Page 1

Bio-Rad® Nuvia™ IMAC
Resin
Instruction Manual
Catalog numbers
780-0800
780-0801
780-0802
Please read these instructions prior to using Bio-Rad Nuvia IMAC
resins. If you have any questions or comments regarding these
instructions, contact your Bio-Rad Laboratories representative.
Page 2

Page 3

Table of Contents
Section 1 Introduction ............................................................ 1
Section 2 Product Description ............................................... 2
Nuvia™ IMAC Resins and
UNOsphere™ Technology .......................................... 2
Chemical Interactions ............................................... 3
Resin Characteristics ................................................ 4
Chemical Compatibilities ........................................... 5
Section 3 General IMAC Procedures ..................................... 7
Protein Binding ......................................................... 7
Washes .................................................................... 8
Elution ...................................................................... 8
Purification under Denaturing Conditions .................. 9
Imidazole Concentrations ........................................10
Section 4 Column Packing — Medium-Pressure
Columns .................................................................11
Slurry Packing an IMAC Column...............................11
Recommended Columns..........................................11
Materials ..................................................................11
Resin Preparation ....................................................12
Method ................................................................... 12
Section 5 Column Packing — Sample Preparation–Sized
Columns ................................................................ 14
Materials ................................................................. 14
Resin Preparation ................................................... 14
Method ...................................................................15
Section 6 Immobilizing Metal Ions ........................................16
Section 7 Medium-Pressure Column Purification of
Histidine-Tagged Proteins....................................17
Materials .................................................................17
Method ...................................................................18
Materials and Method for On-Column
Renaturation ........................................................... 20
Page 4

Section 8 Medium-Pressure Column Purification — Using
an Imidazole Gradient to Determine Optimal
Purification of Histidine-Tagged Proteins ........... 21
Materials ................................................................. 21
Method ................................................................... 22
Section 9 Sample Preparation–Sized Spin-Column
Purification of Histidine-Tagged Proteins ........... 25
Materials .................................................................25
Method ...................................................................26
Section 10 Regenerating, Cleaning, Sanitizing,
and Storage ..........................................................28
Regenerating the Medium .......................................28
Cleaning in Place ...................................................28
Sanitization..............................................................30
Storage .................................................................. 30
Section 11 Troubleshooting Guide ......................................... 31
Section 12 Ordering Information ........................................... 34
Section 13 References ........................................................... 34
Section 14 Legal Notices ...................................................... 34
Page 5

Section 1
Introduction
Immobilized metal affinity chromatography (IMAC) is a powerful
purification technique that relies on a molecule’s affinity for certain
metals immobilized onto a chelating surface. The chelating
ligand, nitrilotriacetic acid (NTA) in this case, may be charged with
transition metals such as Cu2+, Ni2+, Co2+, or Zn2+. This results in
high selectivity for proteins with clustered histidine residues to be
strongly retained on a porous chromatographic support.
The use of IMAC to separate an expressed recombinant protein
fused with a hexahistidine peptide tag was demonstrated
by Hochuli (1988) to yield a highly purified protein in a single
chromatographic step under both denaturing and native
conditions. The strong affinity of a histidine-tagged molecule for
metal ions often makes extensive optimization unnecessary while
also allowing chromatography under conditions that denature
proteins. For this reason, expression and IMAC purification of
histidine-tagged proteins is frequently used for structural and
functional studies of proteins.
Nuvia IMAC Ni-Charged Resin 1
Page 6

Section 2
Product Information
Nuvia™ IMAC Resins and UNOsphere™ Technology
Nuvia IMAC resin, a unique affinity support, is based on Bio-Rad’s
innovative UNOsphere beads, which use proprietary polymerization
and derivatization technologies.* The UNOsphere technology
enables the polymeric high-capacity IMAC resin to exhibit excellent
flow properties without compromising protein binding, recovery, or
purity.
Nuvia IMAC uses NTA as its functional ligand. The tertiary amine
and carboxylic acid side chains of NTA serve as the chelating
ligands for divalent metal ions. The structure offers selective
binding of recombinant histidine-tagged proteins when this resin is
charged with Ni2+ or other transition metals. As a result, the desired
proteins can often be purified close to homogeneity in a single
step.
Structural characteristics such as the polymeric nature, optimized
ligand density, and open-pore structure of Nuvia IMAC beads
result in superb mechanical strength with high stringency, low
nonspecific effects, and the ability to provide separations at fast
flow rates. These unique features of the UNOsphere base matrix
lend a number of performance benefits to the Nuvia IMAC resin.
Nuvia IMAC is also stable across a wide pH range (2–14) and
is compatible with most reagents commonly used in protein
purifications, such as denaturants, detergents, and reducing
agents. It is amenable to separations under native or denaturing
conditions using liquid chromatography instrumentation, gravity
flow columns, or sample-preparation spin columns.
Note: UNOsphere media, from which Nuvia IMAC is derived,
was designed to achieve the highest productivity (grams of
drug or target per operational hour per liter of support) possible.
UNOsphere media may be run at the highest rates and loading
capacities and will stay within the pressure limits of the column and
chromatography system.
* U.S. patent 6,423,666.
2 Nuvia IMAC Ni-Charged Resin
Page 7
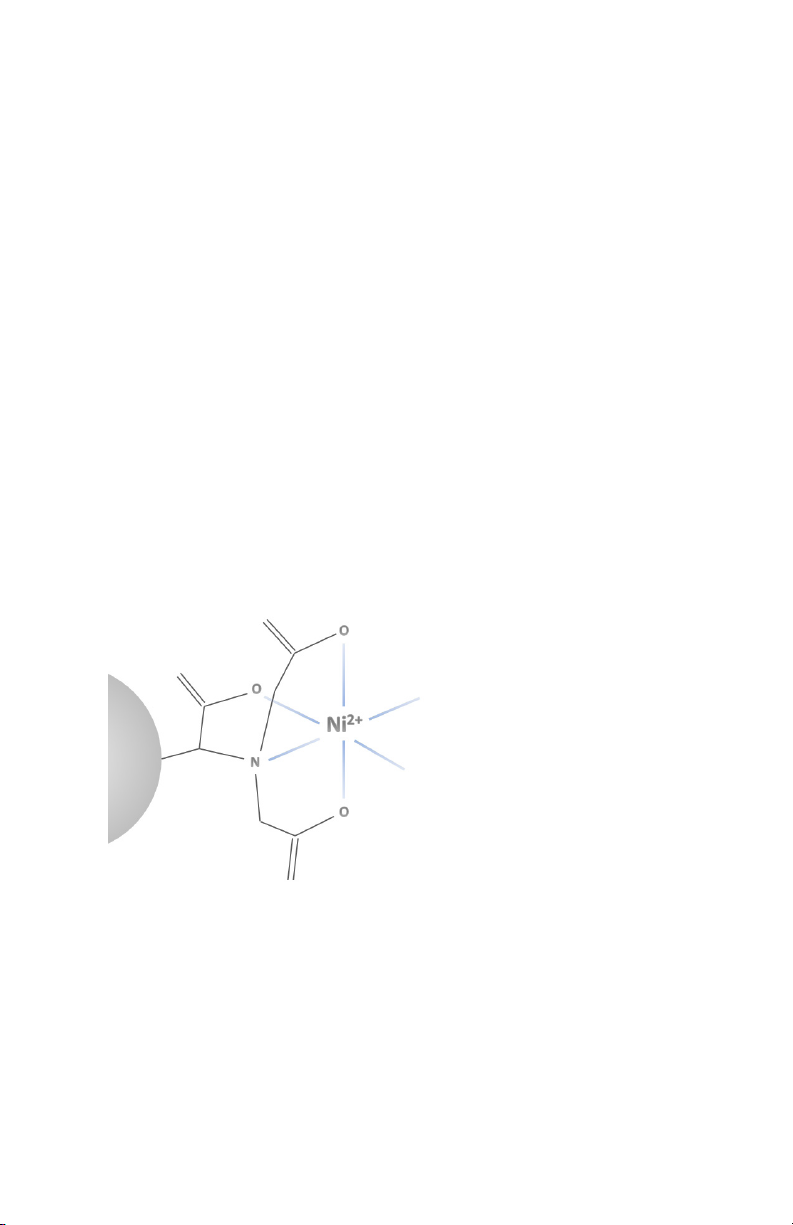
Under optimized conditions, the binding capacity for 6x histidinetagged proteins is >40 mg/ml resin (see Table 1). The product is
a 50% (v/v) slurry of resin, which is suspended in a 20% ethanol
solution. Nuvia IMAC resin is amenable to process and laboratoryscale use and is available precharged with Ni2+ in bottles as well
as prepacked into columns. Table 1 lists key characteristics of the
resin, while Table 2 lists a variety of compounds compatible with
Nuvia IMAC support.
Chemical Interactions
Nuvia IMAC resin is composed of NTA groups coupled to
a UNOsphere base matrix via a proprietary polymerization
derivatization technology. It is well-suited to recombinant histidinetagged purifications and results in high binding capacity and
specificity for the target molecule.
Although the most commonly used metal ion for histidine-tagged
purifications is Ni2+, other metals may be used to increase efficacy
of purification. Therefore, choosing another type of immobilized
metal ion can change the selectivity of an IMAC resin.
Fig. 1. Partial structure of Nuvia Ni-charged IMAC resin. Image illustrates UNOsphere base
bead with coupled NTA functional ligand.
Nuvia IMAC Ni-Charged Resin 3
Page 8

