• Minimize the Gene Expression Plate interface by clicking on the “–” button to return
to the standard view of the Gene Expr tab window
7. Select either Normalized expression (ddCt) or Relative quantity (dCt).
NOTE: For Normalized Expression analysis you must first assign reference gene(s). Click
the Settings tab, then select Gene List spreadsheet to set your desired reference gene(s).
8. Click Recalculate to see your results.
9. Normalized Expression results are graphed. The Data Table spreadsheet is accessed by
clicking Data Table. The Data Table spreadsheet lists the Condition and Gene name,
calculated expression values and CT values. Right-click on the spreadsheet to print or
export this data to Excel.
NOTE: Click the Settings tab, then select Gene List to enter a specific user-defined gene
reaction efficiency.
NOTE: Use the Settings tab, then select Condition List to select a particular sample as a
control sample.
2.4.5 Post-Run Plate Setup Editing Quick Guide
For post-run editing of the Plate Setup saved within a data file:
1. From the Workshop module:
• Click Data File above the directory of the home workshop. Navigate the directory
until the desired data file is found. Double-click the file name to bring the file
directly into the Data Analysis module
or
• Click Data File above the directory of the home workshop. Navigate the directory
until the desired data file is found. Click the file name once to open the plate
setup associated with the data file in the bottom right section of the Workshop
window. Click Analyze to bring the data to the Data Analysis module
2. At the top of the Data Analysis window, click Edit Plate.
3. A modified version of the Plate Setup Editor will open. In this modified window, you
cannot add fluorophores to or remove fluorophores from the fluorophore list. Nor may
you add a previously defined fluorophore or remove a previously defined fluorophore
from a well.
4. Edit any notes about the plate setup in the Notes box.
5. Edit the name of the experiment in the Experiment Name box.
6. For most experiments the Whole Plate Loading box will be checked. With Whole Plate
Loading, changes made to any fluorophore within a well are extended to the other
fluorophores within the well and within the replicate group. If you are editing a plate, the
Whole Plate Loading checkbox may be unavailable because it is not appropriate based on
the current definition of the plate.
7. Click a fluorophore.
- 16 -
Page 26

Section 2 Quick Guides
8. Click a sample type icon.
9. Select the type of replicate loading desired.
10. Click or drag across the plate to define wells with the selected fluorophore and sample
type.
11. Continue defining the remaining wells that will contain the first fluorophore by changing
to other sample type icons as appropriate.
To calculate standard concentrations automatically, click Dilution Series and enter the
upper or lower concentrations and units of the standards, set the dilution factor, and
click Apply Dilution Series.
12. Repeat steps 7–11 for any additional dye layers/fluorophores as required. Remember
that if the Whole Plate Loading box is checked, changes made in standard concentrations
will be applied to all dye layers for that well and extended to all replicates in the group.
13. Click Apply Plate Changes to make the changes. To see the effect on analysis, go to one
of the other Data Analysis windows.
NOTE: To delete a previously defined well, click the Delete All icon, and then click the
well. To delete the selected fluorophore from a previously defined well, click the Delete
Fluorophore icon, and then click the well.
NOTE: Use the Next # checkbox to enter a particular number to assign to the next
standard or sample you define.
NOTE: The original plate setup is retained with the data file and may be restored at any
time by clicking Restore Original Plate.
- 17 -
Page 27
Section 2 Introduction to the iQ5 Optical System Software
Administrator to add
Section 3. Introduction to the iQ5 Optical
System Software (Version 2.1)
The iQ5 software is divided into five sections, called modules. Icons representing each of the
modules are always shown on the left side of the screen. The active or selected module has an
orange background, whereas unselected modules have a gray background. Each module is
subdivided into windows that perform a specific function for that module.
The five modules in the iQ5 software are the:
Workshop Module: This module is used to select a Plate and Protocol and
Run an experiment. It is also where experimental files are selected and
opened for Data Analysis. The Workshop module consists of a Setup and
Plate Summary window. The Protocol, Plate, Run Set and Data File tabs can
be used to select, open, edit or create files. In Setup the selected Protocol
and Plate Setup can be run or the selected data file can be opened for
analysis in Data Analysis.
Run-Time Central Module: This module is used to initiate and monitor
experimental runs. It is accessed from the Workshop module, once the
Protocol and Plate Setup have been chosen, by clicking Run or Run End
Point.
Data Analysis Module: This module contains a suite of tools enabling you
to conduct thorough and varied analyses of your experimental data. Within
this module are screens for Quantitative, Melt Curve/Peak, End Point, Allelic
Discrimination, and Gene Expression analyses. The Edit Plate screen permits
post-run editing of the experimental plate setup, allowing you to correct
erroneous sample type assignments. The Data Analysis module is opened
automatically when you open a saved data file from the Workshop.
Calibration Module: In order to extract the best data from your real-time
PCR experiment, the MyiQ2, MyiQ or iQ5 systems must be calibrated. These
simple and easily-performed calibration routines are accessed in the
Calibration module. There are calibration routines for Pure Dyes, Mask
Alignment, Background, and Well Factors collection.
User Profile Module: This module can be used by the
new users and to set users access/restrictions to various functions of the iQ5
software.
All users can use this module to set their personal preferences for the iQ5
software.
- 18 -
Page 28
Section 4 Workshop Module
Section 4. Workshop Module
This section contains information on the following topics:
o Setup Tab (page 19)
o Plate Setup (page 21)
o Protocol (page 31)
o Run Set (pg 37)
o Opening a Data File (page 39)
o Applying Alternate Calibration Files (page 39)
The Workshop module consists of the Setup and Plate Summary tabs. For detailed information
on the information contained in the Plate Summary tab, refer to section 4.2.10
4.1 Setup Tab
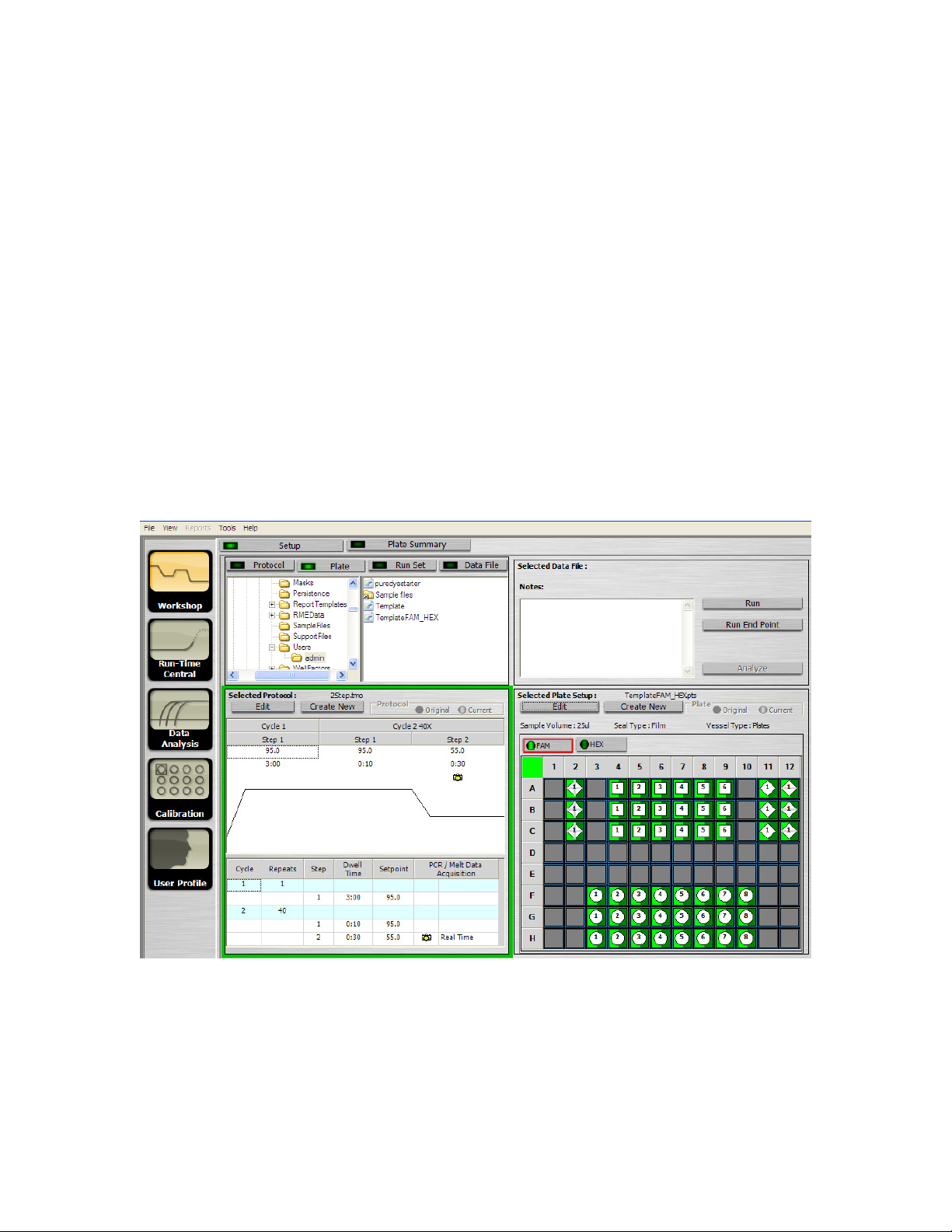
The Setup tab window (Figure 4.1) consists of four sections.
Fig. 4.1. The Setup tab.
- 19 -
Page 29

Section 4 Workshop Module
4.1.1 File Browser Section
Located in the top left of the Setup window, the file browser area contains a folder tree on the
left side and a file list on the right side.
The files displayed in the list depend upon the selected Setup window tab. With the Setup tab
active, the displayed Setup window contains four additional tabs:
• Protocol: Used to select, edit, or create a protocol for running a real-time experiment.
If the Protocol tab is selected, the iQ5 software displays protocol files, file extension
.tmo. The selected protocol appears in the Selected Protocol window
• Plate: Used to select, edit, or create a plate setup for running a real-time experiment. If
the Plate tab is selected, the iQ5 software displays plate setup files, file extension .pts.
The selected plate appears in the Selected Plate Setup window
• Run Set: Consists of a linked protocol and plate setup, useful when you repeat the same
experiment on a regular basis. If the Run Set tab is selected, the iQ5 software displays
run set files with the extension .run. The selected run set, which is a linked protocol and
plate setup, is displayed in the Selected Protocol and Selected Plate Setup windows
• Data File: Primarily used to select and open a data file. You can also use this tab to run
a real-time experiment using the same protocol and plate setup that were used to create
the data file. The selected plate setup can be either the Original or Current (last saved)
plate setup. If the Data File tab is selected, the iQ5 software displays data files created
from previous experimental runs, which have the file extension .opd. The iQ5 software
displays the selected data file name and any associated notes in the Selected Data File
window. The Notes box is editable only after you open the data file. The iQ5 software
displays the protocol and plate setup used in creating this data file in the Selected
Protocol and Selected Plate Setup windows, respectively
4.1.2 Selected Protocol Area
Located in the bottom left of the Setup window, the Selected Protocol area displays the details of
the protocol selected in the file browser area. The selected protocol file name appears at the top
of this window. If a data file is selected, the iQ5 software displays the protocol used to create the
data file and the data file name. The selected protocol is displayed both graphically and in
spreadsheet format.
4.1.3 Selected Plate Setup Area
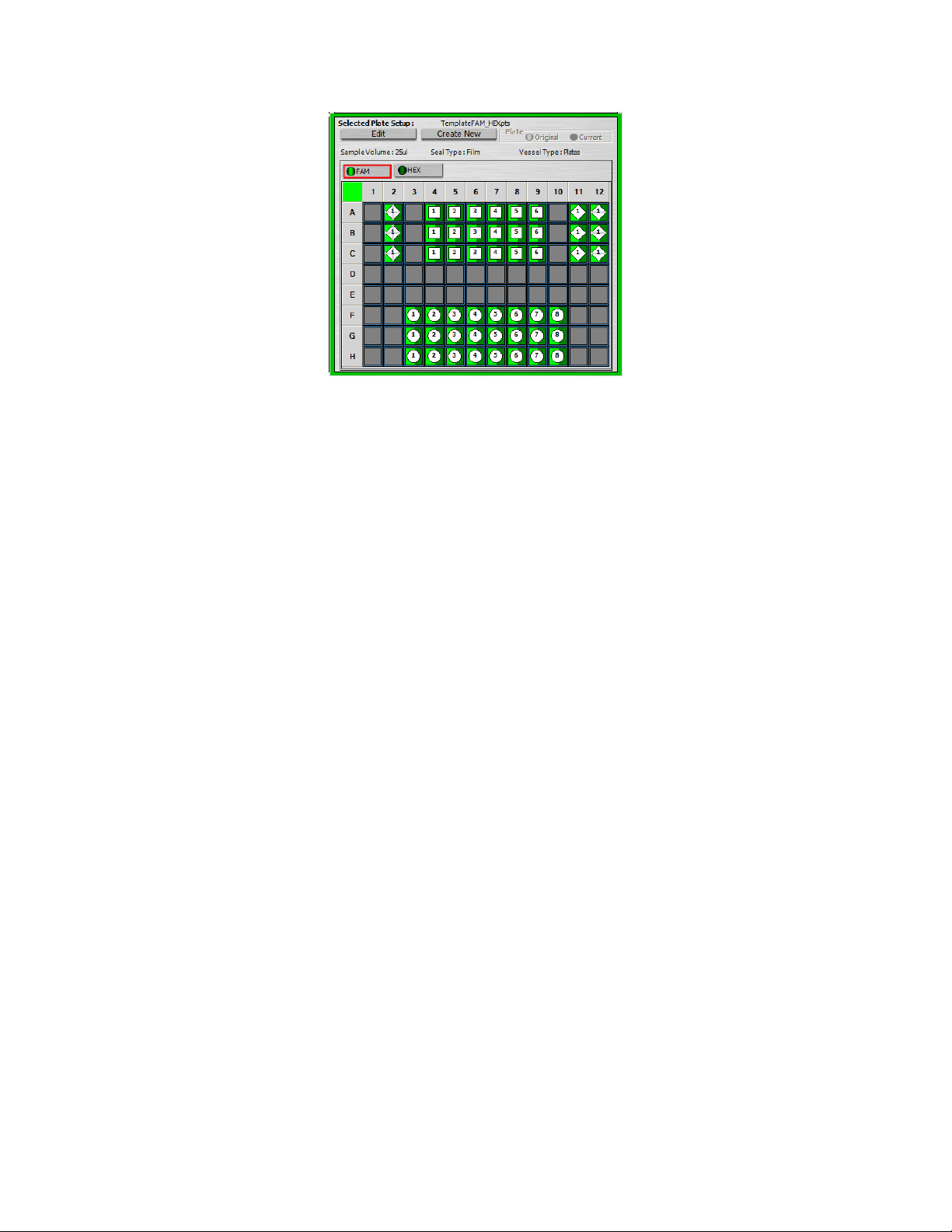
Located in the bottom right of the Setup window, the Selected Plate Setup window (Figure 4.2)
displays the details of the plate setup selected in the file browser area. The selected plate setup
name appears at the top of this window. If you have selected a data file, then the plate setup
within the data file and the data file name are both selected in the iQ5 software.
When you select the Data File tab and a data file, the iQ5 software displays the Original and
Current plate setup. The Original plate setup is the plate setup used to create the data file. The
Current plate setup is the plate setup last saved with the data file. If you are looking at a data file
that contains a plate setup that was edited and saved after the experimental data were collected,
you can revert to the original plate setup by clicking Original in the Plate box. To go back to the
present definition, click Current.
None of the information displayed on the plate in the Workshop home window can be edited
from this screen.
- 20 -
Page 30
Section 4 Workshop Module
Fig. 4.2. The Selected Plate Setup Area.
4.1.4 Selected Data File Area
Located in the top right of the Setup window, the Selected Data File area displays the selected
data file or the run set file name that you selected in the file browser area. The Notes box
displays the notes associated with either the run set or data file. Buttons in this area include:
• Run: Used to initiate a real-time PCR experimental run.
• Begin End Point: Used to initiate an End Point run.
• Analyze: Used to open a data file. You can also open the data file by double-clicking
on the data file name in the file browser area.
4.2 Plate Setup
The MyiQ2, MyiQ, and iQ5 Real-Time PCR Detection systems only display and analyze data from
wells defined in plate setup as containing sample and at least one fluorophore. In the Plate Setup
Editor window, you specify the type of sample and the fluorophores present in each well.
The Plate Setup Editor window is accessed from the main Workshop module window by clicking
on either Edit or Create New in the Selected Plate Setup window (see Figure 4.1), or by doubleclicking on an existing plate setup file from the file browser folder tree.
4.2.1 Plate Setup Editor Window
The Plate Setup Editor window (Figure 4.3) is comprised of a 96-well plate layout, functions for
specifying the sample type and fluorophores in each well, and a spreadsheet displaying the
definition in each fluorophore for any individual well.
- 21 -
Page 31

