Page 1

Gene Pulser Xcell
™
Electroporation System
Instruction Manual
Catalog Numbers
165-2660, 165-2661, 165-2662, 165-2666,
165-2667, and 165-2668
For technical service
call your local Bio-Rad office or
in the U.S. call 1-800-4BIORAD
(1-800-424-6723)
On the Web at discover.bio-rad.com
Page 2

Warranty
The Gene Pulser Xcell electroporation system is warranted against defects in materials and workmanship
for 1 year. If any defects occur in the instruments or accessories during this warranty period, Bio-Rad
Laboratories will repair or replace the defective parts at its discretion without charge. The following
defects, however, are specifically excluded:
1. Defects caused by improper operation.
2. Repair or modification done by anyone other than Bio-Rad Laboratories or an authorized agent.
3. Damage caused by substituting alternative parts.
4. Use of fittings or spare parts supplied by anyone other than Bio-Rad Laboratories.
5. Damage caused by accident or misuse.
6. Damage caused by disaster.
7. Corrosion caused by improper solvent or sample.
This warranty does not apply to the following parts:
Fuses
For any inquiry or request for repair service, contact Bio-Rad Laboratories. Inform Bio-Rad of the model
and serial number of your instrument.
IMPORTANT: This Bio-Rad instrument is designed and certified to meet EN61010* and the EMC
requirements of EN61326 (for Class A) safety standards. Certified products are safe to use when
operated in accordance with the instruction manual. This instrument should not be modified or altered
in any way. Alteration of this instrument will:
• Void the manufacturer's warranty
• Void the EN61010 safety certification
• Create a potential safety hazard
Bio-Rad Laboratories is not responsible for any injury or damage caused by the use of this instrument for
purposes other than those for which it is intended, or by modifications of the instrument not performed
by Bio-Rad Laboratories or an authorized agent.
*EN61010 is an internationally accepted electrical safety standard for laboratory instruments.
Model : Gene Pulser Xcell
Catalog No.: 165-xxxx
Date of Delivery:
Serial No.:
Invoice No.:
Purchase Order No.:
Page 3

Table of Contents
Section 1 Introduction and Safety Information:The Gene Pulser Xcell
System ........................................................................................................1
1.1 General Safety Information ............................................................................1
1.2 Electrical Hazards ........................................................................................2
1.3 Mechanical Hazards......................................................................................2
1.4 Other Safety Precautions ..............................................................................2
Section 2 Unpacking and System Installation ........................................................3
2.1 Unpacking the System Components ............................................................3
2.2 Setting up the System ..................................................................................4
2.2.1 Setting up the Gene Pulser Xcell Main Unit and Connecting
the ShockPod (Cat. #s 165-2660, 165-2661, 165-2662,
165-2666) ........................................................................................4
2.2.2 Connecting the PC Module to the Gene Pulser Xcell Main
Unit (Cat. #s 165-2660, 165-2662, and 165-2668)..........................5
2.2.3 Connecting the CE Module to the Gene Pulser Xcell Main
Unit (Cat. #s 165-2660, 165-2661, and 165-2667)..........................6
2.2.4 ShockPod (Cat. #s 165-2660, 165-2661, 165-2662, and
165-2669) ........................................................................................6
Section 3 Gene Pulser Xcell Operating Instructions ..............................................8
3.1 Section Overview ..........................................................................................8
3.2 Front Panel and Home Screen......................................................................9
3.2.1 Description of Keypad......................................................................9
3.2.2 Home Screen ................................................................................10
3.2.3 Help Screens ................................................................................11
3.3 Manual Operation........................................................................................12
3.3.1 Manual Operation (Guide Guide) ....................................................12
3.3.2 Electroporation using Exponential Decay Pulses ............................12
3.3.3 Electroporation Specifying Time Constant......................................14
3.3.4 Electroporation using Square Wave Pulses ....................................15
3.3.5 Results Screens ............................................................................17
3.3.6 Saving a Program from Manual Operation......................................19
3.3.6A Saving in a Location without a Named User Entry................20
3.3.6B Saving in a Location with a Named User Entry ....................20
3.4 Pre-Set Protocols........................................................................................21
3.4.1 Using a Pre-set Protocol (Quick Guide)..........................................21
3.4.2 Electroporation using a Pre-Set Protocol........................................22
3.4.3 Modifying Pre-Set Protocol Parameters ........................................25
3.4.4 Saving Changes to Pre-Set Protocols ............................................25
3.5 User Protocols ............................................................................................26
Page 4

3.5.1 Using a User Protocol (Quick Guide) ..............................................26
3.5.2 Creating a New User Name ..........................................................26
3.5.3 Creating a New User Protocol........................................................26
3.5.4 Modifying a User Protocol..............................................................30
3.5.5 Deleting a User Name and a User Protocol....................................31
3.5.6 Renaming a User Name or a User Protocol ..................................33
3.6 Last Pulse ..................................................................................................34
3.7 Optimize Operation ....................................................................................34
3.8 Data Management ......................................................................................36
3.9 Measurements ............................................................................................39
3.9.1 Sample Resistance Measurements ................................................39
3.9.2 Calibration and Measurement of Capacitors in the
CE Module..................................................................................................40
3.10 User Preferences ........................................................................................41
3.10.1 Setting the Clock ..........................................................................41
3.10.2 Adjusting the Screen Intensity ........................................................42
3.10.3 Sleep Function Setting ..................................................................42
3.11 The Pulse Trac System ..............................................................................43
3.11.1 Pulse Trac System Description ......................................................43
3.11.2 Pulse Trac Diagnostic Algorithm ....................................................44
Section 4 Overview of Electroporation Theory ......................................................44
4.1 Exponential Decay Pulses ..........................................................................45
4.2 Square Wave Pulses ..................................................................................45
Section 5 Factors Affecting Electroporation: Optimizing
Electroporation ........................................................................................48
5.1 Cell Growth ................................................................................................48