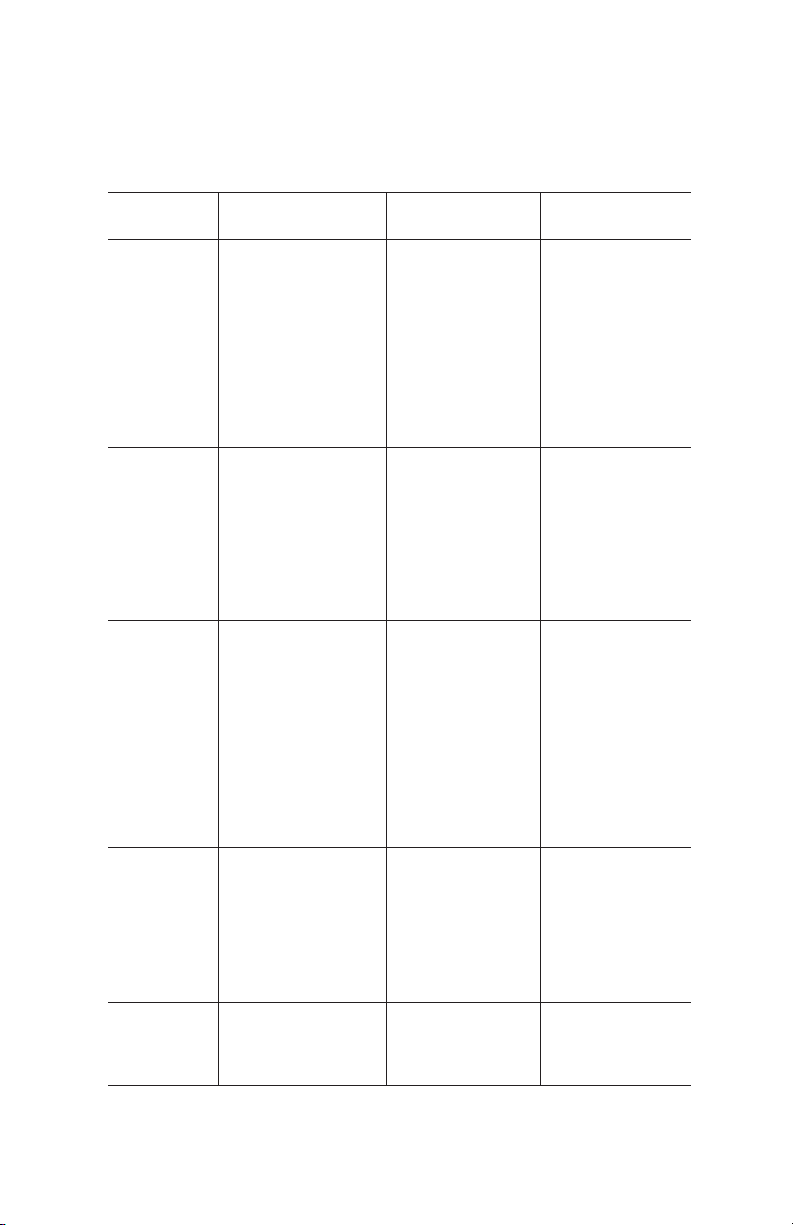
Resin Characteristics
The characteristics of Nuvia IMAC resin are detailed in Table 1.
Table 1. Characteristics of Bio-Rad Nuvia IMAC Resin.
Functional ligand Nitrilotriacetic acid (NTA)
Base bead UNOsphere base matrix
Form 50% suspension in 20% ethanol or pre-
packed into columns; comes precharged
2+
with Ni
Particle size 38–53 µm
Mean particle size 50 µm
Metal ion capacity ≥18 µmol Cu2+/ml Nuvia IMAC resin
Dynamic binding
capacity*
Recommended linear
flow rate
Maximum operating
pressure
pH stability, short-term/
cleaning
Chemical compatibility See Table 2
Storage 4°C to ambient temperature
Shelf life in 20% ethanol ≥3 year at ambient temperature
Operational temperature 4–40°C
Autoclaving conditions 0.1 M sodium acetate at 120°C for 30 min
≥40 mg/ml resin
<500 cm/hr at 25°C
45 psi
2–14
* Dynamic binding capacity conditions (Q10% determination):
Column volume: 1 ml, 5.6 mm x 4 cm (ID x H)
Sample: 1.0 mg/ml 6x histidine-tagged pure protein (40 kD)
Note: Dynamic binding capacity will vary from protein to protein.
4 Nuvia IMAC Ni-Charged Resin
Page 9
Chemical Compatibilities
The chemical characteristics of Nuvia IMAC resin are detailed in Table 2.
Table 2. Chemical Compatibilities for Nuvia IMAC Resins.
Reagent
Group
Buffer
reagents
Chelating
reagents
Sulfhydryl
reagents
Reagent Comments Stability
Tris, HEPES,
MOPS
Sodium or
potassium
phosphate
EDTA, EGTA Strips nickel ions
β-Mercaptoethanol
Used with
proteins more
stable in
nonphosphate
buffers
from the resin
Reduces random
disulfide bonds
preventing
protein
aggregation
during purification
≤50 mM
secondary and
tertiary amines
50 mM sodium
or potassium
phosphate are
recommended
as starting
buffers
≤0.1 mM
successfully
used to remove
trace metal
contaminants
>1 mM can
cause significant
reduction in
binding capacity
≤20 mM
DTT, TCEP
Detergents Nonionic
detergents (Triton,
Tween)
Zwitterionic
detergents
(CHAPS, CHAPSO)
Denaturants Guanidine HCl
(GuHCl)
Urea
Transition metals
at the center of
IMAC resin (Ni2+)
are susceptible to
reduction
Removes
background
proteins
and nucleic acids
Solubilizes
membrane
proteins
Solubilizes
proteins
Nuvia IMAC Ni-Charged Resin 5
≤10 mM DTT
and 20 mM
TCEP
≤2%
≤1%
≤6 M
≤8 M
Page 10
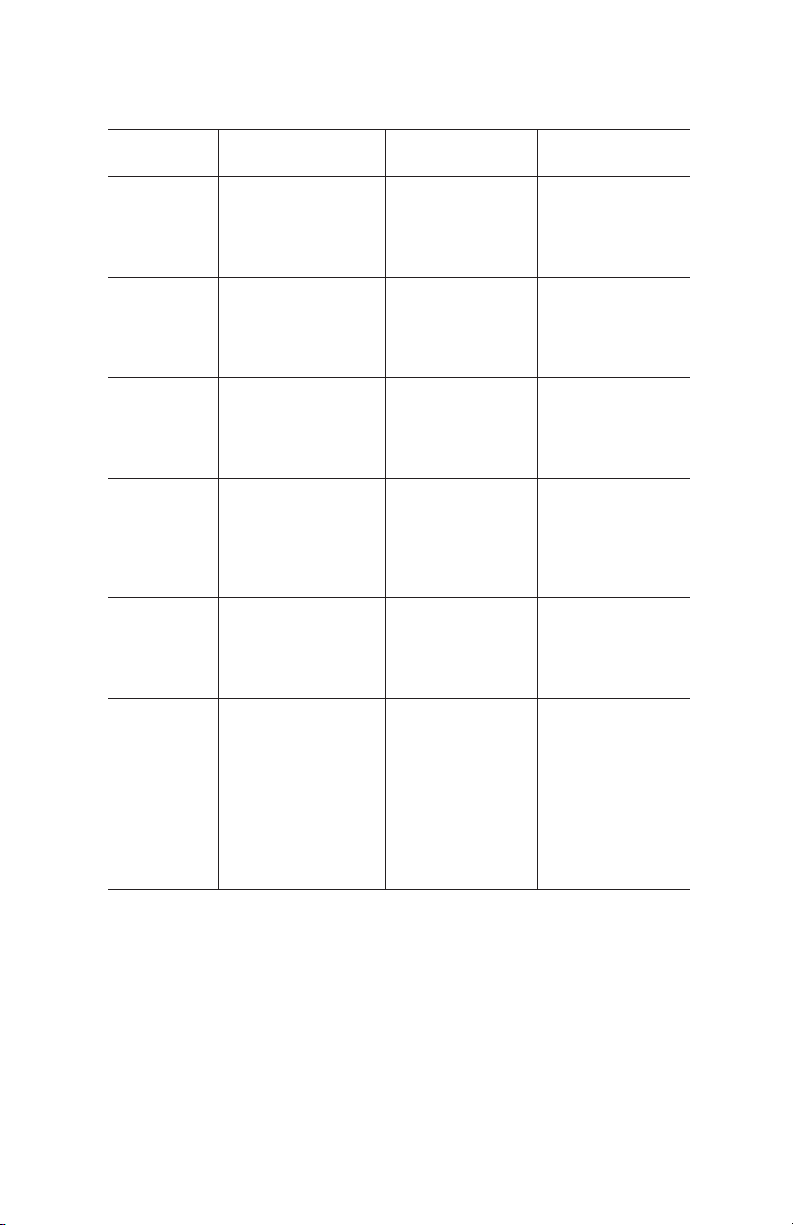
Table 2. Chemical Compatibilities for Nuvia IMAC Resins, Continued.
Reagent
Reagent Comments Stability
Group
Other
additives
NaCl Deters
nonspecific
protein binding
due to ionic
interactions
MgCl
2
Essential
component for
purification of
Ca2+ binding
proteins
CaCl
2
Essential
metal cofactor for
nucleases
≤2 M (at least
300 mM NaCl
should be
included in
buffers)
≤100 mM
(HEPES or Tris
buffers should be
used to prevent
precipitation)
≤5 mM (HEPES
or Tris buffers
should be used
to prevent
precipitation)
Glycerol Included
to prevent
hydrophobic
interactions
between proteins
≤20%
(backpressure
may increase
significantly,
slower flow rates
may be required)
Ethanol Included
≤20%
to prevent
hydrophobic
interactions
between proteins
Imidazole Competes for
binding sites with
histidine-tagged
residues by
interaction
with the metal
residues
May be
used in low
concentrations
in the wash
buffer (<30 mM)
to limit binding
of undesired
proteins; for
elution, ≤500 mM
may be used
6 Nuvia IMAC Ni-Charged Resin
Page 11