Section 4 Workshop Module
Fig. 4.3. The Plate Setup Editor Window.
Exiting the Plate Setup Editor
You can return to the Workshop at any time by pressing Cancel & Exit Plate Editing. Any current
changes made since the file was last saved will be lost.
To save changes before returning to the home screen, click Save & Exit Plate Editing and assign
a name to your new plate setup in a standard Windows Save dialog box. All plate setup files are
automatically assigned an extension of .pts.
NOTE: Plate setup files from earlier versions of the iQ5, iQ, or MyiQ software can be opened iQ5
Optical System software version 2.1, but before they can be saved, you must assign a vessel type
and sealing mechanism if not previously defined.
4.2.2 Well Definition Icons
Fig. 4.4. The Well Definition toolbar.
There are nine icon running across the top of the representation of the experimental plate
(Figure 4.4). These buttons are used to provide the two pieces of information required for each
well: sample type and fluorophore(s) to be monitored.
The active button is always surrounded by a red box.
Standard Pointer for selecting, but not altering, wells.
Defines Standards
Defines Unknowns
Defines No-Template Controls
- 22 -
Page 32

Section 4 Workshop Module
Defines Positive Controls
Paint Bucket icon: When the Paint Bucket icon is active and you click a well in the
plate setup, all wells in that replicate group will be assigned the currently selected
fluorophore.
Delete Fluorophore icon: When the Delete Fluorophore icon is active and you click in
a well in the plate setup, the currently selected fluorophore is removed from all wells in
that replicate group. It does not change the definition of the sample type in that well.
Delete All icon: When the Delete All icon is active and you click a well, all information
from that well is removed. The other members of a replicate group are not affected.
4.2.3 Editing or Creating a New Plate Setup
1. Open the Plate Setup Editor Window within the Workshop module by using one of the
following methods:
• Click Create New in the plate setup display pane to enter the Plate Setup Editor
or
• Click Plate. Select the desired plate setup file from the file tree directory. Double-click
the file name to go directly to the Plate Setup Editor
or
• Click Plate. Select the desired plate setup file from the file tree directory. Click the file
name to open the plate setup in the bottom right section of the Workshop window.
Click Edit to open the plate setup in the Plate Setup Editor
or
• Click Data File. Select the desired data setup file from the file tree directory. Click the
file name to open the plate setup associated with the data file in the bottom right
section of the Workshop window. Click Edit to open the plate setup in the Plate Setup
Editor.
2. Enter or edit any notes about the plate setup in the Notes box.
3. Enter or edit the sample volume, seal type, and vessel type.
4. Enter or edit a name for the experiment.
5. Click Select/Add Fluorophores, and select or edit the fluorophores to be used on the
plate.
6. For most experiments leave the Whole Plate Loading box checked.
• With Whole Plate Loading, changes made to any fluorophore within a well are
extended to the other fluorophores within the well and within the replicate group.
• When Whole Plate Loading is deselected, no assumption is made about the sample
type in the second and subsequent fluorophores. The sample type can vary from
fluorophore to fluorophore and if there are standards you can define individual
dilution series for individual fluorophores.
7. Click a sample type icon.
8. Select the type of replicate loading desired.
- 23 -
Page 33

Section 4 Workshop Module
9. Click a fluorophore.
10. Click or drag across the plate to define wells with the selected fluorophore and sample
type.
11. Continue defining the remaining wells that will contain the first fluorophore by
changing to any other sample type icons required.
To calculate standard concentrations automatically, click Dilution Series and enter the
upper or lower concentrations and units of the standards, set the dilution factor, and
click Apply Dilution Series.
12. Repeat steps 7–11 for any additional dye layers/fluorophores as required. Remember
that if the Whole Plate Loading box is checked, changes made in standard
concentrations will be applied to all dye layers for that well and extended to all
replicates in the group.
NOTE: To delete a previously defined well, click the Delete All icon, and then click the
well. To delete the selected fluorophore from a previously defined well, click the Delete
Fluorophore icon, and then click the well.
13. Click Save & Exit Plate Editing. The extension .pts will be added automatically
NOTE: Selecting or creating plate setup files that contain fluorophores not compatible
with the connected instrument will result in a warning message displayed at the
bottom of the screen (Figure 4.5). This message indicates that while the plate setup
can be viewed and edited, the plate setup contains fluorophores that cannot be used to
collect new data using this plate setup.
Fig. 4.5. Invalid Plate Setup Error.
4.2.4 Fluorophore Selection
When using the MyiQ2, MyiQ, and iQ5 systems, you may specify as many as five different
fluorophores on a single plate provided they are read through the filters available on the
respective systems. Refer to section 9.2.1 for information about system filter specifications and
recommended fluorophores.
With the iQ5 software, as many as five different sets of wells can be read with the same filter
pair (that is, in the same fluorophore) as shown in Figure 4.6. For example, you may have five
different experiments on the same plate, all using SYBR® Green, and each experiment will be
treated independently.
NOTE: The software cannot distinguish between fluorophores read by the same filter pair in the
same well; therefore, in all cases, such dyes have to be in separate wells. For example, you may
not put FAM and SYBR® Green in the same well.
- 24 -
Page 34

Section 4 Workshop Module
Fig. 4.6. List of Five Sets of Wells.
Click Select/Add Fluorophores to open the Fluor Selector box. From this box, choose up to five
fluorophores to be monitored. To remove a fluorophore from the list to be monitored, uncheck
the box next to its name.
To change the default color associated with a fluorophore, click the color box on the row with the
fluorophore name and choose a new color as shown below in Figure 4.7.
Fig. 4.7. The Fluor Selector Window for the MyiQ2 Real-Time PCR Detection System.
The iQ5 Optical System software automatically recognizes which instrument is detected and
displays only the fluorophores appropriate to that system for selection. Figure 4.7 illustrates the
fluor selector window when connected to a MyiQ2 instrument.
You may add new dyes to the list at any time by clicking Add New Fluor. When you add a new
dye, you must specify the filter to be used to monitor that fluorophore and perform a pure dye
calibration for that fluorophore.
- 25 -
Page 35

Section 4 Workshop Module
4.2.5 Whole Plate Loading
There are two pieces of information required for each well: sample type and fluorophore(s) to be
monitored. There are two ways of providing that information to the plate setup editor.
When Whole Plate Loading is selected, you specify the sample type and fluorophores separately.
• In Whole Plate Loading, changes made to any fluorophore within a well are extended to the
other fluorophores within the well upon pressing Enter or clicking in another cell of the
spreadsheet. These changes are then extended to every other well within the replicate
group. This is the most efficient way of defining a plate setup for a multicolor experiment
when all wells have the same sample type at each dye layer (monitored fluorophore). For
example, you might choose to monitor four fluorophores in well A1, and in each dye layer the
sample type is Unknown. Note that if you choose Whole Plate Loading, it is assumed that
standards in each dye layer are at the same concentration, that is, the FAM standard might
be at 500 copies and so, by definition, the HEX standard is also at 500 copies.
• When the sample types vary within a well, or in the special case of standards, when the
concentration of standard varies with the dye layer, deselect Whole Plate Loading. When
Whole Plate Loading is deselected, you specify the type of sample and fluorophore
simultaneously, one dye layer at a time.
• When Whole Plate Loading is toggled off, changes made to one fluorophore are considered
unique to the fluorophore and are not automatically extended to the other fluorophores in
the well. However, these changes are extended in the same fluorophore to every other well
within the replicate group.
4.2.6 Specifying Replicates
The buttons for specifying replicates run down the left side of the representation of the
experimental plate.
This is the default setting and indicates that each sample is identical (that is, a
single replicate). With this button active, if you:
•
Click an unused well, the sample type is incremented by one
•
Click a row letter or column number, the sample type for every
member of the row or column is assigned the identical replicate
number
•
Drag across a selection, the sample type for every member within the
selection is assigned the identical replicate number
- 26 -
Page 36

Section 4 Workshop Module
five wells
the last
lick a column number, the sample number of the first three wells will be
, the second three members are incremented by two,
column within the selection. The last set of replicates may have a smaller
The Horizontal and Vertical Replication buttons, together with the number in the
Size box, specify the direction and number of replicates to be automatically
defined.
With the horizontal direction (row) button active and the Size set at 5, if you:
•
Click a well, the sample type is incremented by one for the next
in the horizontal direction, and each of the five wells will contain an
identical replicate number. If there are not five wells left in the row, the
software will wrap around to the next row
•
Click a row letter, the sample number of the first five wells will be
incremented by one, the next five wells incremented by two, and
two wells will be incremented by three. The software does not
automatically wrap around so that the last set of replicates has the same
size as the others
•
Drag across a selection, the first five members of the selection are
incremented by one, the second five members are incremented by two,
etc. Numbering goes in a horizontal direction and wraps around to the
next row within the selection. The last set of replicates may have a
smaller size than the others, depending on the number of wells within
the selected area
With the Vertical direction (column) button active and the Size set at 3, if you:
•
Click a well, the sample type is incremented by one for the next three
wells in the vertical direction. If there are not three wells left in the
column, it will wrap around to the next column
•
C
incremented by one, the next three wells incremented by two, and the
last two wells will be incremented by three. The software does not
automatically wrap around so that the last set of replicates has the same
size as the others
•
Drag across a selection, the first three members of the selection are
incremented by one
etc. Numbering goes in a vertical direction and wraps around to the next
size than the others, depending on the number of wells within the
selected area
The Next # box allows you to overwrite the default replicate number for the
next sample to be defined. This is an easy way to correct a mistakenly assigned
replicate number. Use the up and down arrows to select the next desired
replicate number, or highlight the field and type the desired number.
4.2.7 Sample Volume, Seal Type, and Vessel Type
In Plate Setup you must specify the sample volume, the type of sealing, and the type of vessel to
be used in the experiment. This is to properly associate the appropriate calibration files unique
for each vessel and sealing combination with the data file. Use the up and down arrow keys to
specify the sample volume, or highlight the field and type in the volume. Select the seal type and
vessel type from the pull-down menus.
- 27 -
Page 37

Section 4 Workshop Module
Fig. 4.8. The Seal and Vessel Type Dialog Box.
4.2.8 Defining a Dilution Series
The iQ5 software can automatically calculate the concentrations of each member of a dilution
series, given the dilution factor and the concentration of either the most or least concentrated
sample (Figure 4.9).
Fig. 4.9. The Dilution Series Dialog Box.
1. Access the dilution series box by clicking on Dilution Series.
2. Define the range of standard replicates to be defined. By default, the beginning and
ending standards are the lowest- and highest-numbered standards, but you may define
only a subset of all standards by clicking on the up or down arrows.
3. Enter the starting concentration, the concentration of the lowest-numbered standard to
be defined. The Scientific Notation button controls only how the data are displayed in the
spreadsheet.
4. Enter a series identifier for the standard if desired. It will be shown in the spreadsheet
and in the reports.
5. Enter the fold dilution factor. The default value is 10.
6. Indicate whether the concentrations decrease or increase as the standard replicates
increase in number. The default setting is Decreasing.
7. Click Apply Dilution Series, and all standard concentrations will be automatically
calculated.
Adding Additional Standards
When Define is selected (at the bottom of the dialog box), standards added to the set after the
Dilution Series box is closed will have their concentrations automatically calculated.
- 28 -
Page 38

Section 4 Workshop Module
To assign values to any newly added standards while Define is not selected, you must open the
Dilution Series dialog box again, make any necessary edits, and click Apply Dilution Series.
4.2.9 Plate Setup Editor Spreadsheets
There are two different spreadsheets available from the Plate Setup Editor window: the Well
Identifier and the Plate Setup Editor Spreadsheets.
Well Identifier Spreadsheet
The Well Identifier spreadsheet (Figure 4.10) opens beneath the representation of the
experimental plate any time that the pointer tool is clicked on a well. This spreadsheet
simultaneously displays information about each dye layer in the active well.
In this spreadsheet you may edit the sample type, identifier, and quantity. Any changes are
applied to all members of a replicate group. It is also possible to change the replicate group
assignment of a well, but doing so will also automatically change the assignment of all other
members of the original replicate group.
NOTE: In order to edit the replicate group assignment of a single well, without changing the
other members of the same replicate group, make the change on the plate, not in the
spreadsheet.
Fig. 4.10. The Well Identifier Spreadsheet.
Plate Setup Editor Spreadsheet
The Plate Setup Editor spreadsheet can be opened by clicking the spreadsheet button in the Plate
Setup Editor window. This spreadsheet displays information about all wells simultaneously, one
fluorophore at a time. Fluorophore selection is made at the top of the spreadsheet.
Fig. 4.11 The Plate Editor Spreadsheet.
Within this spreadsheet you may change the sample type, identifier, and quantity. Note that
changes made to one member of a replicate group are carried through to all members of the
- 29 -
Page 39

Section 4 Workshop Module
replicate group. Replicate group assignment changes should be made on the plate, not in the
spreadsheets.
This single-dye layer spreadsheet has another feature that differentiates it from the single-well
spreadsheet. From this spreadsheet you may Import Well Identifiers from an external comma
separated values (CSV) file.
Importing Well Identifiers
From the Plate Editor spreadsheet you may import well/sample identifiers from an external CSV
file. The number of identifiers must match the number of wells before the import can be carried
out. The simplest way to use the import feature is to fill out the Identifier template file provided
with the software in the sample files folder and then save the file in the CSV format.
4.2.10 Plate Summary
The Plate Summary window shows well definition one fluorophore at a time and provides a more
detailed view of the plate including standard quantities and identifiers, as shown in Figure 4.12.
It is useful to check the Plate Setup after editing or creating the Plate and also before performing
a run. It can also be used to check the plate layout in a data file.
The Plate Summary window can be accessed by:
• Clicking the Plate Summary tab from the Plate Editor window
• Selecting a Plate Setup, Run Set, or Data File, and clicking the Plate Summary tab from
the Workshop module main window. If the Data File tab is selected, the plate shown in
the Plate Summary window depends on whether Original or Current is clicked in the
Selected Plate Setup window. Original is the plate setup that the data file was created
with. Current is the last saved plate setup
Fig.4.12. The Plate Summary Tab.
The spreadsheet displays data one fluorophore at a time. Use the fluorophore buttons to display
the plate summary for a specific dye layer. To print the selected plate setup display, click Print.
To copy and paste the entire plate setup, click Copy to Clipboard.
- 30 -
Page 40

Section 4 Workshop Module
4.3 Protocol
Protocol files direct the operation of the iCycler thermal cycler and also specify when optical data
will be collected during the thermal cycling run. iQ5 software protocol files are stored with the
extension .tmo.
• A protocol is made up of as many as 9 cycles
• A cycle is made up of as many as 9 steps
• A step is defined by specifying a setpoint temperature and the dwell time at that
temperature
• A cycle is a collection of steps that are repeated (up to 600 times)
Every protocol must have at least one data collection step. This may be a real-time data
collection step or a melt curve data collection step.
Temperature and Dwell Time Ranges
• Temperatures between 4.0 and 100.0°C may be entered for any set point temperature
• Finite dwell times may be as short as 1 second (00:01) or as long as 99 minutes and 59
seconds (99:59)
Minimum Dwell Times for Data Collection
• 1 fluorophore = 10 sec
• 2 fluorophores = 20 sec
• 3 fluorophores = 30sec
• 4 fluorophores = 40 sec
Bio-Rad recommends using a slightly higher dwell time than the minimum values, so that more
data points are collected at each repeat.
4.3.1 Selecting a Protocol
1. With the Protocol tab active, use the browser to locate the folder where the protocol is
located, and then click on the protocol name to select it.
2. The selected protocol appears in the Selected Protocol pane (Figure 4.13).
3. The iQ5 software has a number of sample protocol files you can use.
- 31 -
Page 41