5.2 DNA............................................................................................................49
5.3 Electroporation Media ................................................................................49
5.4 Temperature................................................................................................50
Section 6 Electroporation of Bacterial Cells ..........................................................52
6.1 Escherichia coli ..........................................................................................52
6.1.1 Preparation of Electrocompetent Cells............................................52
6.1.2 Electroporation ..............................................................................53
6.1.3 Solutions and Reagents ................................................................53
6.2 Staphylococcus aureus ..............................................................................54
6.2.1 Preparation of Electrocompetent Cells............................................54
6.2.2 Electroporation ..............................................................................54
6.2.3 Solutions and Reagents ................................................................55
6.3 Agrobacterium tumefaciens ........................................................................56
6.3.1 Preparation of Electrocompetent Cells............................................56
6.3.2 Electroporation ..............................................................................56
Page 5

6.3.3 Solutions and Reagents ................................................................57
6.4 Bacillus cereus............................................................................................57
6.4.1 Preparation of Electrocompetent Cells............................................57
6.4.2 Electroporation ..............................................................................57
6.4.3 Solutions and Reagents ................................................................58
6.5 Pseudomonas aeruginosa ..........................................................................58
6.5.1 Preparation of Electrocompetent Cells............................................58
6.5.2 Electroporation ..............................................................................59
6.5.3 Solutions and Reagents ................................................................59
6.6 Streptococcus pyogenes ............................................................................60
6.6.1 Preparation of Electrocompetent Cells............................................60
6.6.2 Electroporation ..............................................................................60
6.6.3 Solutions and Reagents ................................................................61
6.7 Lactobacillus plantarum ..............................................................................61
6.7.1 Preparation of Electrocompetent Cells............................................61
6.7.2 Electroporation ..............................................................................61
6.7.3 Solutions and Reagents ................................................................62
Section 7 Electroporation of Fungal Cells..............................................................62
7.1 Saccaromyces cerevisiae............................................................................62
7.1.1 Preparation of Electrocompetent Cells............................................62
7.1.2 Electroporation ..............................................................................63
7.1.3 Solutions and Reagents ................................................................64
7.2 Schizosaccharomyces pombe ....................................................................64
7.2.1 Preparation of Electrocompetent Cells............................................64
7.2.2 Electroporation ..............................................................................65
7.2.3 Solutions and Reagents ................................................................65
7.3 Pichia pastoris ............................................................................................65
7.3.1 Preparation of Electrocompetent Cells............................................65
7.3.2 Electroporation ..............................................................................66
7.3.3 Solutions and Reagents ................................................................66
7.4 Candida albicans ........................................................................................67
7.4.1 Preparation of Electrocompetent Cells............................................67
7.4.2 Electroporation ..............................................................................67
7.4.3 Solutions and Reagents ................................................................68
7.5 Dictyostelium discoideum............................................................................68
7.5.1 Preparation of Electrocompetent Cells............................................68
7.5.2 Electroporation ..............................................................................69
7.5.3 Solutions and Reagents ................................................................69
Section 8 Mammalian Cells......................................................................................70
8.1 Preparation of Electrocompetent Cells ........................................................70
Page 6

8.1.1 Attached Cells................................................................................70
8.1.2 Suspension Cells............................................................................70
8.2 Electroporation............................................................................................70
8.3 Solutions and Reagents ..............................................................................71
Section 9 References................................................................................................72
Section 10 Specifications and Product Information................................................75
10.1 System Specifications ................................................................................75
10.2 Product Information ....................................................................................76
Page 7

Section 1
The Gene Pulser Xcell
™
Electroporation System
The Gene Pulser Xcell is a pulse generator that uses capacitors to produce controlled exponential or square
wave electrical pulses for cell electroporation. The unit is capable of producing pulses of up to 3000 V on a
high-voltage circuit, and up to 500 V on a low-voltage circuit. For generating pulses on the high voltage
circuit, capacitors of 10, 15, and 25 µF present in the Gene Pulser Xcell main unit are used and generating
pulses on the low-voltage circuit requires use of capacitors in the CE Module. Exponential decay (or
capacitance discharge) and square wave pulses are the most commonly used types of electrical pulse. An
in-depth discussion of these two waveforms can be found in Section 4.
The Gene Pulser Xcell is a modular system, comprising of a main unit and two accessory modules, the CE
module and the PC module, in addition to the shocking chamber and a cuvette with incorporated electrodes.
The CE Module is recommended for use with the Gene Pulser Xcell main unit for electroporation of most
eukaryotic cells, including mammalian cells and plant protoplasts. The CE Module should only be used
with low-resistance media (<1000 ohms). For exponential decay pulses, the CE Module provides a
means of controlling the capacitance of the circuit by increasing the time constant of the pulse. For
square wave pulses, the CE Module provides the large capacitor necessary for delivering a square wave
pulse into low resistance media. This module contains a set of capacitors with a functional range
between 50 and 3275 µF and selectable in 25 µF increments. For square wave pulses, the CE Module
provides the large capacitance, 3275 µF, necessary for delivering a square wave pulse into low
resistance media.