Section 3
General IMAC Procedures
Protein Binding
Protein adsorption to immobilized ions is performed around
neutral to slightly alkaline pH conditions (pH 7.0–8.0). To reduce
nonspecific ionic effects, concentrations of up to 1 M NaCl may
be added to the binding solution. Recombinant 6x histidine tags,
located at either the amino or carboxyl terminus of the protein, can
bind with high affinity to the matrix even when the 6x histidine tag
isn’t completely accessible. In general, the fewer the number of
accessible histidine residues, the weaker the protein binding is to
the affinity matrix. Untagged proteins that have naturally occurring
and noncontiguous histidine residues also bind to IMAC resins, but
with much lower affinity.
Batch mode binding is a good alternative if proteins are expressed
at low levels or if the overall concentration of the recombinant
6x histidine tag is low. In this case, proteins are bound to the
Nuvia™ IMAC resin in solution prior to packing the protein-resin
complex into a liquid chromatography column for wash and elution
steps. Altering the imidazole concentration of the lysis buffer may
also optimize binding. Low concentrations (0–15 mM imidazole)
are recommended and will aid in reducing nonspecific binding of
weakly interacting proteins.
Many additives can be used without affecting the binding of
histidine-tagged proteins to IMAC resins. For example, urea,
GuHCl, nonionic detergents, and organic solvents (refer to
Section 2, Table 2) are all valid options. Chelating agents, such as
EDTA or citrate, should not be included. Reducing agents such as
β-mercaptoethanol and DTT may be used at low concentrations.
Potassium phosphate or sodium phosphate buffers are
recommended solutions for equilibration and binding.
Recommended binding buffer:
• 20–50 mM sodium or potassium phosphate, containing up to
1.0 M NaCl.
Begin with: 50 mM sodium phosphate, 0.3 M NaCl, pH 8.0
Nuvia IMAC Ni-Charged Resin 7
Page 12

Washes
Stringency of 6x histidine-tag binding can be effectively increased
by 1) including low concentrations of imidazole in the binding and
wash solutions, or 2) reducing the pH. Generally, highly expressed
proteins, such as those from a bacterial expression system, have
fewer contaminant proteins that copurify along with the protein of
interest. Endogenous protein contaminants are more abundant
in eukaryotic expression systems and tend to bind to the IMAC
adsorbent more weakly. In these instances, nonspecific binding
of proteins containing neighboring histidine residues becomes
a problem. These endogenous species may be washed from
the resin by either lowering the pH to 6.3 or adding imidazole to
binding and wash solutions in concentrations of 5–30 mM. The
optimal pH and/or imidazole concentration used in wash buffers
is always protein dependent and should always be determined
experimentally.
Recommended wash buffer:
• 5–30 mM imidazole; for example, 50 mM sodium phosphate,
0.3 M NaCl.
Begin with: 5 mM imidazole, 50 mM sodium phosphate,
0.3 M NaCl, pH 8.0
Elution
Desorption of the histidine-tagged protein may be accomplished in
one of three ways: introduction of a competitor ligand, reduction of
the pH, or stripping of the immobilized metal.
In competitive elution, a step or gradient elution with ligands such
as imidazole, histidine, histamine, or glycine may be carried out.
When using a gradient elution with imidazole, it is important to preequilibrate the column with low concentrations of imidazole (1 mM)
and include the same concentration in the sample. This prevents
adsorption of imidazole onto the resin from triggering a drop in pH,
which might prematurely elute bound histidine-tagged proteins.
Lowering the pH of the elution buffer (pH 4.5–5.3) also releases
bound histidine-tagged proteins. In this case, the histidine residues
become protonated and are unable to bind to the immobilized ion.
Protein sensitivity to low pH ranges, however, must be taken into
consideration.
8 Nuvia IMAC Ni-Charged Resin
Page 13