Section 4 Workshop Module
Fig. 4.13. The Protocol Selected Protocol Pane.
4.3.2 Editing or Creating a Protocol
To edit a selected protocol, double-click the protocol file or click Edit in the Selected Protocol
window. To create a protocol from a protocol template, click Create in the Selected Protocol
window. Clicking Edit or Create in the Viewing Protocol window will transfer the software to the
Protocol Editor.
You can edit the protocol in the spreadsheet at the bottom of the Editing Protocol window:
• Editing Dwell Time and Setpoint Temperature: Click in the spreadsheet in the
Dwell Time or Setpoint cell, then enter the dwell time or setpoint temperature
• Data Acquisition: Click in the spreadsheet in the Data Acquisition column, and select
real-time data collection for any step. Choose Melt Curve if data from a melt curve is
required
NOTE: Ensure that every thermal protocol has at least one data acquisition step.
• Inserting Cycles and Steps: To insert a cycle, click in the Insert column on the Cycle
row. Cycles are indicated with a blue background. To insert a step, click in the Insert
column on a Step row. Steps are indicated with a white background.
• Deleting Cycles and Steps: To delete a cycle, click in the Delete column on the Cycle
row. Cycles are indicated with a blue background. To delete a step, click in the Delete
column on a Step row. Steps are indicated with a white background.
• Saving the Protocol: To save a protocol, click Save & Exit Protocol Editing. Enter
the name of the protocol in the Save As dialog box and click Save again
- 32 -
Page 42

Section 4 Workshop Module
The Protocol Editor may only be exited by clicking Save & Exit Protocol Editing or Cancel &
Exit Protocol Editing.
4.3.3 Add Protocol Options
If you want to add any protocol options, first enable them by clicking in the check box next to its
description in the Show Options box. The Protocol Options include:
• Gradient (see section 4.3.4 for details)
• Infinite Hold
• Ramping
• Temperature Change
• Time Change
• Cycle Description
• Step Process
Infinite Hold
When a cycle is not repeated, you can specify the dwell time at any step in that cycle as infinite
by using the Infinite Hold option. This means that the instrument will maintain the specified
temperature until execution is interrupted. When an infinite dwell time is programmed within a
protocol at some step other than the last step, the instrument will go into Pause mode when it
reaches that step and will hold that setpoint temperature until the Continue Running Protocol
button in the Thermal Cycler tab of the Run-Time Central module is selected.
To program an infinite hold:
1. Click Infinite Hold in the Show Options box. A new column titled Hold appears in the
spreadsheet.
2. Select the Hold checkbox for the step that you want to maintain at a constant
temperature and enter the desired temperature in the Setpoint cell of the spreadsheet.
Ramping
The ramp rate is the speed with which the iCycler thermal cycler changes temperatures between
the steps of a cycle, or between cycles. The default condition is for the iCycler thermal cycler to
adjust temperatures at the maximum ramp rate. The iCycler thermal cycler allows you to change
temperatures at a fixed rate less than the maximum.
Ramp rates are adjustable to 0.1°C/second and must fall within the range of 0.1 to 3.3°C per
second for heating and 0.1 to 2.0°C per second for cooling. Invalid ramp rate entries are
adjusted to the nearest valid entry.
To adjust the ramp rate:
Click Ramping in the Show Options box. A new column titled Ramp Rate will appear in the
spreadsheet.
Double-click in the Ramp Rate column on the line of the spreadsheet containing the temperature
toward which you wish to control the ramp rate. Use the pull-down menu to select MIN or MAX
or make a direct entry into the field. If an invalid ramp rates is input, it is adjusted to the nearest
valid ramp rate automatically.
- 33 -
Page 43

Section 4 Workshop Module
Temperature Change
You may program an automatic periodic increase or decrease in the step temperature in a
repeated cycle. Temperature increments or decrements may be as little as 0.1°C per cycle. You
may make the increase or decrease as frequently as every cycle, and the increase or decrease
can begin following any cycle. The temperature increment or decrement may be as large as
desired, as long it does not result in temperatures which are outside the temperature limits of 4–
100°C.
To program a temperature increment or decrement:
1. Click Temperature Change in the Show Options box. Three new columns will appear in
the spreadsheet.
2. For the repeated step you want to affect, enter the incremental change desired in the
Temperature change column. To decrement the temperature enter the decremental
change as a negative number (for example, –0.5).
3. Enter the repeat in which you want the change to occur for the first time in the Begin
Repeat column. Usually it is cycle 2, but it can be any cycle greater than 1.
4. Enter the frequency that you want the change to occur in the “How Often?” column.
Usually you will want the change to occur every repeat, so enter 1 in this column.
Time Change
You may program an automatic periodic increase or decrease in the step dwell time in a repeated
cycle. Time increments or decrements may be as little as 1 sec per cycle. You may make the
increase or decrease as frequently as every cycle, and the increase or decrease can begin
following any cycle. The time increment or decrement may be as large as desired, as long it does
not result in dwell times which are outside the limits of 00:00 and 99:59.
To program a time increment or decrement:
1. Click Time Change in the Show Options box. Three new columns will appear in the
spreadsheet.
2. For the repeated step that you want to affect, enter the change desired in the Time
Change column. To decrement the time, enter the decremental change as a negative
number (for example, –0:05).
3. Enter the cycle in which you want the change to occur for the first time in the Begin
Repeat column. Usually it is cycle 2, but it can be any cycle greater than 1.
4. Enter the frequency that you want the change to occur in the “How Often?” column.
Usually you will want the change to occur every repeat, so enter 1 in this column.
Cycle Description/Step Process
You can choose from a list of descriptive names, or enter one of your own to describe cycle or
step processes.
A cycle description or step process may be entered in the spreadsheet in the following manner:
1. Click Cycle Description or Step Process in the Show Options box.
2. Click the cell of the spreadsheet for the cycle or step you wish to name and either choose
one of the listed names from the pull-down menu or enter one of your own.
- 34 -
Page 44

Section 4 Workshop Module
4.3.4 Gradient
A thermal gradient may be programmed across the reaction block at any step of a protocol. The
gradient runs from the back of the instrument to the front, with the hottest temperature in row A
and the coolest temperature in row H. All wells in each respective row are at the same
temperature so at any time during a gradient step, there will be eight different temperatures
across the block with 12 wells at each temperature. The gradient may be as large as 25°C or as
small as 1°C. The gradient is not linear, but is highly reproducible. No row can be at a
temperature higher than 100°C or lower than 40°C during the gradient step.
NOTE: A gradient cannot be applied to any step, which also has a melt curve.
Programming a Thermal Gradient
To program a thermal gradient:
1. Click Gradient in the Show Options box. Two columns will appear in the spreadsheet and
a representation of the gradient will appear on the right side of the window (Figure
4.14).
2. Click the Gradient checkbox in the spreadsheet for the desired step.
Fig. 4.14. The Protocol Editing Table.
3. The temperature listed in the Setpoint cell of the spreadsheet will be the coolest
temperature on the block during the gradient step (row H). Enter the desired difference
between the coolest and hottest temperatures during the gradient step in the Range cell
of the spreadsheet. The Gradient Display table (Figure 4.15) will update with the
temperatures at each row.
4. You can change the range in the spreadsheet or make a direct entry of the range in the
gradient display. Press Enter and the display will update with the new calculated
temperature for each row.
Fig. 4.15. Gradient Display Table.
- 35 -
Page 45

Section 4 Workshop Module
Programming a Specific Temperature for a Specific Row
To program a specific temperature in any single row:
1. Click Gradient in the Show Options box. Two columns will appear in the spreadsheet and
a representation of the gradient will appear on the right side of the window.
2. Click the Gradient checkbox in the spreadsheet for the desired step.
3. Enter the desired temperature into a specific single row on the gradient display.
4. Press Enter.
5. The temperatures for the other rows will be calculated based on the input desired
temperature and the range.
NOTE: You cannot specify the exact temperature on more than one row at a time.
4.3.5 Melt Curve/Peak
Melt Curve/Peak analysis is a dynamic tool used to measure the melting temperature (Tm) of
double-stranded DNA (dsDNA) molecules. DNA duplexes can be visualized by either incorporation
of DNA-binding dyes (for example, SYBR® Green I) or by hybridization with fluorescently labeled
probes. In the case of DNA-binding dyes and non-cleavable hybridization probes, fluorescence is
brightest when the two strands of DNA are annealed. As the temperature is raised towards the
Tm of the duplex, the fluorescence will decrease at a constant rate (constant slope). At the Tm,
there is a dramatic reduction in the fluorescence with a noticeable change in slope. The rate of
this change is determined by plotting the negative first derivative (–dF/dT) versus temperature.
The greatest rate changes yield visible peaks, representing the Tm of the dsDNA complexes.
A melt curve cycle may follow an amplification cycle or can be conducted independently of the
amplification. The melt curve may be programmed in the following manner:
1. Melt curves are programmed as a repeated cycle containing only one step. The
temperature is programmed to increase or decrease incrementally with each repeat of
the cycle. The increase or decrease combined with the number of repeats may not result
in a temperature that is below 4°C or above 100°C at any time during the protocol.
2. Insert a cycle into the protocol at the point that you want the melt curve. The step
generated in this cycle will be used to generate the melt curve data.
3. Enter the temperature at which you wish to begin the melt curve in the Setpoint cell (4–
100°C).
4. Enter an appropriate dwell time for data collection under the Dwell Time column. Dwell
times for the melt curve will vary based on the number of fluorophores in the
experiment. To enter 10 seconds, type 0 followed by a colon (:), then type 10 (that is, as
the time appears in the spreadsheet). Alternatively, 10 seconds can be entered as 0.10.
5. Click in the PCR/Melt Data Acquisition column and select Melt Curve from the pull-down
menu. A green camera will appear in the Data Acquisition cell, and two additional
columns entitled Temperature Change and End Temperature will be added to the
spreadsheet. By default, the temperature change is set at 0.5°C and the end
temperature is at 95°C (unless the setpoint is 95°C). The number of repeats to achieve
this melt curve is automatically calculated. The temperature change can be as low as
0.1°C increments. Typical temperature change values are 0.3–0.5°C for SYBR® Green I.
- 36 -
Page 46

Section 4 Workshop Module
Fig. 4.16. Melt Curve Protocol Editing Table.
6. Anneal curves can be also created by entering the temperature change as a negative
value (for example, –0.5).
7. Save the protocol by entering a file name and then clicking Save & Exit Protocol Editing.
NOTE: The minimum dwell times necessary for data collection are 10 seconds for 1
fluorophore. We recommend using a slightly higher dwell time than the minimum values
so that more data points are collected at each repeat.
4.3.6 Sample Protocol Files
There are several sample protocol files that can be used as editable templates for custom
protocol design, including:
• 2Step.tmo: This protocol can be used for most single fluorophore or SYBR® Green I
real-time PCR experiments
• 2StepAmp+Melt.tmo: This protocol can be used for most SYBR® Green I real-time
PCR experiments. The melt curve that follows the qPCR experiment can be used to
determine the number of amplicons produced in the qPCR reaction
• Gradient.tmo: This protocol can be used to determine the most optimal temperature to
perform real-time PCR experiments
4.4 Run Set
A Run Set is a linked pairing of a Plate Setup and Protocol files. Run Set files generated by the
iQ5 software are stored with the extension .run. Run sets are useful for experiments that are
repeated on a frequent basis.
When the Run Set tab is selected, run set files are shown in the file browser.
The protocol defined in the run set is shown in the Selected Protocol pane and the plate setup
defined in the run set is shown in the Selected Plate Setup pane (Figure 4.17).
- 37 -
Page 47

Section 4 Workshop Module
Fig. 4.17. The Run Set Selection Window.
4.4.1 Selecting a Run Set
1. Click Run Set.
2. Use the browser below the Run Set button to locate the folder containing the run set file
, and then click on the file name to select it.
3. The selected run set will appear in the Selected Protocol and Selected Plate Setup
windows.
4.4.2 Creating a Run Set
To create a run set:
1. Select the plate setup and protocol that are to be linked.
2. The plate setup and protocol can be stand-alone files (plate setup files have the
extension .pts while protocol files have the extension .tmo) or they can be already
together in a data file (with the extension .opd). A data file has two plate setups: Original
(the Plate Setup the data file was created with) and Current (the plate setup the data file
was last saved with). The run set will be created with the plate setup displayed in the
Selected Plate Setup window.
3. Once you have chosen the protocol and plate setup files, from the File toolbar menu,
select New, Run Set and enter a name for your new run set.
4.5 Data File
This feature is primarily used to select and open a data file. It can also be used to run a real-time
experiment using the same protocol and plate setups that were used to create the data file. The
plate setup that is selected can be either the Original or Current (last saved) plate setup.
- 38 -
Page 48

Section 4 Workshop Module
4.5.1 Selecting a Data File
1. Click the Data File button.
2. Use the file browser below the Data File button to locate the folder that contains the data
file, and then click on the file name to select it.
3. The protocol and plate setup used in the data file will appear in the Selected Protocol and
Selected Plate Setup windows. The plate setup that is shown can be either the Original
or Current (last saved) plate setup. If the data file protocol and plate setup are being
used as the conditions for running a new experiment, then the plate setup that is shown
in Selected Plate Setup window will be the one that is used to run the new experiment.
4.5.2 Opening a Data File
To open an optical data file from an amplification or melt curve experiment, select the name of
the data file in the file browser and click Analyze (or double-click the file name).
NOTE: Data files collected using an iQ5 Real-Time PCR Detection System can be opened when
the computer running iQ5 software (version 2.1) is connected to a MyiQ2 or MyiQ Real-Time
Detection System. A warning message will display at the bottom of the screen indicating that
while the data can be viewed and analyzed, the plate setup contains fluorophores that cannot be
used to collect new data using this plate setup. Similarly, opening a data file collected using a
MyiQ2 system while a MyiQ system is connected to the software will result in the same message
(Figure 4.18).
Fig. 4.18. Invalid Data File Error.
4.5.3 Opening a Pure Dye Calibration file
Opening a pure dye calibration file will not only show the pure dye fluorescent data in the PCR
Quant screen but will also write this calibration file to the appropriate calibration folder. This will
overwrite a calibration file of the same reaction vessel, seal, optical camera and thermal cycler,
and all subsequent experimental data will be collected and analyzed with the new calibration file.
4.5.4 Applying Alternate Pure Dye Calibrations (Alternate RME)
Optical data is saved to the data file together with the appropriate calibration files. For the MyiQ2
and iQ5 Real-Time systems, it is possible to overwrite the pure dye calibration files stored within
the .opd data file. This feature protects you from losing valuable experimental data if, for
example, the calibrations were conducted incorrectly or a run was completed with expired well
factors.
To apply an alternate pure dye calibration RME file to a data file;
1. Select the data file in the file browser.
2. Select Apply Alternate RMEs from the Tools menu.
3. Use the Select RME File Name dialog box to navigate to, then open the RME calibration
file.
- 39 -
Page 49

Section 4 Workshop Module
4. After the RME calibration file has been opened a Save Optical Data File dialog box will
appear. The existing pure dye calibration will be overwritten.
NOTE: It is recommended to save the optical data file with a different name as this will ensure
that the original file, with the original calibration data, is always maintained for reference.
4.5.5 Applying Alternate Background or Well Factors
If well factors have been collected incorrectly, or data has been collected with expired well
factors, you may apply an alternate well factor calibration to a data file.
When selecting an alternate well factors file to apply use Persistent Well Factor files only.
To apply an alternate Well Factor file to a dataset:
1. Select the data file in the file browser.
2. Select Apply Alternate Well Factors from the Tools menu.
3. Use the Select Well Factors File Name dialog box to navigate to, then open the desired
Well Factors calibration file
4. After the Well Factors calibration file has been opened, a Save Optical Data File dialog
box will appear. The existing well factor calibration for this data file will be overwritten.
The same approach can be used to apply alternate Background Factors.
NOTE: It is recommended to save the optical data file with a different name from the original to
ensure that the original file, with the original well factor calibration data, is maintained for
reference.
NOTE: Do not use the well factor file containing dynamic well factors (identified by the name
Dynamic_.....), since this file contains only the well factors for the wells that were used in the
experiment that generated this file. If the well list does not match, the operation will fail. Hence,
it is a good practice to only use the Persistent Well Factor files when applying alternate well
factor.
- 40 -
Page 50