The PC Module is recommended for the electroporation of bacteria and fungi using exponential decay, as
well as in other applications where high-voltage pulses are applied to samples of small volume and high
resistance. The PC Module selects resistance of 50 ohms 1000 ohms in 50-ohm increments. The unit is
used to control the resistance of the circuit by placing resistors in parallel with the sample, thereby providing a means of reducing the time constant of an exponential decay pulse. This provides an effective
means of controlling the time constant when using high-resistance media but has little effect on the time
constant when using low-resistance media. The PC Module greatly reduces the likelihood of an arc
occurring at high voltage. It is not recommended that the PC module be used for square wave pulses due
to the increase in droop of the pulse that can occur (see Section 4).
Both the PC Module and CE Module have integral leads that connect to the main unit (see Section 2
for installation) and both units are controlled directly from the user interface on the front panel of the
main unit.
1.1 General Safety Information
This Bio-Rad instrument is designed and certified to meet the safety requirements of EN61010 and the
EMC requirements of EN61326 (for Class A) and conforms to the “Class A” standards for electromagnetic
emissions intended for laboratory equipment applications. This instrument is intended for laboratory
application only. It is possible that emissions from this product may interfere with some sensitive appliances
when placed nearby or in the same circuit as those appliances. The user should be aware of this potential
and take appropriate measures to avoid interference.
No part of the Gene Pulser Xcell system should be used if obvious external case damage has occurred
or the electronic displays are not functioning as described in the manual. This instrument is only to be
used with the components provided (or their authorized additions or replacements) including, but not
limited to, supplied cables and ShockPod. The operating temperature range for the Gene Pulser Xcell
system and its associated components is 0–35°C.
1
Page 8

There are no user serviceable parts within the unit. The operator should make no attempt to open any
case cover or defeat any safety interlock. This instrument must not be altered or modified in any way.
Alteration of this instrument will
• Void the manufacturer’s warranty
• Void the IEC 1010 safety certification
• Create a potential safety hazard
Bio-Rad is not responsible for any injury or damage caused by the use of this instrument for purposes other
than those for which it is intended or by modification of the instrument not performed by Bio-Rad or an
authorized agent.
1.2 Electrical Hazards
The Gene Pulser Xcell produces voltages up to 3,000 volts and is capable of passing very high currents.
When charged to maximum voltage, the instrument stores about 400 joules. A certain degree of respect
is required for energy levels of this order. System safety features prevent operator access to the recessed
input jacks and to the recessed electrode contacts inside the sample chamber. These mechanical
interlocks should never be circumvented.
The pulse button is active whenever the character space in the lower right corner is flashing. There is
high voltage present whenever the pulse button is depressed and “Pulsing” is shown on the LCD
display on the front of the instrument. Because of the built-in safety interlock in the ShockPod, no pulse
is delivered to the cuvette when the ShockPod lid is opened. If the capacitor has been partially charged
but not fired (for example, when the charging cycle has been interrupted before the pulse is delivered),
some charge may remain on the internal capacitor. This charge will dissipate over 1–2 minutes.
However, the user cannot make contact with any charged electrical components due to the system
safety features.
1.3 Mechanical Hazards
The Gene Pulser Xcell contains a patented arc-protection circuit that dramatically reduces the incidence of
arcing in the cuvette when high voltage is delivered into the sample. The unit incorporates a circuit that
senses the beginning of an arc and diverts current from the sample within <10 µsec, preventing, or
greatly reducing, mechanical, visual, and auditory phenomena at the ShockPod. Should an arc occur,
the sample chamber is effective in containing these small discharges, but nonetheless we strongly
recommend wearing safety glasses when using the instrument.
1.4 Other Safety Precautions
Avoid spilling any liquids onto the apparatus. Use only a paper towel or a cloth wet with either water or
alcohol to clean the outside surfaces of the Gene Pulser Xcell.
Use only the Bio-Rad cables supplied with the Gene Pulser Xcell.
Use the ShockPod only in the assembled condition. Do not attempt to circumvent the protection of the
ShockPod or use it while disassembled.
Verify the display segments periodically.
Read the instruction manual before using the Gene Pulser Xcell Electroporation System. For technical
assistance contact your local Bio-Rad office or, in the US, call technical services at 1-800-4BIORAD
(1-800-424-6723).
2
Page 9

Warning: The Gene Pulser Xcell generates, uses, and radiates radio frequency energy. If it is not used in
accordance with the instructions given in this manual, it may cause interference with radio communications.
The Gene Pulser Xcell has been tested and found to comply with the limits for Class A computing devices
(pursuant to Subpart J of Part 15 of FCC Rules) which provide reasonable protection against such
interference when operated in a commercial environment. Operation of this equipment in a residential area
is likely to cause interference. In this case the user will be required, at their expense, to take whatever
measure may be required to correct the interference.
Section 2
Unpacking and System Installation
The Gene Pulser XCell™ can be purchased as three systems as well as component parts:
165-2660 Gene Pulser Xcell Total System for eukaryotic and microbial cells, 100–240 V, 50/60
Hz, exponential decay and square wave delivery, includes main unit, CE Module, PC
Module, ShockPod, 15 sterile cuvettes (5 each of 0.1, 0.2, and 0.4 cm gap),
instruction manual
165-2661 Gene Pulser Xcell Eukaryotic System, 100/240 V, 50/60 Hz, exponential decay
(25–3,275 µF range) and square wave delivery, includes main unit, CE Module,
ShockPod, 5 sterile cuvettes (0.4 cm gap), instruction manual
165-2662 Gene Pulser Xcell Microbial System, 100/240 V, 50/60 Hz, exponential decay and
square wave delivery, includes main unit, PC Module, ShockPod, 10 sterile cuvettes
(5 each of 0.1 and 0.2 cm gap), instruction manual
165-2666 Gene Pulser Xcell main unit, 100/240 V, 50/60 Hz
165-2667 Gene Pulser Xcell CE Module, 25–3,275 µF range controlled by main unit, includes
integral leads, 5 sterile cuvettes (0.4 cm gap), instruction manual
165-2668 Gene Pulser Xcell PC Module, 50–1,000 ohm range controlled by main unit, includes
integral leads, 10 sterile cuvettes (5 each of 0.1 and 0.2 cm gap)
165-2669 Gene Pulser Xcell ShockPod shocking chamber, includes integral leads for connection
to Gene Pulser Xcell, Gene Pulser II, or MicroPulser
2.1 Unpacking the System Components
Remove all packing material and connect components on a flat, dry surface near an appropriate
electrical outlet.