If lower pH is used to elute bound proteins, tubes filled with a
strong neutralizing buffer such as 1 M Tris-HCl, pH 8.0 may be
used to collect acidic eluates (100–200 µl eluate/ml neutralizing
buffer). The recommended range is pH 3–5 with acetate buffers
being a preferred choice. Weakly bound contaminants may be
washed off in an intermediate wash at around pH 5.5–6.5.
Strong chelating agents such as EDTA and EGTA strip immobilized
ions from the column and cause the bound histidine-tagged
protein to elute as a protein-metal complex. This results in metal
ions appearing in the protein fractions.
Recommended elution buffer for Nuvia IMAC resin
Though a number of conditions can be used to elute the target
protein from the Nuvia resin, adjusting the concentration of
imidazole is recommended.
Recommended elution buffer:
• 20–500 mM imidazole; for example, 50 mM sodium
phosphate, 0.3 M NaCl.
Begin with: 0.5 M imidazole, 50 mM sodium phosphate,
0.3 M NaCl, pH 8.0
Purification under Denaturing Conditions
When overexpressed in E. coli, some proteins may aggregate,
forming what are known as inclusion bodies. These inclusion
bodies need to be solubilized in strong denaturants such as
6 M guanidine HCl or 8 M urea in order to purify the histidinetagged protein. Usually proteins expressed as inclusion bodies
are not in their native conformation, so high concentrations of
denaturants may be used during lysate preparation and protein
purification.
In order to restore the native conformation and activity of the
protein, the denaturant must be removed by dilution, dialysis, or
size exclusion chromatography. Renaturation of the protein while it
is still bound to the IMAC column is a good alternative and offers
several advantages. Aggregation may be kept to a minimum if the
protein refolds on the column when the denaturant is removed.
Higher concentrations of the refolded protein may therefore be
collected.
Nuvia IMAC Ni-Charged Resin 9
Page 14

Finally, the use of a liquid chromatography system ensures that
the adjustment of denaturants, detergents, salts, and pH will be
effectively controlled.
Imidazole Concentrations
For optimal protein purification results, it is crucial that the
imidazole concentrations in lysis, binding, and wash buffers, as
well as elution buffers, be empirically established. Determine
optimized conditions using a small amount of sample. These
conditions may then be used to design the purification protocol
for larger samples on the same column or on a larger column. As
each protein behaves differently, it is helpful to keep the following
points in mind during lysate preparation and purification:
• Low concentrations of imidazole (1–30 mM) in lysis, binding,
and wash buffers are recommended if the potential for
background contaminants exists. The ability for nontagged
contaminating proteins to bind to the resin is generally higher
under nondenaturing conditions than under denaturing
conditions
• Low concentrations of imidazole (1–30 mM) help minimize
nonspecific binding of proteins containing noncontiguous
histidine residues by competing with them for available binding
sites on the transition metal. Competition occurs because
the imidazole ring is also found in the histidine-containing
compound
• If the recombinant histidine-tagged protein does not bind
under higher concentrations, the imidazole concentration can
be reduced for binding and wash steps
• A gradient elution test may be used to determine
concentrations of imidazole for wash and elution steps. Once
the imidazole concentration to elute the protein is established,
large samples and/or columns may be used
10 Nuvia IMAC Ni-Charged Resin
Page 15

Section 4
Column Packing — Medium-Pressure Columns
Slurry Packing an IMAC Column
Slurry packing is preferred for small columns. This method
describes packing a Nuvia™ IMAC resin with aid from a pump. For
best results, use 5–50 mm ID columns and a bed height of
5–30 cm.
Recommended Columns
Bio-Rad’s Bio-Scale™ MT high-resolution columns may be used
for the following column-packing procedure. These columns are
empty and may be packed with the support of choice.
Bio-Scale MT columns are convenient to use with Bio-Rad’s
NGC™ chromatography system or any medium- or high-pressure
system:
• Bio-Scale MT2 column (7 x 52 mm) for bed volumes
up to 2 ml
• Bio-Scale MT5 column (10 x 64 mm) for bed volumes
up to 5 ml
• Bio-Scale MT10 column (12 x 88 mm) for bed volumes
up to 10 ml
• Bio-Scale MT20 column (15 x 113 mm) for bed volumes
up to 20 ml
Materials
• Empty column (1–5 cm ID x 30 cm) with flow adaptors, inlet
and outlet ports
• Glass filter
• Nuvia IMAC Ni-charged resin
• Packing reservoir
• Pump
Nuvia IMAC Ni-Charged Resin 11
Page 16

Resin Preparation
Nuvia IMAC Ni-charged resins come in a 20% ethanol solution,
supplied for resin storage. The column can be slurry packed in
this solution; simply resuspend the resin in the ethanol solution
provided. Before applying sample, ensure that all ethanol is
removed during the equilibration step.
Method
1. Eliminate air from the column dead spaces. Attach the inlet
of a peristaltic or other pump to the outlet of the MT column.
Fill the column with distilled water to about 10% of its volume.
Flush end pieces with distilled water to ensure that the bottom
of the bed support is fully saturated and free of air bubbles.
Allow a few centimeters of distilled water to remain when
closing the outlet valve.
2. Suspend the 50% v/v slurry by gently swirling or stirring with a
glass or plastic rod.
3. Carefully transfer about a third of the slurry down the side into
the column using a glass or plastic rod to avoid introducing air
bubbles.
4. Start the pump at a low flow rate (eg. 0.5 ml/min). The resin
will begin to pack in the column. As the liquid level in the
column drops, continue to transfer the rest of the slurry until
the packed bed reaches 1 cm from the top. Stop or slow the
pump flow rate as necessary.
Gently add more distilled water down the side of the column
to make sure the liquid does not fall below the resin level.
Continue adding distilled water until the bed seems to have
stabilized. Then gently fill the column to the top with distilled
water.
5. Stop the pump and prepare to insert the top adapter.
6. Insert the top flow adaptor. Use care to avoid introducing any
air bubbles. Insert the adaptor into the column at an angle.
Make sure the exit tubing is open so that distilled water can
flow out the top adaptor along with any air.
12 Nuvia IMAC Ni-Charged Resin
Page 17

7. Adjust the adaptor to sit directly on top of the resin bed.
Open the column outlet and pump distilled buffer through the
column at a) a packing flow rate of ~400–600 cm/hr for 5 to
10 min, or b) the maximum pressure allowed by the column
hardware and resin. The resin bed will compress while packing
at high flow rates. Mark the compression level with a pen.
Stop the flow. At this point, the resin bed height may readjust
and rise. If this happens, adjust the flow adaptor to compress
the bed another 0.1–0.5 cm past the level marked with the
pen.
8. Reconnect the pump and equilibrate. Pass eluent (distilled
water) through the column at the packing flow rate. During
equilibration, the bed may compress even further. When a
constant bed height is reached, mark the compression level at
this flow rate. Again, adjust the adaptor to compress the bed
an additional 0.1–0.5 cm past the level marked.
Note: Chromatographic steps during purification should not be
run at greater than 75% of the packing flow rate.
Nuvia IMAC Ni-Charged Resin 13
Page 18