Section 5 Run-Time Central Module
Section 5. Run-Time Central Module
This section contains information of the following topics:
o Initiating a Run (page 41)
o Well Factors (page 42)
o Real-Time PCR experiments using binding dyes (page 43)
o Running End Point experiments (page 43)
o Monitoring a Run (page 45)
o Run-Time Protocol Editing (page 45)
The Run-Time Central module is entered automatically after clicking Run or Run End Point from
the Workshop Setup window (Figure 5.1). There are three tabs within the Run-Time Central
module: Initiate Run, Show Plate and Monitor Run.
Fig. 5.1. Accessing the Run-Time Central Module.
5.1 Initiate Run Window: Run Selected
Use the Initiate Run window (Figure 5.2) to confirm run conditions and then to initiate the optical
data run. The protocol to be run is in the bottom left corner of the window, and the plate setup
that will be used appears in the bottom right corner. The type of well factors to be used in the
run can be selected in the top left section of the window. Record details of the experiment in the
Notes box. These notes will be incorporated into the experimental file.
To begin a run, select your desired well factor collection method and click Begin Run. The Save
dialog box opens. Type a unique name for the optical data file. The iQ5 software saves data
automatically during the experiment.
- 41 -
Page 51

Section 5 Run-Time Central Module
Fig. 5.2. The Initiate Run Window.
5.2 Well Factors
Well Factors are used to compensate for any system or pipeting non-uniformity in order to
optimize fluorescent data quality and analysis. Well Factors may be collected directly from the
experimental plate (dynamic well factors) or from an external well factor plate (persistent well
factors).
The best source of well factors, for correcting non-uniformity, are dynamic well factors collected
from the actual experimental plate. However, in order to collect well factors from the
experimental plate, the plate must meet certain requirements and be cycled for approximately 5
extra minutes.
5.2.1 Dynamic Well Factors
In order to collect dynamic well factors one of the following must be true:
• All wells must have the same combination of fluorophores AND the same concentration
of fluorophores
• No fluorophore is used both alone and in combination with other fluorophores AND no
fluorophore is used in more than one combination of fluorophores
The collection of dynamic well factors is a completely automated process, which begins as soon
as Begin Run is clicked and the file name saved. When collecting dynamic well factors, the plate
is held at 95°C for 2.5 minutes prior to the first cycle with a setpoint of 90.0°C or higher.
5.2.2 Persistent Well Factors
When setting up a plate setup to run the experiment specific reaction vessel and sealing methods
are chosen. This plate setup will not be able to be used to run an experiment unless persistent
well factor and background calibration files with matching vessel and sealing type have already
been collected.
- 42 -
Page 52

Section 5 Run-Time Central Module
Persistent well factors are stored in the Well Factors folder in the iQ5 Folder. Refer to section 7
for information on the calibration and generation of persistent well factors.
In general, persistent well factors can be used for approximately three months, but should be regenerated anytime something pertinent to the optical system is changed such as the optical
filters or the camera lamp. A warning reminder to re-calibrate to generate new persistent well
factors will occur when Persistent Well Factors are older than 90 days.
5.3 Real-Time PCR Experiments Using DNA-Binding Dyes
In most real-time PCR experiments using DNA-binding dyes, like SYBR® Green I or ethidium
bromide, dynamic well factors may not be collected. When the template DNA is denatured, the
fluorescence of these dyes is not sufficiently high to calculate statistically valid well factors using
the experimental plate.
There are three possible solutions to this problem:
• Use Bio-Rad iQ™ SYBR® Green supermix which contains fluorescein, enabling dynamic
well factors to be collected directly from the experimental plate
• Use persistent well factors
• Spike reaction mixes that do not come premixed with fluorescein with fluorescein to a
final concentration of 10 nM. The addition of fluorescein provides sufficient fluorescence
at 95°C for the collection of well factors from the experimental plate while not interfering
with the PCR reaction
5.3.1 Spiking Real-Time PCR Experiments Using DNA binding Dyes
With Fluorescein
The iQ SYBR® Green supermix is already spiked with a small amount of fluorescein that permits
the collection of well factors from the experimental plate. It is also possible to collect well factors
from the experimental plate with other commercial SYBR® Green mixes or with home-brew mixes
by adding sufficient fluorescein to bring the reaction mixture to 10 nM fluorescein.
Prepare a 1 mM solution of fluorescein by making a 1:1000 dilution of the 1 mM stock fluorescein
calibration dye in PCR buffer (10 mM Tris, pH 8.0, 50 mM KCl, 3 mM MgCl2). Then mix 1 part of
the 1 mM fluorescein with 990 µl of master mix to yield a final concentration of 10 nM
fluorescein. Once well factors are collected from the experimental plate, they are written to the
.opd file and the software continues to execute the programmed protocol.
5.4 Initiate Run Window: Run End Point Selected
Initiate an End Point run by selecting a Plate Setup, then click Run End Point. The software
transfers to the Initiate Run window in the Run-Time Central module. An End Point run uses a
predefined canned protocol in which only the temperature of the data collection step can be
modified. Enter the desired temperature of the data collection step into the Setpoint box as
shown in Figure 5.3. Click Enter. Changes entered into the Setpoint box appear in the Selected
Protocol region of the Initiate Run window.
- 43 -
Page 53

Section 5 Run-Time Central Module
Fig. 5.3. The End Point Setpoint Box.
To begin a run, click Begin Run. The Save dialog box opens. Type a name for the optical data
file. The iQ5 software saves data automatically during the experiment.
5.5 Show Plate Window
The Show Plate window (Figure 5.4) can be used to visualize samples loaded into the reaction
block of the system. Use the Show Plate window to verify the position and orientation of samples
prior to starting a run in the MyiQ2, MyiQ or iQ5 instruments.
Fig. 5.4. The Show Plate Window.
The following steps describe typical use of the Show Plate window:
1. Collect an exposure from the camera by clicking on Expose. An image of the
fluorescence from the sample block area will be displayed.
NOTE: If the image displayed is too bright or too dim, it may be necessary to adjust the
exposure time using the pull-down box in the right hand corner of the Show Plate
window.
2. Confirm that the small green boxes are visible around each well where sample
fluorescence will need to be detected. If necessary, open the instrument and correct
sample positions to match the plate setup for sample detection.
- 44 -
Page 54

Section 5 Run-Time Central Module
3. If a multiplex assay is being performed, repeat steps 1 & 2 for all fluors present in Fluor
Selection list.
4. When finished, return to the Initiate Run screen.
5.6 Monitor Run Window
Open the Monitor Run window by clicking the Monitor Run tab. The window appears as shown in
Figure 5.5. Traces are displayed in real-time and run progress is monitored in the top left hand
section of the window.
Fig. 5.5. The Monitor Run Window.
5.7 Run-Time Protocol Editing
During a run, you can access real-time protocol editing options by selecting Run-time Protocol
Editing as shown below.
Selecting this checkbox activates the Next Cycle and Add 10 Repeats buttons as shown below.
• Next Cycle: Click Next Cycle to complete the current repeat of the present cycle
before skipping to the next cycle. For example, you could use this feature when your
samples have clearly crossed threshold and you want to skip to the melt cycle of your
protocol
- 45 -
Page 55

Section 5 Run-Time Central Module
• Add 10 Repeats: Click Add 10 Repeats to add additional repeats to the current cycle.
You can click this button multiple times, however, the total number of repeats is limited
to 600. For example, it may be necessary to add repeats to a run in an experiment
amplifying low copies of DNA to allow all samples to cross threshold. Click Add 10
Repeats to an amplification cycle of 30 repeats to make it 40 repeats. Modifications to
the protocol are updated on the protocol displayed on the iCycler base module
• Pause/Stop/Resume Run: The Pause/Stop Run allows you to pause a thermal
cycling protocol. If you click Pause/Stop Run when the iCycler thermal cycler is at a
setpoint temperature, the instrument will hold at the setpoint temperature and stop
counting down the dwell time. If you click Pause/Stop Run when the iCycler cycler is
ramping to the temperature, the instrument will continue ramping until it reaches the
next setpoint temperature, then pause at that step. Clicking Pause/Stop Run activates
two new buttons. Click Resume Run to resume the thermal cycling protocol. Click End
Run to terminate the experiment
- 46 -
Page 56

Section 6. Data Analysis Module
This section contains information on the following topics:
o PCR Quant tab (page 48)
o Amplification chart (page 49)
o Standard Curve chart (page 53)
o Results section (page 55)
o Melt Curve and Peak charts (page 65)
o Allelic Discrimination module (page 69)
o End Point analysis tab (page 75)
o Gene Expression analysis tab (page 78)
o Gene Study: Multi file Gene Expression Analysis (page 99)
o Post-Run Plate Editing (page 105)
o Reports (page 107)
Section 6 Data Analysis Module
The Data Analysis module is where data is presented and analyzed. When the iQ5 software
opens, the Data Analysis module is gray out and inactive. To analyze a data file, open the file in
the Data File tab of the Workshop module by selecting the file and then clicking Analyze.
The Data Analysis module consists of six tabs:
• PCR Quant: The PCR Quant tab is used to set the analysis conditions for the data file.
The analysis conditions include setting the PCR baseline and threshold, and the wells to
be excluded or included in the experiment. The analysis conditions should be set before
using the Gene Expression, End Point, or Allelic Discrimination tabs. For experiments with
standards of known quantities, the PCR Quant tab is also where absolute quantities can
be determined. The efficiency of the PCR reaction can also be determined using standard
curves with either known quantities used to produce the standard curve or by using a
serial dilution of the template under investigation
• Melt Curve/Peak: Melt Curve/Peak is a dynamic tool used to measure the melting
temperature (Tm) of double-stranded DNA molecules. DNA duplexes can be visualized by
either incorporation of DNA-binding dyes (for example, SYBR® Green I) or by
hybridization with fluorescently labeled probes. Three major applications for Melt
Curve/Peak are peak identification (number of amplified products), characterization of
molecular beacons, and allelic discrimination
• End Point : The End Point tab provides a convenient method of analyzing final RFU
(relative fluorescence unit) values. This can be useful when PCR Analysis is to determine
if a given sample is positive or negative for a particular nucleic acid sequence
• Allelic Disc: The Allelic Disc tab is useful for assigning genotypes to unknown samples
by making comparisons to known genotypes. It can be used to distinguish among
homozygous wild types, homozygous mutants, and heterozygous individuals based either
on threshold cycle or on RFU. Allelic Discrimination analysis can only be performed on
datasets that contain multiplexed PCR data
- 47 -
Page 57

Section 6 Data Analysis Module
• Gene Expr: The Gene Expression screen has flexible tools for the determination of the
fold induction of one gene relative to another gene or relative to itself under different
circumstances, for example, temporally, geographically, or developmentally different
points
• Edit Plate: In the Edit Plate screen, you may make changes to the sample type
assignment or to the quantities of the Standards in the plate setup used to run the
experiment. This is a simple way to salvage your experiment should you make mistakes
in the sample type assignment. While you may make changes to the plate setup, the
original plate setup is never discarded and always remains with the data file so that you
may revert to it at any time
6.1 PCR Quant Tab
The PCR Quant tab is the tab that is first displayed after opening a data file that contains
amplification data (Figure 6.1). For experiments that lack amplification data, such as Melt Curve
only or End Point Only experiments, the PCR Quant tab is grayed out and unavailable for
analysis.
Use the PCR Quant tab to set the analysis conditions for the data file. The analysis conditions
include setting the PCR baseline, setting the threshold, and determining which wells to exclude or
include in the experiment. The analysis conditions should be set before using the Gene
Expression, End Point, or Allelic Discrimination tabs.
For experiments with standards of known quantities, the PCR Quant tab is also where you can
determine absolute quantities for unknown samples. You can also determine the efficiency of the
PCR reaction using standard curves with either known quantities used to produce the standard
curve, or by using a serial dilution of the template under investigation.
Fig. 6.1. The PCR Quant Tab.
- 48 -
Page 58

Section 6 Data Analysis Module
The PCR Quant tab consists of three sections:
• Amplification chart
• Standard Curve chart
• Results section
6.1.1 Customizing the PCR Quant Display
You can customize the size of the sections in the PCR Quant tab in a number of ways. In the
upper corner of each section is a + (plus) button that enlarges the section when clicked. The
enlarged section has a – (minus) button that reduces the section when clicked. You can move
divider bars between each section by clicking and dragging on the divider bar to resize panes to a
specific size.
6.2 Amplification Chart
The amplification chart (Figure 6.2) displays the relative fluorescence for each well at every cycle.
Each trace represents the fluorescence of a given fluorophore for a single well and at each cycle
a single data point is plotted which is the calculated mean of the data collected for that well
during the particular cycle. The data that is used to determine this mean point is set by the Set
Data Analysis window dialog box. The data can be plotted in Background Subtracted, PCR Base
Line Subtracted, or PCR Base Line Subtracted Curve Fit mode.
Fig. 6.2. The Amplification Chart.
6.2.1 Fluorophore Selector Buttons
Selecting Fluorophores to Display
You can use the fluorophore selector buttons, which are located under the amplification chart, to
display which fluorophores appear in the amplification and standard curve charts. Selecting a
single fluorophore is useful for determining the analysis parameters for that fluorophore.
Selecting all fluorophores can be useful for ensuring that the efficiencies of each fluorophore set
are approximately equal.
- 49 -
Page 59

Section 6 Data Analysis Module
Use the fluorophore selector buttons to select and deselect which fluorophores to display. Deselecting a fluorophore removes it from the display, but not from the analysis, selected
fluorophore buttons are displayed with a red border around their perimeter. De-selected
fluorophores are displayed without this red border. In Figure 6.3, the FAM fluorophore has been
selected, and HEX has been de-selected.
Fig. 6.3. Fluorophore Selector Buttons.
6.2.2 Select Analysis Mode
You can select from three options in the Analysis Mode drop-down list box (Figure 6.4).
Fig. 6.4. The Analysis Mode Drop-Down List.
The three analysis modes include:
• Background Subtracted: The background subtracted data is the relative fluorescence
of each fluorophore after normalizing for exposure time, background factors, and well
factors. No further analysis is possible on background subtracted data; therefore, the End
Point, Allelic Disc, and Gene Expr tabs are unavailable
• PCR Base Line Subtracted: To determine threshold cycles, construct standard curves,
and determine the concentration of unknown samples, the data must be PCR baseline
subtracted. The iQ5 software determines each PCR baseline subtracted trace by fitting
the best straight line through the recorded fluorescence of each well during the baseline
cycles. The iQ5 software then subtracts the best fit data from the background subtracted
data at each cycle to generate the PCR baseline subtracted trace. By default, the
software automatically chooses the beginning and end baseline cycles. You can override
this default and manually give each trace a beginning and ending baseline cycle. User
specified settings for PCR baseline subtraction may also be specified in the User
Preferences module
• PCR Base Line Subtracted Curve Fit: The iQ5 software fits the PCR baseline
subtracted data to a smoothed curve using a balanced flank, centroid-finding digital filter.
The curve fit process is performed in such a way that the CT is left invariant for all traces
6.2.3 Log View Button
You can click Log View, shown below, to change between a semi-logarithmic and linear display
of the amplification chart data.
- 50 -
Page 60