Upon receiving your instrument, please check that all items listed were shipped. If any items are missing
or damaged, contact your local Bio-Rad office.
3
Page 10
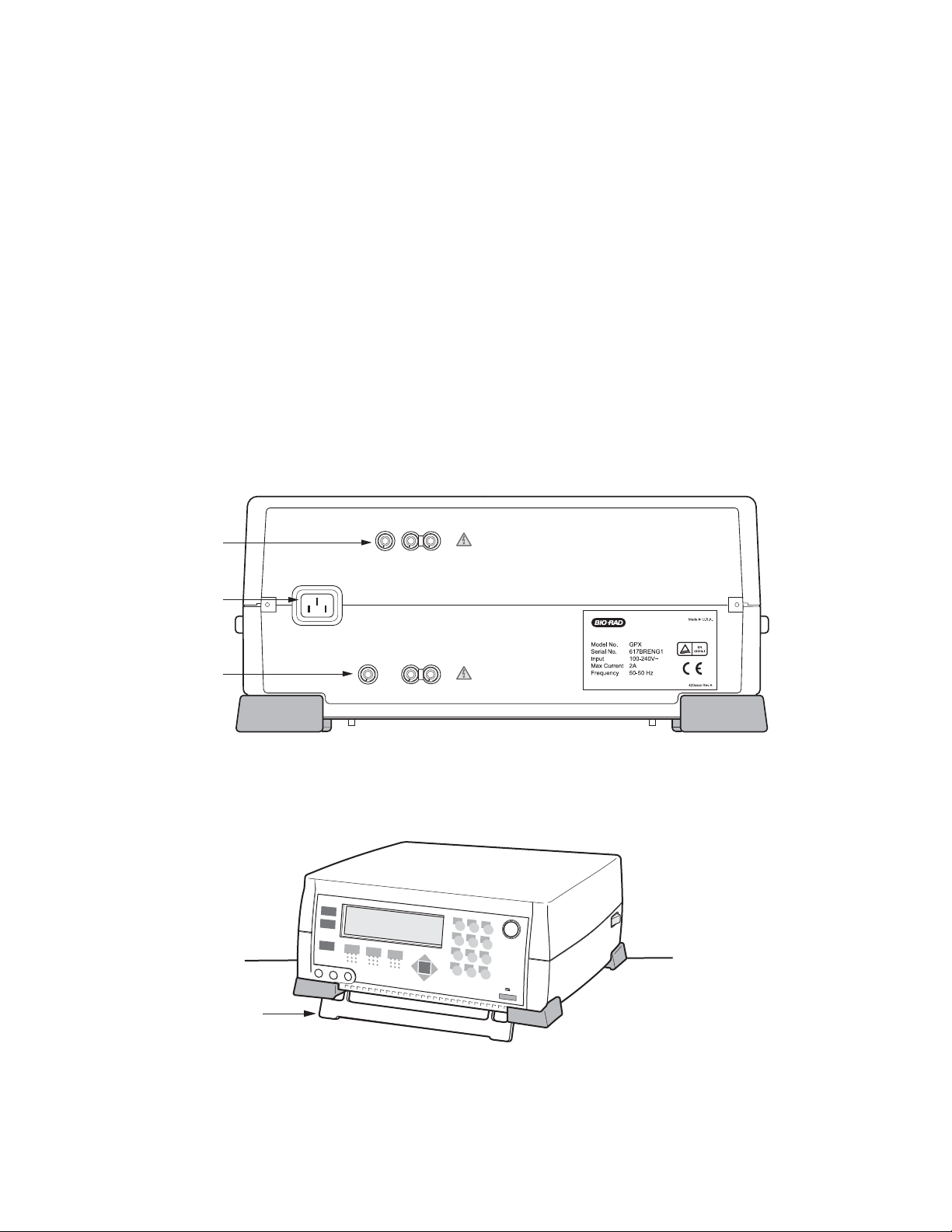
2.2 Setting Up the System
2.2.1 Setting Up the Gene Pulser Xcell Main Unit and Connecting the ShockPod
(Cat. #s 165-2660, 165-2661, 165-2662, and 165-2666)
Follow this procedure for setting up the Gene Pulser Xcell and connecting the ShockPod. Be sure to
turn off the Gene Pulser Xcell main unit before connecting or disconnecting the power cord or any
cables.
1. Attach the power cord to the three-pronged receptacle on the back of the Gene Pulser Xcell unit
(Figure 2.1A). Plug the unit into an appropriate electrical outlet or power strip. There is a foot on the
bottom of the Gene Pulser Xcell which may be pulled down to change the viewing angle of the
LCD display (Figure 2.1B).
2. Insert the black connector of the ShockPod into the left and center output jacks on the front panel
of Gene Pulser Xcell (Figure 2.2); polarity is not important to the electroporation process. The right
output jack is not used with the ShockPod.