Section 5
Column Packing — Sample Preparation–Sized
Columns
Use this method for packing Nuvia™ IMAC resin into sample
preparation micro spin columns.
Materials
1. Reagents
• 0.1 M NiSO4 (or other suitable metal salt solution)
Note: Charging Nuvia IMAC nickel-charged resin (Ni-charged
resin) is not required for initial use. For subsequent uses it is
recommended that the resin be cleaned of all contaminants,
stripped of metal ions, and recharged with proper metal ions
prior to loading sample.
If needed, the procedure to charge Nuvia IMAC (uncharged)
resin is explained in Section 6, Immobilizing Metal Ions.
2. Equipment
• Sample preparation–sized columns (for example,
Micro Bio-Spin™ columns, catalog #732-6204)
• Plasticware, 2 ml capped and 2 ml capless tubes
• Nuvia IMAC Ni-charged resin
• Tabletop centrifuge
• 1 ml pipet with wide-bore pipet tips
Resin Preparation
Nuvia IMAC Ni-charged resins come in a 20% ethanol solution,
supplied for resin storage. The column can be packed in this slurry
or decanted into distilled water and packed.
14 Nuvia IMAC Ni-Charged Resin
Page 19

Method
1. Thoroughly suspend Nuvia IMAC resin.
2. Place the column into an appropriate collection vessel; for
example, a 2 ml capless collection tube.
3. Using a pipet, transfer the appropriate amount of Ni-charged
Nuvia IMAC resin to a microcentrifuge tube. If using a Micro
Bio-Spin column, transfer 0.2 ml slurried Ni-charged Nuvia
IMAC resin to the column. This is equivalent to ~100 µl of a
packed resin bed.
4. Remove storage solution by centrifugation. Centrifuge at
1,000 x g for 15 sec to pack resin.
5. Wash column with at least 5 column volumes (or ~500 µl) of
distilled water. Centrifuge at 1,000 x g for 15 sec to pack resin.
6. If using Ni-charged resin, equilibrate the column with at least
5 column volumes of binding buffer. The column is now ready
for separation.
Nuvia IMAC Ni-Charged Resin 15
Page 20

Section 6
Immobilizing Metal Ions
Efficacy of protein binding by IMAC depends on two factors: the
number of available histidine, cysteine, and tryptophan residues
on a protein’s surface, and the number of coordination sites on
the immobilized ion that are not occupied by the chelating ligand
and thus available to bind amino acid residues. Nuvia™ IMAC resin
uses a quadridentate ligand (NTA), which leaves two of the six
coordination sites on the nickel ion accessible to the protein of
interest.
Although the most commonly used ion is Ni2+, protein selectivity
may be increased through the choice of metal ion used,
understanding of the structure of the metal-chelate complex
and its interaction with the protein, knowledge of the protein’s
expression level, and the ligand density of the IMAC adsorbent.
While high ligand density usually means higher binding capacity, it
can also translate into lower target protein selectivity. Nuvia IMAC
resin, based on the polymeric UNOsphere™ technology, has been
specifically formulated with an optimal number of chelating ligands
on the resin’s surface and pores to deliver both good capacity and
excellent protein purity.
1. Equilibrate the column with 5 column volumes of 50 mM
sodium acetate, 0.3 M NaCl, pH 4.0.
2. After slurry packing is complete (see Sections 4 and/or 5), the
column is ready for the removal or addition of metal ions.
3. If necessary, strip any metal ions by washing with 10 column
volumes of 50 mM sodium phosphate, 0.3 M NaCl, and
0.05–0.5 M EDTA, pH 7.5.
4. Make a 0.1–0.3 M solution of the metal ion of choice. For best
results, the pH of the solution should be <7 (neutral to weakly
acidic).
5. Apply 3–5 column volumes of the metal ion solution.
6. Wash with 5 column volumes of 50 mM sodium acetate, 0.3 M
NaCl, pH 4.0. Remove excess ions by washing.
7. Wash with 10 column volumes of deionized water.
8. Equilibrate with at least 5 column volumes of starting buffer; for
example, 50 mM sodium phosphate, 0.3 M NaCl, pH 7–8. The
column is now ready for separation.
16 Nuvia IMAC Ni-Charged Resin
Page 21

Section 7
Medium-Pressure Column Purification
of Histidine-Tagged Proteins
For this guideline, the sample is applied to a packed column and
the proteins are eluted using a high concentration of imidazole.
The guideline does not optimize the imidazole concentration, but
instead provides for fast capture of the target protein that may be
used as a quick check for protein expression levels. Higher levels
of purity are achievable by optimizing imidazole concentrations,
which improves protein separation. See Section 8, MediumColumn Purification – Using an Imidazole Gradient to Determine
Optimal Purification of Histidine-Tagged Proteins.
Materials
1. Reagents
Binding buffer
• 50 mM sodium phosphate (NaH2PO4)
• 300 mM NaCl
• Low concentrations imidazole* (0–15 mM)
• Adjust to pH 8.0
Wash buffer
• 50 mM sodium phosphate (NaH2PO4)
• 300 mM NaCl
• Low concentrations of imidazole* (0–30 mM)
• Adjust to pH 8.0
Elution buffer
• 50 mM sodium phosphate (NaH2PO4)
• 300 mM NaCl
• Higher concentrations of imidazole* (250–500 mM)
• Adjust to pH 8.0
* For optimal protein purification results, it is crucial that the
imidazole concentrations in lysis, binding, and wash buffers, as
well as elution buffers, be empirically established. Determine
optimized conditions using a small amount of sample.
Nuvia IMAC Ni-Charged Resin 17
Page 22

2. Equipment
• IMAC column (as prepared in Section 4)
3. Biological Sample
• Clarified lysate
The binding capacity of the Nuvia™ IMAC resin is ~40 mg
histidine-tagged protein per ml resin. Larger amounts of
protein will require use of a larger column.
4. Additional Materials
• Medium-pressure chromatography system (such as
Bio-Rad’s BioLogic™ or NGC™ system)
• Equipment for determining total protein concentration
within the lysate
Method
1. Equilibrate the column with at least 5 column volumes of
binding buffer.
2. Add or dilute sample in binding buffer and load onto the
column using a desired flow rate.
The choice of binding buffer will vary based on the properties
of the sample to be purified. Sodium or potassium phosphate
is recommended as a general starting buffer; for example,
50 mM sodium phosphate, 300 mM NaCl, 5 mM imidazole,
pH 8.0. Binding of histidine-tagged protein on the Nuvia IMAC
resin is optimal in the pH range of 7–8.
The column may be run at flow rates up to 500 cm/hr. Higher
binding of histidine-tagged proteins will be achieved at lower
flow rates. Average binding capacity of the Nuvia IMAC resin is
approximately 40 mg histidine-tagged protein/ml resin.
3. Collect fractions.
These fractions represent unbound proteins.
4. Wash the resin with at least 5 column volumes of wash buffer
to remove unbound sample.
Wash out remaining unbound solutes. Repeat wash steps as
necessary for the A
to be at or near baseline.
280
18 Nuvia IMAC Ni-Charged Resin
Page 23