Section 6 Data Analysis Module
6.2.4 Selecting and Viewing Traces
• Identifying a specific amplification trace: Identify a specific trace by moving the
mouse pointer along the trace until the hand icon appears. The dialog box identifies the
trace, by both well name and fluorophore, in the top left corner of the amplification chart
• Selecting a specific amplification trace: Select a specific trace by moving the mouse
pointer on the trace until the hand icon appears, and then double-click. The dialog box
displays the selected trace. If you click the fluorophore selector buttons from the
amplification chart, traces from other fluorophores in the selected well also appear
• Viewing all traces: To restore a view of all traces right-click on the amplification chart
and select Show All Traces
• Zoom: To zoom in on a section of the plot, click and drag with the mouse on the desired
region. To zoom out, select the plot and then type R, or right-click on the plot and then
click Restore Graph in the menu
Analyze Wells
Click Analyze Wells on the PCR Quant screen to select the wells that you wish to include in data
analysis. The Select Wells to Analyze floating window appears as shown in Figure 6.5.
Fig. 6.5. The Select Wells to Analyze Window.
You can include or exclude wells for analysis in this floating window. The original data are always
preserved, and excluded wells can always be added back to the data analysis in this window.
- 51 -
Page 61

Section 6 Data Analysis Module
Removing wells from data analysis changes the calculation of the threshold location. Changing
the calculation of the threshold location may result in a change in the threshold crossing (CT)
location, thus affecting all subsequent data analyses that depend on CT values. This would
include standard curve calculations and quantification of unknowns, gene expression, and allelic
discrimination using threshold crossing values.
In addition, removing wells from analysis will change the statistics for replicates.
To select the wells to include in data analysis:
1. In the PCR Quant tab, click Analyze Wells. The Select Wells to Analyze floating window
appears. A set of fluorophore selector buttons appears at the top of this window. When
you click a fluorophore button, the sample type present in that well for that fluorophore
is colored.
2. To select or deselect an individual well, click inside that well. Wells included in the
analysis appear with a yellow and black border. Wells excluded from the analysis appear
with a pale border. When a well is excluded from analysis, all fluorophores in that well
are excluded.
3. To select all wells, click Select All.
4. To toggle the current selection so all wells currently selected will be unselected and vice
versa, click the uppermost left cell of the spreadsheet; this cell has the color of the
currently selected fluorophore. You can also perform this action on a subset of the
spreadsheet by clicking the appropriate row or column header. Only the wells in that row
or column are toggled.
5. After you determine which wells to include in analysis, click Apply if you want the Select
Wells to Analyze window to remain open after the iQ5 software re-analyzes the wells. If
you are satisfied with your selection, click OK to close the dialog box and update the
wells to include in the analysis. If you click Cancel, the iQ5 software closes the dialog box
and discards all changes.
NOTE: This procedure does not permanently remove data. This procedure only removes that
data from the current analysis. You can add the excluded wells back to the analysis at any time
by including them in the Select Wells to Analyze dialog box.
Display Wells
Click the Display Wells button on the PCR Quant screen to select the wells that you wish to
include in the data display.
After you click Display Wells, the Select Analyzed Wells for Display floating window appears, as
shown below in Figure 6.6.
You can include or exclude wells for display in the Select Analyzed Wells for Display floating
window. You can only choose from analyzed wells. Wells that have been excluded from analysis
in the Select Wells to Analyze window do not appear in the Select Analyzed Wells for Display
window.
Selecting wells for display does not change the underlying data analysis. Therefore, the
calculation of thresholds and replicate statistics does not change.
- 52 -
Page 62

Fig. 6.6. The Select Analyzed Wells for Display Window.
To select the wells to include in data display:
Section 6 Data Analysis Module
1. In the PCR Quant tab, click Display Wells. The Select Analyzed Wells for Display
window appears. Only those wells that are included in the analysis appear in the window.
2. Select and de-select wells using the same methods described in the analyze wells section
on page 51.
3. After you determine which wells to include in analysis, click Apply if you want the Select
Analyzed Wells for Display window to remain open after the iQ5 software re-analyzes the
wells. If you are satisfied with your selection, click OK to close the window and update
the wells to include in the analysis. If you click Cancel, the iQ5 software closes the
window and discards all changes.
The Select Analyzed Wells for Display window displays traces for fluorophores with parameters
per the selected fluorophore selector buttons on the amplification chart. You can use this dialog
box to select all wells for display. Restore all traces for display by right-clicking on the
amplification chart and clicking Show All Traces in the menu.
6.3 Standard Curve Chart
The standard curve chart appears when, for a given fluorophore, more than 2 standards (with
different quantities) are defined in the plate setup. The fluorophore selector buttons on the
amplification chart determine which fluorophores appear in the standard curve chart shown
below in Figure 6.7.
The bottom of the chart displays a legend that includes:
• The color used to plot each fluorophore
• The name of the fluorophore
• The efficiency of the reaction
• The coefficient of determination (R2)
- 53 -
Page 63

Section 6 Data Analysis Module
• The slope of the line
• The y-intercept
You can enlarge the standard curve chart by clicking on the plus (+) sign in the upper left corner
of the section.
Fig. 6.7. The Standard Curve Chart.
6.3.1 Standard Curve Chart Menu
Right-clicking on the standard curve chart opens a menu (Figure 6.8).
Fig. 6.8. The Standard Curve Chart Menu.
This menu includes the following options:
• Copy Graph: Copy Graph copies the standard curve chart to the clipboard. To copy the
entire graph, enlarge the chart by clicking the + button in the upper left corner of the
standard curve Chart. Then click Copy Graph in the menu
- 54 -
Page 64

Section 6 Data Analysis Module
• Print Data: Print Data prints the Standard Curve/CT Results spreadsheet to your
specified printer in Windows
• Print Graph: Print Graph prints the graph to your specified printer in Windows
• Restore Graph: Restore Graph is active only after you zoom in on the chart. Clicking
Restore Graph returns the standard curve chart to its un-zoomed state
• Show Labels: Show Labels labels standards and unknowns with the well name as
shown in Figure 6.9
Fig. 6.9. The Standard Curve Chart with Labels.
6.4 Results Section
The Results workbook consists of three spreadsheets:
• Plate spreadsheet
• Standard Curve/CT Results spreadsheet
• Amplification Data (RFU) spreadsheet
Click Results to toggle between the Plate Spreadsheet and the Amplification Data (RFU) or
Standard Curve/CT Results spreadsheets.
6.4.1 Plate Spreadsheet
The Plate spreadsheet (Figure 6.10) displays data for each well in a grid fashion that represents
the plate setup used in the experiment. The spreadsheet is viewable at the bottom of the PCR
Quant screen and can be expanded to full screen by clicking on the + button in the top left hand
corner of the spreadsheet.
- 55 -
Page 65

Section 6 Data Analysis Module
Fig. 6.10. The Plate Spreadsheet.
The spreadsheet displays only one fluorophore at a time. You can use the Plate spreadsheet
fluorophore selector buttons found under the Plate spreadsheet to determine which fluorophore
is displayed in the spreadsheet. Each well has a colored bar at the top of the cell, which is the
color used in the amplification chart to display the amplification trace for this well and
fluorophore. The sample type is always displayed in this spreadsheet.
Above the spreadsheet is a set of checkboxes you can use to display the Identifier,
Concentration, Threshold Cycle, and End Point Calls in the Plate spreadsheet.
6.4.2 Standard Curve/CT Results Spreadsheet
The Standard Curve/CT Results spreadsheet displays the well, fluorophore, sample type,
identifier, replicate number, threshold cycle, starting quantity, and statistics for replicate groups.
Access the Standard Curve/CT Results spreadsheet by clicking Results on the PCR Quant screen.
By default the Standard Curve/CT Results tab will open displaying the spreadsheet results (Figure
6.11).
Fig. 6.11. Standard Curve/CT Results Spreadsheet.
- 56 -
Page 66

Section 6 Data Analysis Module
Columns can be sorted by clicking on the column headings and reordered by clicking and
dragging to move columns. Data can be exported by right-clicking on the spreadsheet and
selecting Export to Excel. Data can be printed by right-clicking on the spreadsheet and selecting
Print. None of the data in this spreadsheet may be edited.
6.4.3 Amplification Data (RFU) Spreadsheet
You can view the individual RFU readings for each amplification trace at every cycle in the
Amplification Data (RFU) spreadsheet. Access the Standard Curve/CT Results spreadsheet by
clicking Results on the PCR Quant screen and selecting the Amplification Data (RFU) tab at the
bottom of the spreadsheet view (see Figure 6.11).
The spreadsheet can display data for only one fluorophore. The well number appears at the top
of the spreadsheet, and the cycle number appears down the side of the spreadsheet. In Single
Point mode, each cycle is represented by one point that is the mean of all the data points
analyzed during that cycle. In All Candidates mode, each individual data point displays.
6.5 Amplification Plot Context Menu
You can access PCR amplification plot data analysis and display parameters by right-clicking on
the PCR amplification plot. After you right-click on the plot, the amplification plot menu displays
(Figure 6.12).
Fig. 6.12. The Amplification Plot Menu.
6.5.1 Data Analysis Options
There are three additional data analysis options in the amplification plot menu:
•
Set Data Analysis Window
•
Digital Filter
•
BaseLine Threshold
Set Data Analysis Window
After you click Set Data Analysis Window in the Amplification plot menu, the Set Data Analysis
Window appears as shown in Figure 6.13. The number of data points collected per cycle depends
on the exposure and the dwell time of the cycle. You can change the percentage of data points
used in determining the RFU value, and you can choose to select this percentage from either the
beginning or the end of the cycle or from a window in the middle of the cycle.
- 57 -
Page 67

Section 6 Data Analysis Module
Fig. 6.13. The Set Data Analysis Window.
To set the Data Analysis Window:
1. Right-click on the PCR amplification plot, and click Set Data Analysis Window in the
menu.
2. Click either the Beginning of Cycle or End of Cycle option to select data from the
beginning of the cycle or the end of the cycle, respectively.
3. Set the window width in the Set Window Width scroll box.
4. To use all data points in the cycle, select Use Full Cycle Scan.
5. Click OK to return to the PCR Quant screen.
NOTE: To center the analysis around the data collected with a window in the middle of the cycle,
select Set Window Center and Width. Use the up or down arrows of the Set Window Width scroll
box to select the percentage of data points. These data points will be chosen around the value
set in the Set Window Width (% Full Cycle) text box. Click OK to return to the PCR Quant screen.
Digital Filter
After you click Digital Filter in the amplification plot menu, the Set Digital Filters window appears
(Figure 6.14). There are two intra-cycle data filtering options available: Rolling Boxcar and
Weighted Mean. The default filter is the weighted mean, as this filter is the only one available
during data acquisition.
Fig. 6.14. The Set Digital Filters Window.
The weighted mean is determined by the equation:
Oi = (Ri + c*M)/(1 + c)
- 58 -
Page 68

Section 6 Data Analysis Module
Where:
• Oi is the filtered value for a given data point, i
• Ri is the unfiltered value for data point i
• c is a weighted factor with a value of 2
• M is the arithmetic mean of all data points for the well within the given cycle.
The rolling boxcar filter is the arithmetic mean of the data readings i – (w–1), where w is the
filter width. For example, if you want to calculate the 20th data point (i=20) and the width is 4
(w=4), then you take the mean of the data points 17–20, data point 21 is the mean of the data
points 18–21, etc. The rolling boxcar filters only apply within a cycle. A global filter that smoothes
data from cycle to cycle is also available by clicking Enable Global Filter. The global filter operates
on the trace for a given well and fluorophore using all cycles together in a single pass. Global
filtering should be reserved for data that appears very noisy and should not be applied routinely.
Base Line Threshold Settings
In the amplification chart, the iQ5 software uses the baselines for each individual trace and the
threshold that is set for the fluorophore to set conditions to determine the threshold cycle.
Baseline cycles and threshold are only calculated by the software when in PCR Base Line
Subtracted or PCR Baseline Subtracted Curve Fit mode. You can override the automatic
conditions set for baseline and threshold in the Base Line Threshold window.
1. Ensure that only one fluorophore from the Amp Chart Fluorophore Selector buttons is
selected
2. Right-click on the amplification chart
3. Click Base Line Threshold. After you click Base Line Threshold in the amplification plot
menu, the Base Line Threshold Parameter window appears and displays the fluorophore
of the traces to be adjusted.
4. Select User Defined. In Figure 6.15, the Base Line Threshold Parameter window shows
the FAM traces are to be adjusted.
Fig. 6.15. The Base Line Threshold Parameter Window.
- 59 -
Page 69

Section 6 Data Analysis Module
Start and Ending Cycle Mode
To change the baseline range for all wells, click Select All. Then click Edit Range to enter the
start and end cycle for baseline calculation (Figure 6.16).
To change the baseline for individual wells, click on a well and then click Edit Range to enter
the start and end cycle for baseline calculation.
Fig. 6.16. The Both Start and Ending Cycle Mode.
Start Cycle Only Mode
By default both the start and ending cycle for baseline calculation are altered. If you select the
Start cycle only mode, you can only edit the Start Cycle box. This mode is useful if you want to
retain the automatically determined end cycle but wish to change the value of the start cycle.
Enter the desired value for the start cycle for the selected traces and then click OK (Figure 6.17).
Select individual wells or all wells as described above.
Fig. 6.17. The Start Cycle Only Mode.
Ending Cycle Only Mode
In the Ending cycle only mode, you can only edit the Ending Cycle box. This mode is useful if
you want to retain the automatically determined start cycle but wish to change the value of the
end cycle. Enter the desired value for the end cycle for the selected traces and then click OK
(Figure 6.18). Select individual wells or all wells as described above.
- 60 -
Page 70

Section 6 Data Analysis Module
Fig. 6.18. The Ending Cycle Only Mode.
Setting the Threshold Manually
By default, the iQ5 software automatically calculates the threshold. You can override autocalculation in one of two ways:
•
On the amplification chart, move the cursor over the displayed threshold line until the
cursor icon becomes a hand. Left-click on the displayed threshold line and drag the
threshold to the desired position
•
From the Base Line Threshold Parameter dialog box, select User Defined in the Crossing
Threshold section at the bottom of the dialog box. Enter the desired value for the
threshold position, and then click OK
6.5.2 Amplification Plot Data Display Options
There are seven additional data analysis options in the amplification plot menu:
•
Single Point
•
All Candidates
•
Adjust Graph
•
Define Trace Style
•
Display Data
•
Restore Graph
•
Show All Traces
Single Point
The Single Point mode of data presentation, in which the iQ5 software averages all data
collected at a particular step and then plots the average, is the default mode. For example, if the
iQ5 software collects four data points during the third repeat of an amplification cycle, the mean
of those 4 points is plotted at cycle 3. The alternative to viewing the data in Single Point mode is
to view the data in All Candidates mode.
All Candidates
In All Candidates mode, the iQ5 software plots every single data point that is collected. You
cannot use automated data analysis features when the data are presented in All Candidates
mode. This mode is unavailable when the analysis mode is PCR Base Line Subtracted Curve Fit.
- 61 -
Page 71

Section 6 Data Analysis Module
The alternative to viewing the data in All Candidates mode is to view the data in Single Point
mode.
Adjust Graph
Rescale or change the amplification plot from linear to log, or vice versa, by selecting Adjust
Graph. The Chart Axes Range Definition window appears (Figure 6.19).
Fig. 6.19. The Chart Axes Range Definition Window.
Enter the maximum and minimum values for the x-and y-axes into the Y Max, Y Min, X Min, and
X Max scroll box(es), or use the up and down arrows in each scroll box.
Change the display to a semi-logarithmic view by clicking Log View. Click Log View again to
revert to a linear plot.
Revert the plot settings to the default by clicking Restore Auto Scaled Chart.
NOTE: You can also change the display to a semi-logarithmic view by selecting Log View in the
PCR Quant tab.
Define Trace Style Option
You can customize the display by selecting Define Trace Style option. The Define Trace Style
window appears (Figure 6.20).
Change the trace color and symbol, which are used to display data points, on a well-by-well basis
or in groups of sample types. You can preview all changes before you apply them by clicking
Preview.
- 62 -
Page 72