3. To turn on the Gene Pulser Xcell, press the power switch on the right rear of the instrument
(Figure 2.2).
Fig. 2.1A. Rear panel of Gene Pulser Xcell main unit showing receptacles for the power cord,
CE Module, and PC Module.
Fig. 2.1B. Bottom panel of the Gene Pulser Xcell main unit. The foot may be pulled down to raise
the front of the unit.
4
Connectors
to CE Module
CE MODULE
Power Connector
POWER INLET
PC MODULE
Connectors
to PC Module
1
2
3
4
5
6
7
8
9
0
Tilt-up Foot
Page 11

Fig. 2.2. Connecting the ShockPod to the Gene Pulser Xcell main unit.
2.2.2 Connecting the PC Module to the Gene Pulser Xcell Main Unit (Cat. #s 165-2660,
165-2662, and 165-2668)
The PC Module and CE Module may be connected to the Gene Pulser Xcell main unit in any order and
both may be connected at the same time. Before connecting the PC Module to the Gene Pulser Xcell,
be sure that the Gene Pulser Xcell is turned off. There is no power switch on the PC Module; it is
controlled entirely by the Gene Pulser Xcell main unit.
1. Place the PC Module near the Gene Pulser Xcell main unit. The units, along with the CE Module,
are stackable; either the PC Module or the Gene Pulser Xcell main unit can be placed on the bottom.
Place the feet of the top unit at the corners of the bottom unit to interlock the two.
2. Insert the red/black connector that is permanently attached to the cable on the back of the PC
Module into the appropriately labeled receptacle on the back of the Gene Pulser Xcell main unit
(see Figure 2.3). The connector is keyed so that it will insert only into the proper receptacle and in
the correct red/black orientation.
Fig. 2.3. Rear view (back panel) of PC Module showing the connection to the Gene Pulser Xcell
main unit.
Power Switch
5
ShockPod
1
2
3
4
5
6
7
8
9
0
Connectors
Xcell ShockPod
Xcell PC Module
Foot
CE MODULE
Xcell Main Unit
Connections to
PC Module
Foot
POWER INLET
PC MODULE
Page 12
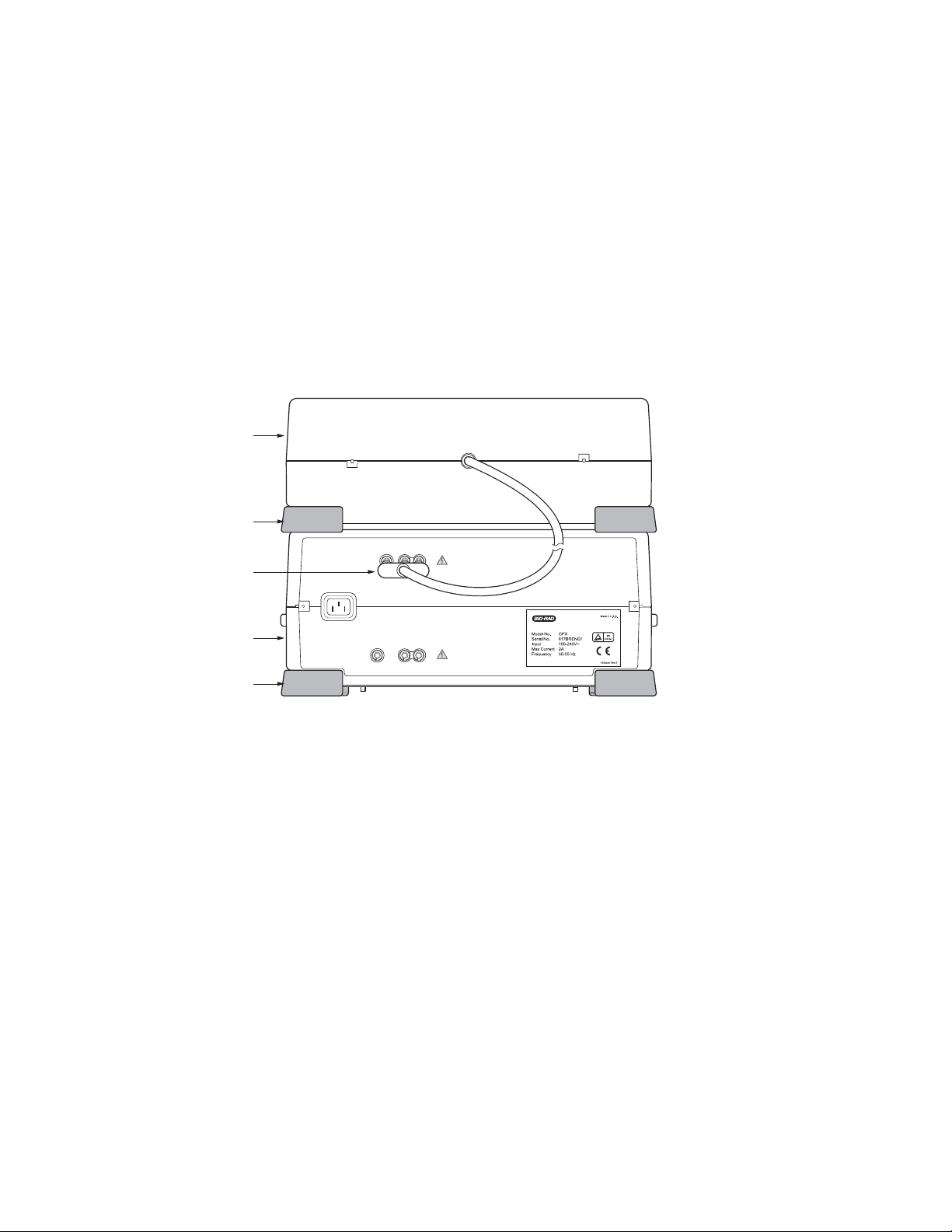
2.2.3 Connecting the CE Module to the Gene Pulser Xcell Unit (Cat. #s 165-2660,
165-2661, and 165-2667)
The PC Module and CE Module may be connected to the Gene Pulser Xcell main unit in any order and
both may be connected at the same time. Before connecting the CE Module to the Gene Pulser Xcell, be
sure that the Gene Pulser Xcell is turned off. There is no power switch on the CE Module; it is controlled
entirely by the Gene Pulser Xcell unit.