5. Collect fractions from wash steps.
Pool recovered unbound proteins with fractions collected in
step 3.
6. Elute bound proteins with 5 column volumes of elution buffer.
Collect 2 ml fractions, or approximately 0.2 column volumes
each.
The choice of elution buffer will vary depending on the
procedure used. For example, a range of imidazole
concentrations (100–500 mM) may be used to elute bound
protein from the Nuvia IMAC resin.
7. Repeat elution steps 2 to 4 more times.
Save the eluates for further analysis (A
, SDS-PAGE, ELISA,
280
etc.).
IMAC Purification under Denaturing Conditions
In some cases it may be necessary to use denaturants such as
urea to solubilize inclusion bodies, which are not generally in their
native conformation. To perform this, up to 8 M urea (or 6 M
guanidine HCl) may be used in the binding, wash, and elution
buffers listed above. Elution is still achieved by increasing the
imidazole concentration.
Note: If using guanidine HCl (GuHCl), it must be removed from
purified samples prior to loading onto SDS-PAGE gels to prevent
precipitation. Proteins that have been lysed and adsorbed onto
the column with guanidine HCl may be washed and eluted with a
urea-based buffer.
Often the protein can be restored to its native form. To do this, the
denaturant used to lyse and purify the sample must be removed
using dilution, dialysis, or size exclusion chromatography.
Renaturation of the protein while it is still bound to the IMAC
column is a good alternative and offers several advantages.
Aggregation may be kept to a minimum if the protein refolds on the
column when the denaturant is removed. Higher concentrations of
the refolded protein may therefore be collected. Guidelines for
on-column renaturation are suggested on the following page.
Nuvia IMAC Ni-Charged Resin 19
Page 24

Materials for On-Column Renaturation
1. Reagents
Refolding buffer
• 20 mM sodium phosphate (NaH2PO4) (pH 8.0)
• 20 mM imidazole
• 300 mM NaCl
• 1 mM β-mercaptoethanol
Urea binding buffer (refolding buffer with 8 M urea)
Phosphate elution buffer (with high imidazole)
• 20 mM sodium phosphate (NaH2PO4) (pH 8.0)
• 500 mM imidazole
• 300 mM NaCl
• 1 mM β-mercaptoethanol
Note: The β-mercaptoethanol should be added to solutions
only immediately before use.
Method for On-Column Renaturation
1. Wash the column containing the bound protein with 10 column
volumes of urea binding buffer.
2. Apply a linear gradient from 100% urea-binding buffer to 100%
refolding buffer over 60 min at 0.5 ml/min. Refolding is initiated
by a descending gradient from 8 to 0 M urea.
3. Apply a linear gradient from 100% refolding buffer to 100%
phosphate elution buffer with high imidazole.
4. If necessary, add another chromatography step.
Size exclusion chromatography may be a good choice
because aggregates of unrefolded protein can be removed
and the buffer composition of the purified material can be
changed simultaneously.
20 Nuvia IMAC Ni-Charged Resin
Page 25

Section 8
Medium-Pressure Column Purification — Using
an Imidazole Gradient to Determine Optimal
Purification of Histidine-Tagged Proteins
Gradient elution tests are useful because they do not require
optimization of imidazole concentrations, but instead may be
used to determine suitable imidazole concentrations for wash and
elution steps. Once a suitable concentration has been determined
using a gradient elution such as the one outlined below, often an
easier protocol using step elution may be used for subsequent
purification of larger sample volumes.
With step elution, the protein can be collected in smaller
volumes and at higher concentrations. Using this protocol, the
concentration of imidazole that elutes the target protein may be
calculated and used for a step protocol.
This protocol requires the use of a gradient mixer coupled to a
chromatography system such as Bio-Rad’s NGC™ system to
establish a linear gradient.
Materials
1. Reagents
Binding buffer
• 20 mM sodium phosphate (NaH2PO4)
• 300 mM NaCl
• 20 mM imidazole
• Adjust to pH 8.0
Elution buffer
• 50 mM sodium phosphate (NaH2PO4)
• 300 mM NaCl
• 500 mM imidazole
• Adjust to pH 8.0
2. Equipment
• Chromatography system with dual pump and gradient
capability
• Fraction collector
• IMAC column (as prepared in Section 4)
Nuvia IMAC Ni-Charged Resin 21
Page 26

3. Biological Sample
• Clarified lysate
Note: Keep the sample as small as possible during
optimization of binding and elution conditions.
4. Additional Materials
• Equipment for assessing protein purity and recovering of
the histidine-tagged protein
Method
Part 1: Optimizing the Imidazole Concentration
1. Purge the entire flow path of the chromatography system with
water according to the manufacturer’s instructions.
Connect the column and wash it with 10 column volumes
of water. Disconnect the column either by valve switching
or manually. Purge the flow path before the column with
elution buffer from inlet B and, with the column offline, then
with binding buffer from inlet A. Purge the entire system with
binding buffer. Reconnect the column to the system.
2. Equilibrate the column with 10 column volumes of binding
buffer (0%B).
3. Begin collecting fractions of 1 column volume.
If a 1 ml IMAC column is used, 1 ml fractions are
recommended. For larger columns, reduce the fractions
collected to amounts from 0.2 to 0.5 column volumes.
4. Load sample and collect the flowthrough in fractions
appropriate to the size of the column (as recommended
above).
Note: Monitor the backpressure while sample is being applied.
If the sample is insufficiently clarified, backpressure will
increase.
5. Wash the unbound material with 10 column volumes of
binding buffer (0%B).
6. Elute the sample with a linear gradient of 0–50% elution buffer.
7. Wash the column with 100% of the elution buffer for 5 column
volumes.
22 Nuvia IMAC Ni-Charged Resin
Page 27

8. Equilibrate the column with 10 column volumes of binding
buffer (0%B). Stop collecting fractions.
9. Identify the fractions that contain the histidine-tagged protein
using an activity assay (such as one of Bio-Rad’s protein assay
kits), UV absorbance, SDS-PAGE, or western blot analysis
with antihistidine antibodies or antibodies specific to the target
protein.
10. Calculate the concentration of imidazole that corresponds to
the elution peak of the histidine-tagged protein.
Note: The delay due to column volume and the tubing in
the chromatography system need to be considered when
comparing the actual gradient trace to the programmed
gradient.
11. Based on calculated imidazole concentration, a stepwise
experiment may now be designed.
The following tips are useful to keep in mind when designing the
experiment:
• Maintain the concentration of imidazole in the binding
(also called equilibration) buffer at 20 mM. If large amounts
of contaminants are also adsorbed onto the resin, the
concentration of imidazole in the sample and equilibration
buffer may be increased. This may reduce the overall amount
of target protein bound and should be carried out with care.
However, it will also increase the column’s binding capacity
for the target protein due to the reduction in contaminating
proteins
• Include a wash step with an imidazole concentration slightly
lower than the concentration necessary to elute the target
protein. This will increase purity by removing unbound
contaminants without eluting the bound histidine-tagged
protein. The optimized wash step should include 50 mM
sodium phosphate, 0.3 M NaCl, and an appropriate
concentration of imidazole
• The elution buffer should contain a concentration of imidazole
greater than the calculated concentration corresponding to the
eluted peak of target protein
• Perform a trial run
Nuvia IMAC Ni-Charged Resin 23
Page 28

Part 2: Using an Optimized Imidazole Concentration for
Purification
1. Prepare 500 ml binding buffer and 500 ml elution buffer for a 5
ml column.
Purge the pumps with the fresh buffers. Adjust buffer volumes
for larger scale purifications.
2. Equilibrate the column with 10 column volumes of
equilibration/binding buffer.
3. Begin collecting fractions.
If a 1 ml IMAC column is used, 1 ml fractions are
recommended. For larger columns, reduce the fractions
collected to amounts ranging from 0.2 to 0.5 column volumes.
4. Load sample and collect the flowthrough in fractions
appropriate to the size of the column, as recommended
above.
Note: Monitor the backpressure while the sample is
being applied. If the sample is insufficiently clarified, the
backpressure will increase.
5. Wash the column with a minimum of 5 column volumes of
binding buffer to remove unbound contaminants.
6. Wash the column with a minimum of 5 column volumes of
binding or starting buffer that contains imidazole in a quantity
that will not elute the target protein.
7. Elute the histidine-tagged protein with 5 column volumes of
elution buffer with an optimized imidazole concentration.
8. Wash the column with 5 column volumes of elution buffer.
Stop collecting fractions.
9. Re-equilibrate the column with 10 column volumes of
equilibration (binding) buffer.
10. Assess the purity and recovery of the target protein using an
activity assay (such as one of Bio-Rad’s protein assay kits),
UV absorbance, SDS-PAGE, or western blot analysis with
antihistidine antibodies or antibodies specific to the target
protein.
24 Nuvia IMAC Ni-Charged Resin
Page 29