Section 6 Data Analysis Module
Fig. 6.20. The Define Trace Style Window.
To modify a trace style:
1. Select the Sample Types Group to be modified (for example, Standards). (Note that
green is the default color that is automatically assigned to the Standards group when this
button is active.)
2. To specify a different color for a given sample type group on the plate, click on the
colored button in the Select Color column.
3. Select the symbol type (the default style is “None”).
4. Click Preview to see your changes, and if those changes are satisfactory, click OK.
To choose on a well–by-well basis, click the Selected Well(s) button, select a color and symbol
type, and then select the desired wells in the plate grid at the bottom of the Define Trace Style
dialog box. Click a column or row header to change an entire row or column. The dialog box
outlines the edge of each well in the grid in the selected trace color.
Select All Replicates to apply the selected color to all selected replicates of the wells.
To reset original trace style settings, click Random Colors and Remove Symbols, then click OK.
Display Data Option
Click Display Data to change the view of the results spreadsheet as follows:
• When Display Data is unselected, the results spreadsheet displays the plate spreadsheet
view
• When Display Data is selected, the results spreadsheet displays two tabs: the
Amplification Data (RFU) tab and Standard Curve/CT Results tab. You can perform the
same function by clicking Results
- 63 -
Page 73

Section 6 Data Analysis Module
Restore Graph Option
Click Restore Graph to redraw the graph to its original size after you zoom in on the graph.
Show All Traces Option
Click Show All Traces to show all traces after you single out one or more using Display Wells or
by clicking on a trace in the amplification plot.
6.5.3 Data Export Options
The amplification plot menu offers four additional options for printing and exporting graphs or
data.
• Copy Graph: Click Copy Graph to copy the amplification plot into the clipboard to import
into other programs
• Print Graph: Click Print Graph to print the graph to your default Windows printer
• Print Amplification Data: The Print Amplification Data option is only available when
the iQ5 software displays the Amplification Data (RFU) spreadsheet. You can display this
spreadsheet by clicking Results in the PCR Quant tab, or by clicking Display Data in the
amplification chart menu. When you click Print Amplification Data, the Print Preview
dialog box appears so you can print the amplification data
• Print Std Curve Data: The Print Std Curve Data option is only available when the iQ5
software displays the Standard Curve/CT Results spreadsheet. You can display this
spreadsheet by clicking Results in the PCR Quant tab, or by clicking Display Data in the
amplification chart menu. When you click Print Std Curve Data, the Print Preview dialog
box appears so you can print the amplification data
NOTE: If you alter the width of the columns in the spreadsheet in the Standard Curve/CT
Results spreadsheet, the printed spreadsheet will reflect that change.
6.5.4 Exporting Results to Microsoft Excel
Exporting Data from the Results Table
1. Make sure the Results button is activated. If not, click Results.
2. Right-click on the spreadsheet. A shortcut menu appears (Figure 6.21).
3. Select Export to Excel.
4. Enter a name and file destination for the Excel file generated with your data export.
- 64 -
Page 74

Section 6 Data Analysis Module
Fig. 6.21. Exporting PCR Quant Data Tables to Microsoft Excel.
The Export to Excel command is useful for exporting exact values from the spreadsheet. When
the Export to Excel command is selected from the menu, an Export to Excel file save box is
displayed. Choose a location where the Excel file is to be saved and click Save. The iQ5 software
automatically exports the selected data into a protected workbook.
•
The protected workbook generated by the iQ5 software contains the text values of what
is represented on the spreadsheet. For example, checkboxes from the software
application are replaced by “True” or “False” text in Excel.
•
The numeric values contained in the protected workbook are exact values from the
software application that include several non-significant figures beyond the decimal point.
This is important to note when considering whether to transfer iQ5 data spreadsheets by
a copy and paste command or the Export to Excel command. With the copy and paste
command only the significant digits displayed in the iQ5 software interface are
transferred to Excel.
6.5.5 Printing Results
The Print command on the Results table menu will print the displayed spreadsheet (Amplification
Data or Standard Curve/CT Results). When selected, a Print Preview box is opened which
contains an illustration of the spreadsheet as it will appear once printed. Clicking the printer icon
opens the Windows print dialog box. Click OK to complete the printing task.
6.6 Melt Curve and Melt Peak Charts
The RFU data collected during the melt curve part of the experiment are plotted as a function of
temperature, as shown in Figure 6.22.
The vertical temperature bar may be dragged to any position on the plot. The temperature bar
on the melt peak plot moves in sync to the same relative position on the melt curve plot and
. If you place the cursor over a trace, so that the pointer turns into a hand, the trace will be
versa
identified in the small box placed in the top right corner of the plot. The chart can be expanded
to fill the window by clicking the + box in the top left corner of the plot. Once expanded, it can
be returned to its original size by clicking the – box in the top left corner.
vice
- 65 -
Page 75

Section 6 Data Analysis Module
Fig. 6.22. Melt Curve Chart.
The data in the melt peak chart are derived from the melt curve chart (Figure 6.23).
Fig. 6.23. Melt Peak Chart.
The data in the melt peak chart show the negative rate of change in fluorescence with changing
temperature, that is:
–d(RFU)/dT
Where T is temperature.
In analyzing the melt peak data, the software assigns Begin and End temperatures to each peak
and then calculates an area beneath that curve. The floor of the peak area is specified by the
position of the threshold bar.
In order to be identified as a valid peak, a peak must have a minimum height relative to the
distance between the threshold bar and the height of the highest peak. Therefore, if the
threshold bar is dragged downward, increasing the distance between the threshold bar and the
highest peak, previously unidentified peaks may show up in the spreadsheet as their height
becomes significant in relation to the highest peak. Similarly, dragging the threshold bar up can
cause previously identified minor peaks to drop off the spreadsheet.
- 66 -
Page 76

Section 6 Data Analysis Module
The vertical temperature bar may be dragged to any position on the plot. The temperature bar
on the melt curve plot moves along to the same position as the temperature bar on the melt
peak plot is moved. If you place the cursor over a trace, so that the pointer turns into a hand,
the trace will be identified in the small box placed in the top right corner of the plot. The chart
can be expanded to fill the window by clicking the + box in the top left corner of the plot. Once
expanded, it can be returned to its original size by clicking the – box in the top left corner.
6.6.1 Melt Curve and Melt Peak Chart Menu
There are a number of features of the plot that may be modified as well as control of data
filtering. All changes specified by the context menu accessed on the melt peak plot are also
carried out on the melt curve plot. Those features are accessed by a right click on the chart,
which brings up the menu shown in Figure 6.24.
Fig. 6.24. Melt Curve/Peak Chart Context Menu.
6.6.2 Delete Selected Peaks
To remove a peak from the analysis, highlight the peak in the spreadsheet and then click Delete
Selected Peaks. You can press the Shift key to highlight multiple peaks and delete them all at
once.
6.6.3 Edit Melt Peak Begin/End Temperature
You may edit the begin and end temperatures for a melt peak by clicking Edit melt peak
begin/end temps as shown below.
After you click this option button, the currently active peak displays alone with a Begin Temp bar
and an End Temp bar. Drag the bars to the desired begin and end temperatures. As you drag the
begin and end bars along the Melt Peak plot, the iQ5 software tracks the movement on the melt
curve chart and updates the movement in the spreadsheet. When the Edit melt peak begin/end
temps option is activated, the Display Wells and Analyze Wells dialog boxes are unavailable.
In Figure 6.25, the peak begin temperature is 59°C, the peak end temperature is 79°C, and the
threshold is 1.
- 67 -
Page 77

Section 6 Data Analysis Module
Fig. 6.25. A Melt Peak Chart.
6.6.4 Melt Curve/Peak Control Area
The control area appears between the spreadsheet and the plots. This area contains buttons to
access the Display Wells and Analyze Wells dialog boxes and the Restore Defaults function
(Figure 6.26).
Fig. 6.26. The Control Area.
The position of the temperature bar, threshold bar, and the height and well of the currently
selected peak are also displayed here. You cannot edit any of these fields. When the Edit melt
peak begin/end temp option is active, the most-recently modified temperature (begin or end) is
displayed in the Temperature Bar field.
6.6.5 Melt Curve/Peak Spreadsheet
The spreadsheet below the plots displays information about the melt peaks, the RFU data, or the
–d(RFU)/dT data for each well (Figure 6.27). None of the fields may be edited, though you may
eliminate peaks from the spreadsheet by either highlighting the peak in the spreadsheet and
clicking Delete Selected Peak, or by dragging the threshold bar up to a position above the peak.
Fig. 6.27. The Melt Peaks Spreadsheet.
- 68 -
Page 78

Section 6 Data Analysis Module
The spreadsheet displays the following information for each peak:
• Peak ID: A unique identification number in the format RCC.N, where R is a row letter, CC
is a column number, and N is a number beginning with 0, 1, 2, etc. In Figure 6.27, the first
peak for well E3 is identified as E03.0 and the second peak as E03.1
• Melt Temp: The temperature at the highest point of the melt peak
• Peak Height: The highest point of the melt peak
• Begin and End Temp: Starting and ending point for melt peak. Area calculations are
based on these starting and ending points. These values may be edited, but not in the
spreadsheet
• Area: The area beneath the melt peak curve, bounded by the default peak begin and end
temperatures and the default position of the threshold bar. Because this area calculation is
defined by the default values of begin and end temperature and threshold bar, it does not
change as the threshold bar is moved or if the begin or end temperatures are edited
• Area Fraction %: When there is more than one peak associated with a well, the
contribution of each well to the total area beneath all melt peaks for that well is calculated.
The iQ5 software makes this calculation using default begin and end temperatures and the
default position of the threshold bar, so data are not affected by edits to any of those
parameters
• Edited Begin Temp: When the peak begin temperature bar is dragged to a new position,
it is reflected here
• Edited End Temp: When the peak end temperature bar is dragged to a new position, it is
reflected here
6.7 Allelic Discrimination Module (For Multiplex Data Only)
The Allelic Discrimination module is useful for assigning genotypes to unknown samples by making
comparisons to known genotypes. It can be used to distinguish among homozygous wild types,
homozygous mutants and heterozygous individuals based either on the CT or RFU value. RFU data
may be chosen from any cycle in the experiment. The assignments can be made automatically if
controls are specified, or you can make the assignments manually.
NOTE: The Allelic Discrimination module is not available with working with single color data files or
any data file generated by the MyiQ Real-Time PCR Detection System.
6.7.1 Allelic Discrimination Plot
The Allelic Discrimination plot shows either RFU (Figure 6.28A) or C
two different fluorophores at the same time. Choose which plot to view using the RFU or
Threshold Cycle radio button in the Display Mode box.
In Automatic Call mode, two bars, one vertical and one horizontal, divide the plot into four
sections: one for each homozygous state, one for the heterozygous state and a non-reactive
section. The positions of these bars may be adjusted in the Automatic Call mode. The bars do not
appear in Manual Call mode.
(Figure 6.28B) data from
T
- 69 -
Page 79

Section 6 Data Analysis Module
A B
Fig. 6.28. Allelic Discrimination Plot; RFU View (A) and Threshold Cycle View (b).
Genotype assignments for unknown samples are determined by plotting the RFU for one
fluorophore (allele 1 on the x-axis) against the RFU for the other fluorophore (allele 2 on the yaxis) on the allelic discrimination plot.
• If the unknown RFU values are greater than the horizontal and vertical bars, then the
genotype is heterozygous
• If the unknown RFU values are greater than the horizontal bar and less than the vertical
bar, the genotype is homozygous for allele 2 (allele 2 RFU is plotted on the y-axis)
• If the unknown RFU values are less than the horizontal bar and greater than the vertical
bar, the genotype is homozygous for allele 1 (allele 1 RFU is plotted on the x-axis)
• If the unknown RFU values are less than the horizontal and vertical bars, then no
genotype can be assigned
Genotype assignments for unknown samples may also be determined by plotting the threshold
cycle for one fluorophore (allele 1 on the x-axis) against the threshold cycle for the other
fluorophore (allele 2 on the y-axis) on the allelic discrimination plot.
• If the unknown threshold cycle values are greater than the horizontal and vertical bars,
then the genotype is “none”, or not assigned. Samples not crossing threshold during the
protocol are placed in this quadrant
• If the unknown threshold cycle values are greater than the horizontal bar and less than
the vertical bar, the genotype is homozygous for allele 1 (allele 1 threshold cycle is
plotted on the x-axis)
• If the unknown threshold cycle values are less than the horizontal bar and greater than
the vertical bar, the genotype is homozygous for allele 2 (allele 2 threshold cycle is
plotted on the y-axis)
• If the unknown threshold cycle values are less than the horizontal and vertical bars, then
the genotype is heterozygous
- 70 -
Page 80

Section 6 Data Analysis Module
In Manual Call mode, the plot displays the RFU or threshold cycle data only. The threshold bars
do not appear, and modifications to calls are made manually by using the drop-down menu in the
Call column of the data spreadsheet. Alternatively, modifications to calls are made by choosing
the call type from the radio buttons in the Allele Call box and then clicking and dragging to select
desired samples in the graph.
You may zoom in on the allelic discrimination plot by first selecting Zoom Enabled from the plot
menu, then left-clicking and dragging over the area you wish to enlarge. Zoom out by choosing
Restore Graph from the context menu.
6.7.2 Allelic Discrimination Plot Menu
Open the allelic discrimination plot menu by right-clicking on the allelic discrimination plot (Figure
6.29).
Fig. 6.29. The Allelic Discrimination Plot Menu.
• Adjust Graph: This feature allows you to change the way that the plot is presented.
The maximum and minimum values for either axis may be entered directly into the text
boxes. Alternatively the up and down arrows can be used to set the maximum and
minimum values
• Restore Graph: Use this to redraw the graph after zooming
• Zoom Enable: Use this to enlarge the desired plot area
• Show Labels: Labels each sample on the Allelic Discrimination plot with its well name
• Copy Graph: This will copy the displayed Allelic Discrimination plot to the clipboard for
import into other programs
• Print Graph: This prints only the graph
• Print Data: This prints the Allelic Discrimination spreadsheet data
6.7.3 Allelic Data Spreadsheet
A six-column spreadsheet is displayed in the window (Figure 6.30). The first column is the well
number, and the second and third are the identifier entered at plate setup for the two
fluorophores. The fourth and fifth column present the RFU or threshold cycle data for both
fluorophores, and the last column is the genotype call made by either the software or the user.
- 71 -
Page 81

Section 6 Data Analysis Module
Fig. 6.30. The Allelic Data Spreadsheet.
• In Automatic Call mode, the iQ5 software creates the assignments based on the positions
of the vertical and horizontal bars
• In Manual Call mode, the Call column becomes editable through a menu. This can be
very useful if you want to change the definition of a sample from an unknown to a
positive control, for example
• The data in the spreadsheet may be copied to the clipboard for export to another
program by clicking in the top left corner of the spreadsheet and typing CTRL+C
6.7.4 Automatic/Manual Call
Select the type of analysis by clicking Automatic Call or Manual Call (Figure 6.31).
Fig. 6.31. The Automatic and Manual Call Options.
Automatic Call is the default option.
• Choose Automatic Call for the software to make genotype assignments for every
unknown based on the positions of the vertical and horizontal bars and presents those
assignments in the data spreadsheet. If at least three positive (homozygous) controls are
specified for each fluorophore, the positions of the bars are based on the mean and
standard deviation of the threshold cycles or RFU values of the controls. If insufficient
numbers of controls are specified, then the position of each bar is determined by the
range of threshold cycles or RFU values in the selected fluorophores. The positions of the
bars may be manually adjusted by dragging them. Click Recalculate after any change in
the position of the vertical and horizontal bars and the software will make new genotype
assignments based on the adjusted positions.
• Choose Manual Call to make the genotype assignments directly into the data spreadsheet
or on the plot. You can use the manual call feature to change the definitions of one or
- 72 -
Page 82

more wells on the plot or in the spreadsheet and then return to Automatic Call and the
software will position the vertical and horizontal bars based on the new definitions, and
then make genotype assignments.
6.7.5 Manual Calls
To make manual calls on the plot:
1. Click Manual Call. A new box appears in the window (Figure 6.32).
Fig. 6.32. The Manual Call Specification Options Box.
Section 6 Data Analysis Module
2. Select one of the allelic call types by clicking its radio button.
3. On the plot, drag the cursor around the well or wells to be assigned the call. When the
mouse button is released, the new assignment will be made on both the plot and the
data spreadsheet.
To make manual calls in the data spreadsheet:
1. Click Manual Call.
2. Click the cell for the desired well in the Call column of the data spreadsheet. Select the
new call from the pull-down menu.
3. Note that “unknown” may only be selected in Manual Call mode.
6.7.6 Display Mode
Choose the aspect of the data that will be analyzed by clicking either Threshold Cycle or RFU
(Figure 6.33).
Fig. 6.33. The Display Mode Option Buttons.
When you click RFU, the Select Cycle option (Figure 6.34) appears so you can analyze RFU data
from any cycle. Change the cycle by entering a new value in the Select Cycle drop-down list.
- 73 -
Page 83