1. Place the CE Module near the Gene Pulser Xcell main unit. These units, along with the PC Module
are stackable in any order. Place the feet of the top unit at the corners of the bottom unit to interlock
the two.
2. Insert the red/black connector that is permanently attached to the cable on the back of the CE
Module (Figure 2.4) into the appropriately labeled receptacle on the back of the Gene Pulser Xcell
main unit (see Figure 2.1). The connector is keyed so that it will insert only into the proper receptacle
in the correct red/black orientation.
Fig. 2.4. Rear view (back panel) of the CE Module showing the connection to the Gene Pulser Xcell
main unit.
2.2.4 ShockPod (Cat. #s 165-2660, 165-2661, 165-2662, and 165-2669)
The ShockPod is designed for one-hand operation. Press the tab on the front of the chamber to
release the latch and to open the top. Push the top down gently to close the chamber. The safety
design of the system requires that the top be closed before pulsing can occur. Additionally, the top
must also be closed to use the ShockPod to measure resistance of electroporation media (see Section
3.9.1). Because of the built-in safety interlock in the ShockPod, no pulse is delivered to the cuvette
when the ShockPod lid is opened.
The cuvette slot will accept any commercially available cuvette. Bio-Rad cuvettes are designed with a
notch on one of the non-electrode sides so they may be inserted into the cuvette slot only in the
proper orientation.
6
Xcell CE Module
Foot
Connections to
CE Module
Xcell Main Unit
Foot
POWER INLET
CE MODULE
PC MODULE
Page 13
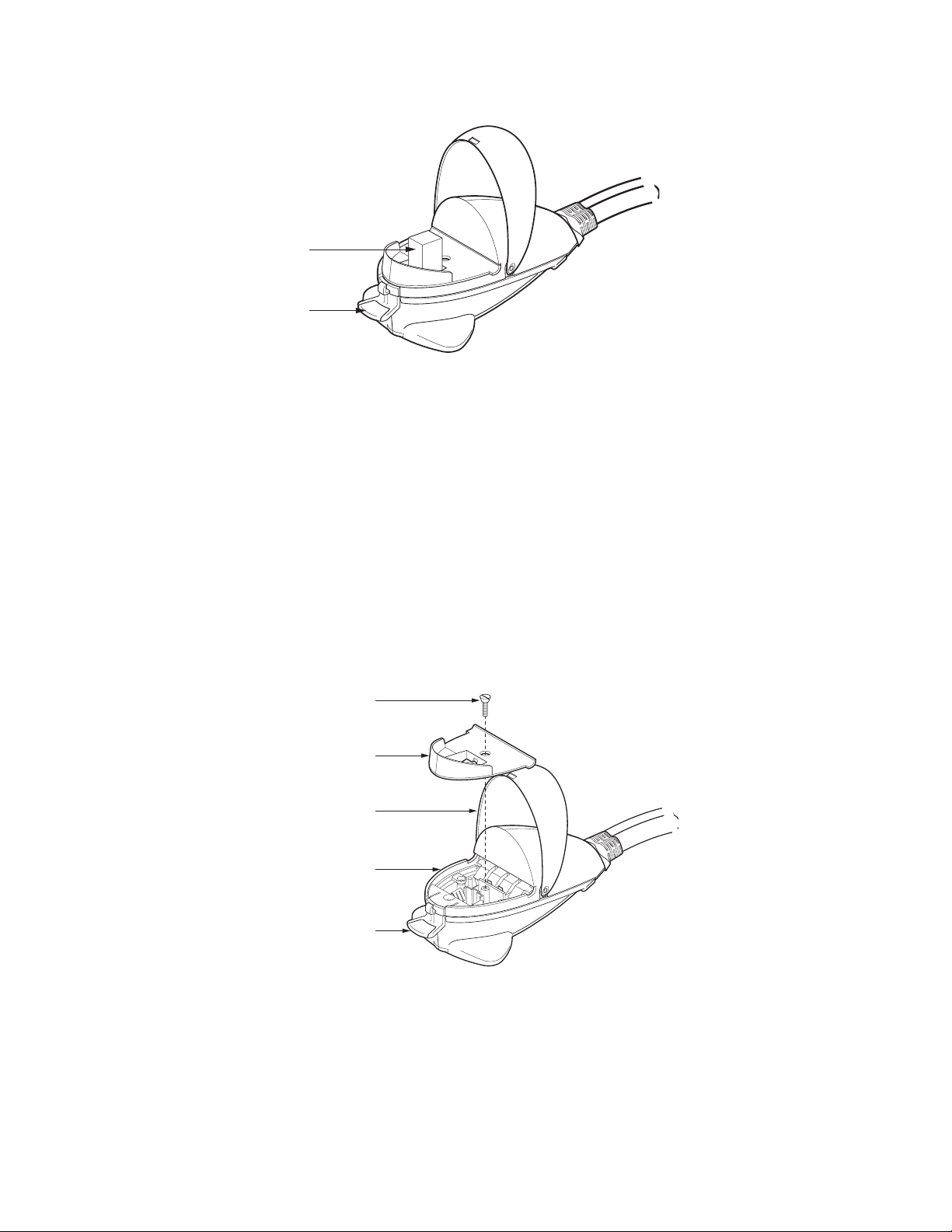
Fig. 2.5. ShockPod with cuvette.