Section 9
Sample Preparation–Sized Spin-Column Purification
of Histidine-Tagged Proteins
Materials
1. Reagents
Binding/wash buffer
• 50 mM sodium phosphate (NaH2PO4)
• 300 mM NaCl
• 5 mM imidazole
• Adjust to pH 8.0
Optimized wash buffer with imidazole (optional, see Section 8)
• A wash buffer containing slightly less imidazole than
necessary to elute the target protein may be used
to increase the stringency of the wash step. Refer to
Section 8, Medium-Pressure Column Purification —
Using an Imidazole Gradient to Determine Optimal
Purification of Histidine-Tagged Proteins
• Once the concentration of imidazole in the wash
step is determined using medium-pressure column
chromatography, a stepwise elution step may be
carried out as indicated in this protocol
Elution buffer
• 50 mM sodium phosphate (NaH2PO4)
• 300 mM NaCl
• 500 mM imidazole
• Adjust to pH 8.0
Note: If necessary, up to 8 M urea may be added to these
buffers in order to solubilize proteins in inclusion bodies. See
Section 7 for more information.
2. Equipment
• Sample preparation–sized IMAC spin column (as prepared
in Section 5) (for example, Micro Bio-Spin™ columns, cat.
#732-6204)
• Plasticware, 2 ml capped and 2 ml capless tubes
Nuvia IMAC Ni-Charged Resin 25
Page 30

3. Biological Sample
• Clarified lysate
The binding capacity of Nuvia™ IMAC resin is ≥40 mg histidinetagged protein per ml resin. Larger amounts of protein will
require a larger column
4. Additional Materials
• Equipment for assessing protein purity and recovery of the
histidine-tagged protein
Method
Reserve a small amount of lysate prior to loading sample onto
the column. This will serve as sample for the lysate lane for later
analysis with, for example, SDS-PAGE.
Part 1: Binding the Sample
1. Start with a prepacked spin column, charged with the metal
ion of choice.
See Section 5, Column Packing — Sample Preparation–Sized
Columns for protocol.
2. Place prepacked spin column in an appropriate spin collection
tube.
3. Pre-equilibrate the spin column with 5 column volumes of
binding buffer.
The choice of binding buffer will vary based on the properties
of the sample to be purified. Potassium phosphate and
sodium phosphate are recommended as general starting
buffers; for example, 50 mM sodium or potassium phosphate,
300 mM NaCl, pH 8.0. Binding of histidine-tagged protein on
the Nuvia™ IMAC resin is optimal in the pH range of 7–8.
4. Add an appropriate amount of lysate (≤0.5 ml) to the micro
spin column.
5. Mix by pipetting up and down 5 times.
Incubate for up to 5 min in micro spin column.
6. Centrifuge at 1,000 x g for 1 min.
Remove the unbound proteins by centrifuging.
26 Nuvia IMAC Ni-Charged Resin
Page 31

Part 2: Washing the Resin
1. Insert micro spin column into new collection vessel.
2. Wash the resin with at least 5 column volumes of binding
buffer containing imidazole.
Pipet up and down at least 5 times.
Note: If previously determined, you may use an optimized
concentration of imidazole that is slightly less than the
concentration necessary to elute the target protein. See
Section 8.
3. Centrifuge at 1,000 x g for 1 min.
Remove remaining unbound proteins by centrifuging. The
wash step can be repeated if necessary.
Part 3: Eluting the Histidine-Tagged Protein
1. Insert micro spin column into a new, clean collection vessel.
2. Elute bound proteins with 5 column volumes of elution buffer.
Pipet up and down at least 5 times and incubate for up to
5 min.
The choice of elution buffer will vary depending on the
procedure used. For example, a range of imidazole
concentrations (150–500 mM) may be used to elute bound
protein from the Nuvia IMAC resin.
3. Analyze fractions from above steps by A
, SDS-PAGE,
280
ELISA, etc.
Nuvia IMAC Ni-Charged Resin 27
Page 32

Section 10
Regenerating, Cleaning, Sanitizing, and Storage
Nuvia™ IMAC columns are well-suited for reuse. The polymeric
nature and open pore structure of the resin allow the column to be
run at high flow rates during regeneration, cleaning, and sanitizing
steps. Protein separations are unaffected, even after numerous
cycles, as reproducibility is extremely high.
Unless otherwise stated, the following steps may be carried out at
2 ml/min.
Regenerating the Medium
Regeneration cleans the medium adequately to start the next
cycle. In general, IMAC columns may be used a number of
times before they need to be recharged with metal ions. When it
becomes necessary, regenerate metal-charged Nuvia IMAC resins
by first stripping with an EDTA solution. Wash the column with 10
column volumes of 20 mM sodium phosphate, up to 1 M NaCl,
and 0.2 M EDTA, pH 7.4. Ensure that residual EDTA is completely
removed from the column by washing it with 3–5 column volumes
of binding buffer followed by 3–5 column volumes of distilled water.
Recharge the IMAC resin as recommended in Section 6,
Immobilizing Metal Ions, or proceed with cleaning-in-place
measures (below).
Cleaning in Place
A column used to purify protein from cell extract usually contains
some soluble substances and cell debris that are nonspecifically
adsorbed onto the matrix. Cleaning in place eliminates material
not removed by regeneration and prevents progressive buildup of
contaminants. If the column is to be reused, these contaminants
should be cleaned from the column if they were not completely
removed during the sample clarification steps.
The following steps may be followed to clean IMAC columns. This
protocol also includes a regeneration step. For optimal results, the
column can be run at 2–5 ml/min.
28 Nuvia IMAC Ni-Charged Resin
Page 33

1. Strip metal ions.
Wash with 10 column volumes of 50 mM sodium phosphate,
0.3 M NaCl, and 0.05–0.5 M EDTA, pH 7.5.
2. Wash the column with one of the following solutions at
2 ml/min:
a. 1% acetic acid
• This solution may be used as a cleaning, sanitizing,
and storage solution with Nuvia IMAC resins
• 10–15 min exposure time
• Rinse with 10 column volumes of distilled water
b. 2 M NaCl (removes ionic contaminants)
• 10–15 min exposure time
• Rinse with 10 column volumes of distilled water
c. 1 M NaOH up to 3 hr (removes precipitated, hydrophobic,
and lipoproteins)
• Exposure time is usually 1–3 hr
• Rinse with 10 column volumes of distilled water
d. 70% ethanol or 30% isopropyl alcohol (removes
precipitated, hydrophobic, and lipoproteins)
• 15–20 min exposure time
• Alternatively, use 0–30% gradient isopropyl alcohol
over 5 column volumes, followed by 2 column
volumes 30% isopropyl alcohol
• Rinse with 10 column volumes of distilled water
3. Remove cleaning solution(s) from column by rinsing with 10
column volumes of binding buffer.
• Rinse column with 50 mM sodium phosphate,
0.3 M NaCl, pH 8
• Ensure the eluate is ~pH 8 and the UV signal has returned
to baseline
4. Recharge the column with metal ion of choice.
• Refer to Section 6, Immobilizing Metal Ions
Nuvia IMAC Ni-Charged Resin 29
Page 34