Section 6 Data Analysis Module
Fig. 6.34. The Select Cycle Drop-Down List Box.
6.7.7 Vertical Threshold
The Vertical Threshold box (Figure 6.35) displays the current position of the vertical bar. This box
is not editable. The positions of the vertical bar along with the position of the horizontal bar
determine the genotype assignment for unknown samples.
Fig. 6.35. The Vertical Threshold Box.
6.7.8 Horizontal Threshold
The Horizontal Threshold box (Figure 6.36) displays the current position of the horizontal bar.
This box is not editable. The position of the horizontal bar along with the position of the vertical
bar determines the genotype assignment of unknown samples.
Fig. 6.36. The Horizontal Threshold Box.
6.7.9 Normalize Data
When the RFU option button is selected, the Normalize Data option (Figure 6.37) appears as
shown below if the plate setup contains a no template control (NTC) sample type for both Allele
1 and Allele 2.
Fig. 6.37. The Normalize Data Button.
The RFU data may be normalized and displayed on a plot that ranges from 0 to 1 on both axes.
This display is sometimes a very effective presentation of RFU data. The RFU data is normalized
to the NTC value as a linear combination of Allele 1 and Allele 2 specific signals. The iQ5 software
uses the following formula initially introduced by Livak et al. (1995):
- 74 -
Page 84

Section 6 Data Analysis Module
Reference: Livak JL, Marmaro J and Todd JA, Towards fully automated genome-wide
polymorphism screening,
Nature Genetics
, 9, 341-342 (1995)
6.7.10 Restore Default
At any time, all modifications to the allelic discrimination data can be reversed by clicking
Restore Default. The iQ5 software will reload the original x-axis allelic 1 fluorophore and y-axis
allelic 2 fluorophore data, original display mode and Show Labels setting that were originally
saved in the .opd file and will also set the Automatic Call method to recalculate the data, plot the
chart, and display the results in the allelic discrimination data spreadsheet.
6.8 End Point Analysis
The End Point analysis module is a convenient method of analyzing final relative fluorescence
unit (RFU) values.
End point analysis can be performed in two ways:
• Selecting the End Point tab in the Data Analysis module for an existing data file
• Click Run End Point to initiate the collection of End Point data from a sample plate
Newly collected End Point data is displayed immediately following an End Point run. Any existing
.opd data file can also be viewed in the end point module however, the corresponding PCR
quantification data must be analyzed in either the PCR Base Line Subtracted Mode or the PCR
Base Line Subtracted Curve Fit mode before the End Point tab becomes available. Although any
file with amplification data may be analyzed post-run in both the PCR Quant and End Point tabs,
an End Point run may only be analyzed in the End Point tab. Melt Curve-only experiments, which
neither contain amplification data nor End Point data, may not be analyzed in the End Point tab.
6.8.1 The End Point Analysis Settings
The End Point tab is comprised of several sections, each one described in detail below. The
Display Wells and Analyze Wells buttons have the same functions as described in section 6.2.4.
Method Box: End Point Analysis Methods
Below the file and fluorophore information is the Method box. The Method box allows you to
select the method of assigning positive and negative values to your unknowns based on RFU
values. The Method box consists of the following three choices:
• Negatives: This is the default method. Select this method to use negative controls to
define or call unknown samples. Samples are considered positive if they are greater in
RFU value than the negative control average plus the tolerance
• Positives: Select this method to use positive controls to define or call unknown samples.
Samples are considered positive if they are greater in RFU value than the positive control
average minus the tolerance
- 75 -
Page 85

Section 6 Data Analysis Module
• Positives & Negatives: Select this method to use positive and negative controls to
define/call unknown samples. Samples are considered positive if they are greater in RFU
value than the positive control average minus the tolerance. Samples are considered
negative if their RFU value is less than the negative control average plus the tolerance
End Point Tolerance
End Point Tolerance defines the margins for sorting unknowns as positives or negatives. How the
tolerance variable and the type of tolerance are applied depends on which method is selected.
The End Point Tolerance drop-down list box consists of two choices:
• RFUs: This is the default Tolerance choice and should be selected if you would like to
use an absolute RFU value for the tolerance value. The minimum RFU tolerance value is
2, whereas the maximum is the absolute value of the highest RFU value minus the
absolute value of the lowest RFU value. The default RFU tolerance value is 10% of the
total RFU range. The definition of the RFU range is dependent on the method chosen
• For the negative method, the range is the highest RFU value minus the negative control
average.
• For the positive method, the range is the positive control average minus the lowest RFU
value.
• For the positives & negatives method, the range is the positive control average minus the
Negative control average.
• Percent of Range: Select this setting if you would like to use a percentage of the RFU
range for the tolerance value. The minimum percent of range is 1 percent, whereas the
maximum percent of range is 99 percent. The default percent of range tolerance is 10
percent.
End Cycles to Average
End Cycles to Average is the number of cycles from the last cycle, which will be used to calculate
an average End Point RFU value. The End Cycles to Average field defaults to 2 for End Point Only
runs and 5 for non-End Point Only runs. In order for the iQ5 software to analyze non-End Point
Only data in the End Point tab, at least 6 repeats of data acquisition must be performed.
Number of Ranks
The Number of Ranks allows assignment of samples into distinct groups based on their RFU
values. The absolute range (highest RFU minus lowest RFU) is divided by the number of ranks
selected. The default rank value is 10 and the minimum number of ranks is 3.
Colored rank boxes, displayed below Number of Ranks and Sort Data by Call, symbolize the
number and order of ranks in the end point analysis. To the right of the colored rank boxes are
five color-gradation buttons that allow a change in the color scheme of the rank boxes, once the
data are analyzed.
Sort Data by Call
This function is used to sort End Point samples into Positive and Negative Calls (as well as No
Calls wells which do not fall into the previous two categories). Sort Data by Call is essentially the
Positives & Negatives method but also includes sorting and color coding the Positive and
Negative Calls. The ranking function is disabled in this mode.
- 76 -
Page 86

Section 6 Data Analysis Module
Results
The Results box lists the following information:
• Source of Data: Displays the analysis mode of the source data. For End Point only data,
this is Background Subtracted. For end point analysis of PCR quantification data, the
source of data must be either PCR Base Line Subtracted or PCR Base Line Subtracted
Curve Fit. You may alter the analysis mode of the source data in the PCR Quant tab
within the Data Analysis module
• Current method: Displays the currently selected method for assigning positive and
negative values to unknowns
• Tolerance: Displays the currently selected mode of tolerance calculation. Immediately
beneath the displayed mode is the tolerance value. If RFUs are chosen, then the Set
Tolerance in RFUs is given. If Percentage of Range is chosen then the Calculated
Tolerance in RFUs appears
• Range: The value displayed here is dependent on the method chosen
• If the Positives method is chosen, the Range is the Positive Controls Average RFU
minus the lowest RFU.
• If the Negatives method is chosen, the Range is the highest RFU minus the
Negative Controls average RFU.
• If the Positives & Negatives method is chosen, the Range is the Positive Controls
average RFU minus the Negative Controls average RFU.
• + Controls Average RFUs: Displays the average RFU of all positive controls. No value
is shown if there are no positive controls defined.
• + Control – Tolerance: In the Positives or Positives & Negatives methods, samples
with values equal to or higher than this amount will be called as Positive. No value is
shown if there are no positive controls defined.
• – Controls Average RFUs: Displays the average RFU of all negative controls. No value
is shown if there are no negative controls defined.
• – Control + Tolerance: In the Negatives method, samples with values equal to or
higher than this amount will be called as Positive. In the Positives & Negatives method,
samples with values less than this amount will be called Negatives. No value is shown if
there are no negative controls defined.
6.8.2 End Point Analysis Spreadsheet
The End Point analysis spreadsheet contains the necessary data to perform end point analysis,
and also displays the values assigned to unknowns after Recalculate is clicked. The table
headings include the following:
• Well: Lists the well ID of each sample. You may click on the Well column header to sort
the table by wells. Note that you may also include or exclude certain wells from end point
analysis via the Analyze Wells button above the spreadsheet
• Sample Type: Lists the sample type of every well, as defined in the plate setup. You
may click on the Sample Type column header to sort the table by sample type
• End RFUs: Lists the absolute RFU averages for each well, as calculated from the end
cycles, which are specified in the End Cycles to Average field. You may click on the End
RFUs column header to sort the table by end RFUs
- 77 -
Page 87

Section 6 Data Analysis Module
• Define Controls: Lists any positive or negative controls defined in the original plate
setup file. You may also add new controls or edit existing controls in this column by
clicking on the drop-down menu on the right side. The options are (+) Positive, (–)
Negative, or blank. Alternatively, you may type the letter ‘p’ or the plus sign (+) to select
a positive control, and you may also type the letter "n" or the minus sign (–) to select a
negative control. The controls specified in this column are used in the end point analysis
calculations that assign positive or negative values to the unknowns. You may click on
the Define Controls column header to sort the column by the controls
• Unknowns Call: If a well is not defined as a positive control or a negative control in the
Define Controls column, it is considered an unknown for end point analysis. This column
displays the value assigned to each unknown after Recalculate has been clicked.
Unknowns Call may be (+) Positive, (–) Negative, or blank. You may click on the
Unknowns Call column header to sort the table by Unknowns Call
• Unknowns Ranking: Lists the rank into which each unknown falls. An unknowns
ranking depends on the total number of ranks and the end RFUs value. You may click on
the Unknowns Ranking column header to sort the table by Unknowns Ranking
• Identifier: Lists the identifier for every well, as defined in the plate setup. You may click
on the Identifier column header to sort the table by identifier
6.8.3 Recalculate Button
Clicking Recalculate performs the final calculations used to determine unknown sample calls.
The Recalculate button is active when sufficient controls have been specified for the selected
method and End Point Tolerance settings. The following conditions result in an error message:
• If the Positives method is selected and there is not at least one positive control defined in
the End Point analysis spreadsheet
• If the Negatives method is selected and there is not at least one negative control defined
in the end point analysis spreadsheet
• If the Positives & Negatives method is selected and there is not at least one positive
control and at least one negative control defined in the End Point Analysis spreadsheet
• If no Controls are defined
• If the range is less than one (that is, the negative control RFU average is larger than the
positive control RFU average)
End point analysis cannot be performed if any of these error conditions is present when you click
Recalculate.
Once you are satisfied with your end point analysis, you may choose to view, save, or print a
customized End Point Analysis report. Click the Reports menu to obtain customized reports for
the end point data. Click Print to print the End Point analysis spreadsheet.
6.9 Gene Expression Analysis
The Gene Expression (Gene Expr) tab is used to evaluate relative differences in any target
concentration. For example, you can evaluate relative numbers of viral genomes or stably
transfected sequences. The most common application is evaluating target concentration in cDNA
samples to infer steady state messenger RNA levels.
- 78 -
Page 88

Section 6 Data Analysis Module
Gene expression analysis is the most common real-time PCR application. Therefore, this manual
uses terminology that is specific to this kind of study. You can safely substitute “gene” with the
word “sequence” or “target” when you read about how to use the gene expression module.
Without stringently quantified controls, you cannot use this part of the iQ5 software to evaluate
target concentrations of particular sequences relative to each other. The iQ5 software can only
evaluate relative differences of a sequence between a group of samples.
The gene expression analysis screen has flexible tools for the determination of the fold induction
of one gene relative to another gene, or relative to itself under different circumstances; that is,
temporally, geographically, or developmentally different points. Additionally, within the gene
expression analysis screen, it is possible to create a Gene Study from multiple plates of data
collected at different times.
The gene expression analysis screen (Figure 6.38) consists of four main areas:
• The graphical data view section: Displays graphed data and graph settings
• The Settings and Data Table options: Located to the right of the graph section. Used
respectively to specify analysis settings and access the data spreadsheet
• The gene expression plate interface section: Located at the bottom of the screen. A
spreadsheet view of the plate setup used to modify and edit gene and condition names
• Analysis method radio buttons: Located above the gene expression plate interface
section. Used to select between normalized and relative quantity gene expression
analysis methods
Fig. 6.38. The Gene Expression Analysis Tab Window.
- 79 -
Page 89

Section 6 Data Analysis Module
Basic Workflow Steps for Gene Expression Analysis
1. In the PCR Quant screen with a .opd data file open, assess threshold and baseline
information for the data file and make changes if necessary.
2. Click the Gene Expr tab.
3. Make any changes to the Gene and Condition (for example, Sample and Treatment)
assignments in the gene expression plate interface.
4. Set reference genes if required, and choose an analysis method:
• Normalized Expression (ddCT) is the default; or
• Relative Quantity (dCT)
6. OPTIONAL: Assign attributes (sample color, show graph, etc.) and user-defined
reaction efficiencies in the Gene List.
7. OPTIONAL: Assign attributes (sample color, show graph, etc.) and set control samples
in the Condition List.
8. Click Recalculate to see your results.
If you want to compare these data to results obtained in other .opd files, you will need to enable
this file for Multi-file Gene Expression analysis, also called a Gene Study. To enable your file for
Gene Study:
1. Click Enable for Gene Study.
2. Go to the File menu to save your file.
Normalized expression is graphed in the gene expression graph interface. The data can be
accessed in the Gene Expression Settings interface within the Data Table.
6.9.1 Normalized Gene Expression (ddC
Rather than using some other method to normalize data, you may use the measured expression
level of one or more reference genes as a normalization factor. Reference genes should be genes
which are not regulated in the biological system being studied.
Normalized expression is the relative quantity of your gene normalized to the relative quantities
of the reference gene(s). Simply stated, the quantities of the reference genes are used to
normalize the values of your genes of interest.
Provided the reference genes are not regulated in your system, normalized expression
calculations will account for loading differences or variations in cell number represented in each
of your samples. This value is sometimes referred to as ddCT or ∆∆CT because of the equation
initially introduced by Livak et al. (1995) to evaluate normalized expression. The software uses a
modification of this equation.
Normalized expression can be viewed as normalized target sequence quantity if your assay has
included controls for this purpose.
To calculate Normalized Expression (ddCT):
∆∆CT)
T,
1. In the PCR Quant screen with a .opd data file open, assess Threshold and Baseline
information for the data file and make changes if necessary.
2. Click the Gene Expr tab.
- 80 -
Page 90

Section 6 Data Analysis Module
3. Make any changes to Gene and Condition (for example, Sample and Treatment)
assignments in the gene expression plate interface.
• Expand the gene expression plate interface view by clicking on the “+” button to
make well identification and selection easier. Highlight the wells in the gene
expression plate interface you wish to edit
• Change the gene assignments in these wells by typing your desired name into the
gene pull-down men, then click enter to apply the name to the selected wells
• Change the condition assignments in these wells by typing your desired name into
the condition pull-down menu, then click enter to apply the name to the selected
wells
NOTE: The Gene and Condition Names have a character entry limit of 15 characters.
Minimize the gene expression plate interface by clicking on the “–” button to return to
the standard view of the Gene Expr tab window.
4. Click the Settings tab, then select Gene List to set your desired reference gene(s).
5. Click Normalized expression (ddCt).
6. Click Recalculate to see your results.
7. Normalized expression results are graphed. The Data Table spreadsheet is accessed by
clicking Data Table. The Data Table spreadsheet lists the Condition and Gene name,
calculated expression values and CT values. Right-click to print or export this data to
Excel.
NOTE: Use the Settings tab, then click Gene List to enter a specific user-defined gene
reaction efficiency. Use the Settings tab, then click Condition List to select a particular
sample as a control sample.
6.9.2 Relative Quantity (dC
Relative quantity is the target sequence concentration relative to other samples in the
experiment. This is sequence quantity without taking into account reference genes for
normalization. No calculations are made to account for loading differences or differences in the
number of cells represented in each of your samples. Relative quantity can be viewed as nonnormalized expression. This value is sometimes referred to as dCT or ∆CT because of the equation
used to calculate relative quantity.
By definition, relative quantity data is not normalized. Typically researchers that do not use
reference genes are confident in one of the two following considerations:
• Each condition (sample) assayed represents the same amount of biological sample.
Typically they choose to load the same mass of RNA or cDNA in each well and feel that
mass of nucleic acid is an effective way of normalizing the resulting data. No modification
of relative quantity data is needed to obtain normalized data. The data are normalized by
experimental design
or;
• Any variance in the amount of biological sample loaded will be normalized in post-PCR
analysis by some method. For example, a researcher might choose to simply divide the
relative quantity value by the normalizing factor indicated after each of the examples
listed below. Options may include but are not limited to:
T, ∆CT
)
• Mass of nucleic acid loaded for each sample. Rel Quant/ng RNA represented in each
sample
- 81 -
Page 91