The plastic panel covering the electrode may be removed for cleaning should the electrode become
dirty. To clean the unit:
1. Disconnect the ShockPod from the Gene Pulser Xcell main unit.
2. Press the tab to open the chamber.
3. Using a phillips screwdriver, loosen the screw holding the safety panel (Figure 2.5). Lift the screw
and the safety panel out of the ShockPod by the lip on the front of the panel.
4. The area around the electrodes may be cleaned with a cotton swab and warm, soapy water. Dry
the electrodes using a dry cotton swab or Kimwipe.
5. Reassemble the unit by centering the safety panel on the base of the ShockPod and tightening the
screw.
Fig. 2.6. Exploded view of the Gene Pulser Xcell ShockPod.
Cuvette
7
Press to Open
Screw
Safety Panel
To p
Base Unit
Ta b
Page 14

Section 3
Gene Pulser Xcell
™
Operating Instructions
3.1 Section Overview
This section describes the operation of the Gene Pulser Xcell. The following summarizes the organization
of this section.
Section 3.2 below describes the functions of the keys on the front panel, the Home screen on the LCD
display, and the Help functions built into the Gene Pulser Xcell.
• The keys on the front panel of the main unit control the Gene Pulser Xcell. Section 3.2.1 describes
the uses of these keys.
• The Home screen provides easy access to programs built into the Gene Pulser Xcell as well as a
direct method of manually entering pulse parameters to electroporate a sample. Section 3.2.2
describes these programs.
• On-screen help is built into the software of the Gene Pulser Xcell. This may be accessed from any
screen as described in Section 3.2.3.
The Gene Pulser Xcell has three modes of operation: manual operation, pre-set protocols, and user protocols.
Section 3.3 describes the Manual mode, which may be used to rapidly program the parameters necessary
for delivering either an exponential decay or a square wave pulse.
• Section 3.3.2 describes delivering an exponential decay pulse.
• Section 3.3.3 describes delivering an exponential decay pulse but specifying a time constant
rather than a capacitance and resistance value.
• Section 3.3.4 describes delivering a square wave pulse.
• Section 3.3.6 explains how programmed settings may be saved as user protocols.
Section 3.4 describes the Pre-set protocols in which the pulse parameters have been optimized for a
number of commonly used bacterial and fungal species and mammalian cell lines.
• Pre-set Protocols may be called up and used directly (Section 3.4.2) or may be modified prior
to being used (Section 3.4.3).
• A modified Pre-set Protocol may be saved as a User Protocol (Section 3.4.4).
Section 3.5 describes a custom mode (User Protocols) in which users may store optimized pulse
parameters that they use in their own work.
• User Protocols may be created in any of four ways:
• In the User Protocols menu as a new protocol (Section 3.5.3).
• In the User Protocols menu as an edited (modified) program (Section 3.5.4)
• In the Manual menu as a new protocol (Section 3.3.4).
• In the Pre-set Protocol Menu as a modified protocol (Section 3.4.4).
• User Protocols, once created and saved, may be called up and used directly like Pre-set
Protocols (Section 3.5.1).
8
Page 15

3.2 Front Panel and Home Screen
3.2.1 Description of the keypad
See Figure 3.1 for a view of the Gene Pulser Xcell front panel.
Alpha-numeric keys This array of keys permits entering numbers and letters into the Gene Pulser
Xcell. Pressing the Shift key toggles between alphabetic and numeric input. To
type an alphabetic character, press the Shift key to enter alpha mode, then
press the key with the appropriate letter. To type an a, press the 2 key once; to
type a b, press the 2 key twice; to type a c, press the 2 key three times. To use
the same key twice, for example to type a then b, advance the cursor using
the Right Arrow Key. The firmware on Gene Pulser Xcell will automatically
change between alpha and numeric input depending on the parameter being
entered. In Protocol screens and Directory screens where a two-digit entry
must be made, the second digit must be entered within 2 seconds of the first
entry, otherwise the screen will default to the single-digit entry.
Home key Returns the user to the Home screen from anywhere in the program.
Back key Returns the user one level back in hierarchy toward the Home screen.
Help key Displays on-screen help text.
Save key Saves User Names and User Protocols.
Delete key Removes only the last entry in the field; also used to remove User Name and
User Protocol files.
Clear key Removes the entire line of the field.
Enter key Indicates that a choice has been made and moves the cursor to the next
location.
Arrow keys The Up and Down Arrow keys move the cursor up or down one row at a time.
Depending on the screen and location of the cursor, the Right and Left Arrows
may (1) move the cursor right or left one space at a time, (2) toggle forward
and backward one screen when there are multiple screens for the same
menu, or (3) increase or decrease numerical input values.
Pulse button: Results in discharging a pulse. During this time “Pulsing” is shown on the LCD
display. A tone sounds to indicate that the pulse has been delivered. When
multiple pulses are delivered, a tone sounds after the last pulse has been
delivered. The Pulse is discharged to the electrodes if the ShockPod is
connected and the lid is closed. Otherwise, it is discharged safely within the
instrument.