Sanitization
Sanitization inactivates microorganisms and prevents buildup of
endotoxins. For optimal results, the column should be run at
300 cm/hr.
The column may be washed with the following solution:
• 1 M NaOH
Rinse solution from column with 3–5 column volumes of distilled
water. Re-equilibrate the column with 3–5 column volumes of
binding buffer.
Storage
Nuvia IMAC resin is stable at room temperature across a broad
pH range (2–14). The medium may also be stored in either of the
following solutions:
• 2% benzyl alcohol
• 20% ethanol
30 Nuvia IMAC Ni-Charged Resin
Page 35

Section 11
Troubleshooting Guide
Problem Possible Cause Solution
Sample is too viscous High concentration
of host nucleic acids
in lysate
Insufficient amount of
homogenization
buffer
Viscosity of extract can be reduced by
adding Benzonase nuclease (1.7 U/ml)
with 1 mM MgCl2 to fragment bacterial
DNA. Incubate on ice for 15 min
Dilute sample by adding more
homogenization buffer
Sample application
causes column
to clog
No protein is eluted Expression of target
Insufficient
clarification of sample
protein in extract is
very low and is not
found in the eluate
Target protein is
found in inclusion
bodies or possible
insufficient lysis
Target protein
is found in the
flowthrough
Prevent cell debris from clogging the
column by increasing the centrifugation
speed or filtering the sample
Check expression level of protein by
estimating the amount in the extract,
flowthrough, eluted fraction, and pellet
upon centrifugation. Use western blotting
with anti-6x histidine antibodies, target
protein-specific antibodies, ELISA, or
enzyme activity determination
Apply larger sample volume
Minimize contact with hydrophobic
surfaces (that is, polystyrene tubes).
Proteins at low concentration may bind to
the surface of the tube
Increase intensity/duration of disruption
and homogenization
If protein is insoluble, use 6 M guanidine
HCl or 8 M urea to lyse denatured proteins
(see Sections 3, and 7)
Reduce imidazole concentration in
sample, binding, and wash buffers.
An imidazole gradient may be used to
determine optimal amounts for wash and
elution conditions
Check pH levels of sample. A decrease in
pH may result during the homogenization
step or during growth of the culture
medium. Adjust pH to 7–8
The histidine tag may not be accessible.
Use denaturing conditions to purify protein
or reclone the plasmid construct with the
histidine-tagged sequence placed at the
opposite terminus
Nuvia IMAC Ni-Charged Resin 31
Page 36

Problem Possible Cause Solution
No protein is eluted
(continued from
previous page)
Protein precipitates
during purification
Poor recovery
of target protein
Histidine-tagged
protein is not pure
Target protein is found
in the flowthrough
(continued from
previous page)
Elution conditions
are too mild or
protein may be in
an aggregated or
multimer form
Temperature is too
low
Aggregate forms
Protein is found in the
flowthrough
Binding capacity of
the column has been
exceeded
Target protein was
not detected in the
flowthrough
Strong nonspecific
adsorption of the
target protein to the
matrix
Contaminants elute
with target protein
Strongly bound
contaminants elute
with protein
Association of
contaminating
proteins with target
protein via disulfide
bonds
Proteolytic cleavage during fermentation
or purification has caused the histidine tag
to be removed. Add protease inhibitors or
make a new construct with histidine tag
attached to other terminus
Elute with pH or imidazole step elution
Perform the purification at room
temperature
Add solubilization agents to samples and/
or buffers: 0.1% Triton X-100, Tween 20,
20 mM β-mercaptoethanol and ≤20%
glycerol to maintain protein solubility
See recommendations in No protein is
eluted section
Increase the column size or reduce the
sample volume application
Capillary sample loop is too small
Reduce hydrophobic adsorption by
including detergents or organic solvents,
or by increasing the concentration of NaCl
Make binding and wash steps more
stringent. Include 10–20 mM imidazole in
binding and wash buffers
Prolong the wash step containing
imidazole
Column is too large; reduce amount of
Nuvia™ IMAC resin used
Very high concentrations of imidazole
will cause strongly bound contaminants
to elute as well. Reduce the imidazole
concentration during the elution
Include ≤30 mM β-mercaptoethanol.
Exercise caution if using DTT
32 Nuvia IMAC Ni-Charged Resin
Page 37

Problem Possible Cause Solution
Histidine-tagged
protein is not pure
(continued from
previous page)
Association between
the histidine-tagged
protein and protein
contaminant
Potential degradation
of fusion protein by
proteases
Contaminants exhibit
similar affinity to target
protein
Add nonionic detergent or alcohol (that
is, ≤2% Triton X-100, 2% Tween 20, or
≤20% glycerol) to reduce hydrophobic
interactions. Concentration of NaCl may
be increased to minimize electrostatic
interactions
Include 1 mM PMSF or other protease
inhibitor in lysis buffer to reduce partial
degradation
Add a chromatography step; that is, ion
exchange, hydrophobic interaction, or
size exclusion
Nuvia IMAC Ni-Charged Resin 33
Page 38

Section 12
Ordering Information
Nuvia IMAC™ Ni-Charged Resin
Catalog # Description
780-0800 Nuvia IMAC Ni-Charged Resin, 25 ml bottle
780-0801 Nuvia IMAC Ni-Charged Resin, 100 ml bottle
780-0802 Nuvia IMAC Ni-Charged Resin, 500 ml bottle
Bio-Scale™ Mini Nuvia™ IMAC Ni-Charged Cartridges
Catalog # Description
780-0811 Bio-Scale Mini Nuvia IMAC Ni-Charged Cartridge,
1 x 5 ml column
780-0812 Bio-Scale Mini Nuvia IMAC Ni-Charged Cartridge,
5 x 5 ml columns
Section 13
References
Hochuli E (1988). Large-scale chromatography of recombinant
proteins, J Chromatogr 444, 293–302.
Porath J et al. (1975). Metal chelate affinity chromatography, a new
approach to protein fractionation. Nature 258, 598–599.
Section 14
Legal Notices
Triton is a trademark of Union Carbide. Tween is a trademark of ICI
Americas, Inc. Benzonase is a trademark of Merck KGaA Corp
34 Nuvia IMAC Ni-Charged Resin
Page 39

Page 40

Bio-Rad
Laboratories, Inc.
Life Science
Group
Web site ww w.bio-rad.com USA 800 424 6723
Australia 61 2 9914 2800 Austria 01 877 89 01 Belgium 09 385 55 11
Brazi l 55 11 3065 7550 Canada 905 364 3435 China 86 21 6169 8500
Czech Republic 420 241 430 532 Denma rk 44 52 10 00
Finland 09 804 22 00 France 01 47 95 69 65 Ger many 089 31 884 0
Greece 30 210 9532 220 Hon g Kong 852 2789 330 0
Hungary 36 1 459 6100 India 91 124 4029300 Israel 03 963 6050
Italy 39 02 2160 91 Japan 81 3 6361 7000 Kore a 82 2 347 3 4460
Mexico 52 5 55 488 7670 The Netherlands 0318 540666
New Zealand 64 9 415 2280 Norway 23 38 41 30
Poland 48 22 331 99 99 Portugal 351 21 472 7700
Russia 7 495 721 14 04 Singapore 65 6415 3188
South Africa 27 861 246 723 Spain 3 4 91 590 5200
Sweden 08 555 1270 0 Switzerland 026 674 55 05
Taiwan 88 6 2 2578 7189 Thailand 1800 88 22 88
United Kingdom 020 8328 200 0
Sig 121310044307 Rev A US/EG
 Loading...
Loading...