Section 6 Data Analysis Module
• Number of cells from which nucleic acid was isolated. Rel Quant/number of cells
represented in each sample
• Mass of tissue from which nucleic acid was isolated. Rel Quant/mass of tissue
represented in each sample
To calculate Relative Quantity (dCT):
1. In the PCR Quant screen with a .opd data file open, assess threshold and baseline
information for the data file and make changes if necessary.
2. Click the Gene Expr tab.
3. Make any changes to Gene and Condition (for example, Sample and Treatment)
assignments in the gene expression plate interface.
• Expand the gene expression plate interface view by clicking on the “+” button to
make well identification and selection easier. Highlight the wells in the gene
expression plate interface you wish to edit.
• Change the gene assignments in these wells by typing your desired name into the
gene pull-down men, then click enter to apply the name to the selected wells.
• Change the condition assignments in these wells by typing your desired name into
the condition pull-down menu, then click enter to apply the name to the selected
wells.
NOTE: The Gene and Condition Names have a character entry limit of 15 characters
• Minimize the gene expression plate interface by clicking on the “-” button to return to
the standard view of the Gene Expr tab window.
4. Click Relative Quantity (dCt).
5. Click Recalculate to see your results.
6. Relative quantity results are graphed. The Data Table spreadsheet is accessed by clicking
Data Table. The Data Table spreadsheet lists the Condition and Gene names, and
calculated expression values and CT values. Right click to print or export this data to
Excel.
NOTE: Use the Settings tab, then click Gene List to enter a specific user-defined gene
reaction efficiency.
NOTE: Use the Settings tab, then click Condition List to select a particular sample as
a control sample.
6.9.3 Specifying Gene and Condition Labels for Gene Expression
Analysis
Use the gene expression plate interface to edit gene names (for example actin, GAPDH), and
experimental conditions names (for example, treatment types, time course points, etc.). Use the
Settings option to select which of these genes and/or conditions to use for normalization and set
other analysis and display parameters.
Using the Gene Expression Plate Interface to Edit Gene and Condition Names
Open a .opd Real-Time PCR Data file.
- 82 -
Page 92

Section 6 Data Analysis Module
1. In the PCR Quant screen, assess threshold and baseline for the data file and then make
changes if necessary.
2. Click the Gene Expr tab.
3. Highlight and select the wells to be edited in the gene expression plate interface.
NOTE: Expanding the gene expression plate interface view by clicking on the “+” button
makes the entire plate area visible for easier well identification and selection.
4. Change the gene and condition assignments in these wells by typing in the gene and
condition pull-down menus, then click Enter to apply the name to the selected wells.
Entered names will then be added to the pull-down menu, and become available for
selection from this menu.
NOTE: The Gene and Condition Names have a character entry limit of 15 characters.
In Figure 6.39, wells B4, C4, D4 are selected, and are being changed from condition
name 1 Hr to 1.5 Hrs. The new condition name, 1.5 Hrs, was typed in the Condition
Name box.
In addition to the gene expression plate interface, gene and condition names can also be edited
post-run using Edit Plate. Opening this window when analyzing collected data by clicking on the
Edit Plate tab at the top of the screen will open a modified version of this window. In this
modified window you can edit sample type, replicates, standard concentrations and
primer/probe-gene names. For further details on post-run edit plate functionality, refer to section
6.11.
Fig. 6.39. Editing Gene and Condition Names in Gene Expression Plate Interface.
Copying Conditions to All Data Sets
The Copy conditions to all data sets button should be activated if you want all fluorophores
or data sets to have the same Condition Name(s) assigned to selected groups of wells. This is a
useful function if:
- 83 -
Page 93

Section 6 Data Analysis Module
• You have a multiplex experiment. In this case all sample types in the varying
fluorophores must be the same
or;
• You are importing multiple .opd (data files) into a Gene Study
Make sure the Copy conditions to all data sets button is highlighted (active) when you assign
condition (sample) names as shown in Figure 6.40. Active is the default state for this button.
Fig. 6.40. Copying Gene and Condition Settings to all Fluorophores.
Gene Expression Plate Interface Context Menu
The plate interface menu is displayed by right-clicking on the plate interface. When this action is
performed, a menu is displayed with the following items: Copy Gene Name, Copy Condition
Name, Paste Gene/Condition Name, Show Sample Type, Print and Export to Excel (Figure 6.41).
Fig. 6.41. The Context Menu of the Gene Expression Plate Interface.
• Copy Gene Name and Copy Condition Name: These two commands are useful
shortcuts for copying and pasting gene and condition names across many different
samples within a single .opd or Gene Study.
- 84 -
Page 94

Section 6 Data Analysis Module
1. Activate the copy command by first selecting the wells to be copied from the plate
interface.
2. Right-click on the plate interface to reveal the Copy Gene or Copy Condition options,
and select the desired option.
3. Once the Copy Gene or Copy Condition option has been selected, the Paste
Gene/Condition Name text from the menu becomes active (no longer grayed out).
4. Return to the plate interface to select the destination wells for the copied
identification information.
5. Right-click on the plate interface to reveal and select the Paste Gene/Condition Name
option.
• Show Sample Type: The Show Sample Type option is selected by default. This
selection is illustrated by a check mark shown before the text in the menu. Clicking on
Show Sample Type toggles the display of the Sample Type in the plate interface.
Choosing to remove the sample type information from the plate interface simply allows
visibility of a greater number of wells in the plate interface grid.
• Print: The Print command, on the plate interface menu, will print the displayed plate
interface.
• Export to Excel: The Export to Excel command is useful for exporting exact values from
the plate interface. When the Export to Excel command is selected from the menu, the
iQ5 software exports the selected data into a protected workbook that is automatically
open by Microsoft Excel. The protected workbook contains all of the formatting and
fluorophore information separated into worksheets.
NOTE: The numeric values contained in the protected workbook are exact values from
the software application that include several non-significant figures beyond the decimal
point. This is important to note when considering whether to transfer spreadsheets by a
copy and paste command or the Export to Excel command. With the copy and paste
command, only the significant digits displayed in the iQ5 software interface are
transferred to Excel.
6.9.4 Setting Analysis Parameters for Gene Expression Analysis Using
the Settings Tab
The Settings tab is used to assign gene and conditions parameters and efficiency values to be
used in the calculations for gene expression analysis. There are three sections of the Settings
options window:
• Gene List
• Condition List
• Data Set List
6.9.4.1 Gene List: Assigning Reference Genes and Target-Specific Reaction
Efficiency
When the Settings button is active and Gene List is selected, the genes being analyzed in the
experiment are listed along with their full name. This Full Name column is editable so that longer,
more descriptive names may be entered if your Gene Name entry was abbreviated.
NOTE: Gene Name entries have a character entry limit of 15 characters.
- 85 -
Page 95

Section 6 Data Analysis Module
To access the gene list:
1. Make sure the Settings button is activated. If not, click Settings. See the example in
Figure 6.42.
2. Click Gene List.
The list of genes appears in the spreadsheet directly below the Gene List option button as shown
in Figure 6.42.
Fig. 6.42. Selecting the Gene List Option.
Assigning a Reference Gene
To assign a reference gene:
1. With the Settings button active, select Gene List.
2. Select the checkbox in the Ref column within the control condition(s) to which you want
to assign a reference gene. You can assign the role of reference gene to as many genes
as you wish. In Figure 6.43, Actin and Gapdh have been selected as reference genes.
Fig. 6.43. Assigning a Reference Gene.
- 86 -
Page 96

Section 6 Data Analysis Module
Reaction Efficiency and Gene Expression Analysis
Efficiency describes how much of your sequence of interest is being produced with each cycle. An
efficiency value of 100% means that you are doubling your sequence of interest with each cycle.
People focus on efficiency for a number of reasons. There is evidence that using accurate
efficiencies for each of your gene (primer/probe) pairs will give you more accurate results when
using the mathematical modules used in the Gene Expr tab.
You set the efficiency for each of your genes in the Gene List. The default value is 100%. So if no
standards were included on the plate, by default an efficiency value of 100% will be reported.
This efficiency value is fully editable to allow entry of other than a default 100% value
established from previous experiments.
If a standard curve was included on the plate, the software will automatically calculate the
efficiency for that gene and report it in the Efficiency (%) column. To use Auto Efficiency, you
must have standards in your experiment that result in valid standard curves in the PCR Quant
tab. The iQ5 software only requires two standards at different concentrations, though it is
recommended to have at least four samples in triplicate across a relevant dynamic range.
6.9.4.2 Assigning and Naming Control Samples Using the Condition List
When the Settings button is active and Condition List is selected, the conditions being analyzed in
the experiment are listed along with their full name. This Full Name column is editable so that
longer more descriptive names may be entered if your Condition Name entry was abbreviated.
NOTE: Condition Name entries have a character entry limit of 15 characters.
The Condition List option represents particular tests or treatments being evaluated for the
purposes of your experiment. A condition can be as simple as “sample 1” or complex as “mouse
#123558 liver + PMA”, though the latter example is often too long to present on a graph.
You can view conditions as samples in the Condition List spreadsheet as shown in Figure 6.44.
Fig. 6.44. The Condition List.
- 87 -
Page 97

Section 6 Data Analysis Module
Setting Control Conditions
You can assign one condition as the control condition by selecting a checkbox next to the sample
in the Condition List as shown in Figure 6.44.
The iQ5 software assigns the control sample a value of 1 for every gene, and all other conditions
will be presented with values relative to this one. This makes it simple to evaluate fold expression
relative to the chosen (control) sample.
NOTE: The control condition will have a value of 1 for all genes.
To access the condition list:
1. Make sure the Settings radio button is activated.
2. Click Condition List option.
3. Select the checkbox(es) in the Ctrl column within the control condition(s) to which you
want to assign a control condition.
6.9.4.3 Data Set List
The Data Set List displays pertinent information about the data being analyzed in a given Gene
Study or .opd file. The information contained within the cells of the Data Set List is for display
only, and cannot be modified.
Each row of the Data Set List is linked to a single data tab of the gene expression plate interface,
located at the bottom of the Gene Expr window. Selecting a row from the spreadsheet
automatically toggles to its representative dye layer data tab in the plate interface. Selecting a
dye layer data tab results in toggling to its representative row in the Data Set List spreadsheet.
Information displayed in the Data Set List includes:
•
Name: Displays the fluorophore name for a given set of collected data. A number is
placed before the abbreviated fluorophore name. The assigned number helps to
distinguish between data from the same fluorophore when multiple files are used (as in a
Gene Study)
•
Full Name: This cell contains the original name of the opd file as well as the original
location of the opd when it was created
•
Created Date: This cell displays the date and time the .opd file was created
The Name and Full Name column contents can be alphabetically sorted by clicking on the title
header of each cell. This feature is helpful when long list of fluorophores are created as with
multiple files (see Gene Study).
Fig. 6.45. Accessing and viewing the Data Set List.
- 88 -
Page 98

6.9.5 Applying an Analysis Method
1. Choose an analysis method by clicking one of the option buttons:
• Normalized expression (ddCt) is the default and requires you to first choose a
reference gene)
or
• Relative quantity (dCt)
2. Click on Recalculate to apply the new settings.
NOTE: Many changes made to the plate setup and analysis options require that you
recalculate. If you do not immediately see the change you expect to see, click
Recalculate (Figure 6,46).
Fig. 6.46. Applying an Analysis Method.
Section 6 Data Analysis Module
6.9.6 Data Table for Gene Expression Analysis
The Data Table spreadsheet for gene expression analysis is accessed by clicking Data Table. The
Data Table spreadsheet lists the Condition and Gene names, and calculated expression and C
values (Figure 6.47). Columns can be resized and re-ordered by clicking and dragging. Any
condition being used as a control is indicated by an asterisk in the Ctrl column. Columns can be
sorted by Condition or Gene name by clicking on the sort triangle icon next to the column
heading.
T
Fig. 6.47 Gene Expression Analysis Data Table.
- 89 -
Page 99

Section 6 Data Analysis Module
Corrected Values Calculations
The efficiency value (E) used in gene expression calculations has an associated error. If a
standard curve was generated as part of the real-time PCR assay, this error can be calculated
and used to adjust the error associated with the following standard deviation values:
•
Rel Quant SD
•
Unscaled Expression SD
•
Expression SD
Corrected Values for all gene expression results are displayed on the Data Table spreadsheet
when Show Details is selected (Figure 6.48). When Show Details is active, three new columns
appear in the Data Table spreadsheet. The new columns display the error correction propagated
from the standard curve data for:
•
Relative Quantitation Standard Deviation
•
Unscaled Expression Standard Deviation
•
Expression Standard Deviation
The new columns are named:
•
Corrected Rel Quant SD
•
Corrected Unscaled Expression SD
•
Corrected Expression SD
Fig. 6.48. The Gene Expression analysis Data Table – Show Details view.
A difference between corrected and non-corrected values will only be seen if a standard curve
was created as part of the real-time PCR assay. The equations used in determining the error
propagation are the Standard Error, Standard Error for Normalized Expression, and the Standard
Error for the Normalized Gene of Interest (GOI).
The equation for Standard Error is shown below:
SD
SE =
- 90 -
n
Page 100

Section 6 Data Analysis Module
Where n = number of measurements
The Standard Error for Normalized Expression equation is shown below:
2
Re
×=
NFNFSE L
nn
QuantlSE
Re
×
Quantln
)1(Re
fsample
)1(Re
fsample
Re
+
QuantlSE
Re
×
Quantln
2
)2(Re
fsample
)2(Re
fsample
Where NF = Normalization Factor
The Standard Error for Normalized GOI equation is the following:
2
Re
++
QuantlSE
Re
×
Quantln
)(Re
nfsample
)(Re
nfsample
Where
GOI
GOIGOISE
normnorm
= the Normalized Gene of Interest
norm
2
×=
NF
NFSE
n
n
+
GOISE
GOI
2
Exporting Data from the Data Table
1. Make sure the Data Table button is activated. If not, click Data Table.
2. Right-click on the spreadsheet. A shortcut menu appears (Figure 6.49).
3. Select Export to Excel.
4. Enter a name and file destination for the Excel file generated with your data export.
Fig. 6.49. Exporting Data Table Spreadsheets to Microsoft Excel.
Printing Data
The Print command on the spreadsheet menu will print the displayed spreadsheet (Gene List,
Condition List, Data Set List, or Data Table). When selected, a Print Preview box is opened,
which contains an illustration of the spreadsheet as it will appear once printed. Clicking the
printer icon opens the Windows Print dialog box. Click OK to complete the printing task.
The Export to Excel command on the spreadsheet menu is useful for exporting exact values from
the spreadsheet. When the Export to Excel command is selected from the menu, an Export to
Excel file save box is displayed. Choose a location of where the Excel file is to be saved and click
Save. The iQ5 software automatically exports the selected data into a protected workbook.
The protected workbook generated by the iQ5 software contains the text values of what is
represented on the spreadsheet. For example, checkboxes from the software application are
replaced by “True” or “False” text in Excel.
The numeric values contained in the protected workbook are exact values from the software
application that include several non-significant figures beyond the decimal point. This is
important to note when considering whether to transfer data by a copy and paste command or
the Export to Excel command. With the copy and paste command only the significant digits
displayed in the iQ5 software interface are transferred to Excel.
- 91 -
 Loading...
Loading...