9
Page 16
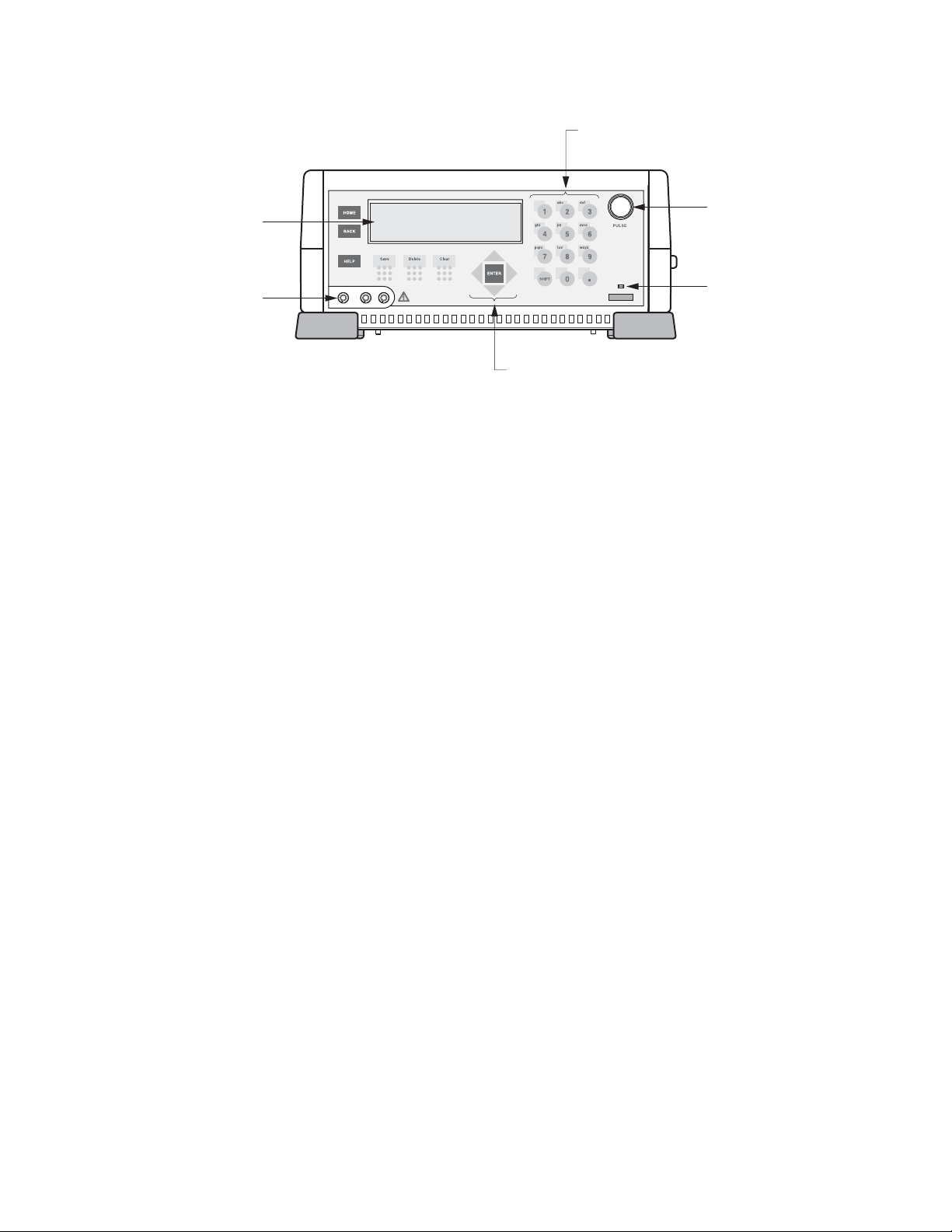
Fig. 3-1. Gene Pulser Xcell front panel. See Section 3.2.1 for an explanation of the key functions.
3.2.2 Home Screen
Upon turning on the power to Gene Pulser Xcell, the unit will run a series of test algorithms, including a
check of the Pulse Trac system (see Section 3.11) and the firmware. During this time the LCD will display the Bio-Rad logo, the product name, and the version of firmware. After this instrument initialization,
the Home Screen is displayed (Figure 3.2). From this screen it is easy to access the programs
described below. You may return to this screen from anywhere in the program by pressing the Home key
on the keypad.
Home consists of two screens. The first contains the most commonly used programs. The second
contains advanced functions. The Left and Right Arrow keys can be used to toggle between them.
To select a program, when the Home screen is displayed on the LCD screen, press numbers 1–10 on
the Alpha-Numeric keypad followed by the Enter key. Alternatively, use the Up and Down Arrow keys to
scroll to the desired selection; press Enter to select the program.
1. Exponential protocol: for delivery of an exponential decay pulse: enables the user to
program a set of conditions and to deliver the pulse
2. Time constant protocol: for delivery of an exponential decay pulse by specifying a time
constant: enables the user to program a set of conditions and to
deliver the pulse
3. Square wave protocol: for delivery of a square wave pulse: enables the user to program
a set of conditions and to deliver the pulse
4. Pre-set protocols: pre-programmed optimized settings for electrotransformation of
commonly used bacterial, fungal, and mammalian cells
5. User protocols: a program which allows the user to store up to 144 protocols
including all instrument settings and recall these protocols to
deliver a pulse
6. Last pulse: allows the user to recall the electroporation parameters for the
last pulse and to deliver a pulse using the same conditions
7. Optimize: allows the user to program electroporation parameters and an
increment for voltage that may be changed after each pulse in order
to optimize electroporation conditions
LCD Screen
10
Keypad
Pulse
Button
Connectors to
Shocking Chamber
Arrow and
Enter Keys
Power LED
Indicator
Page 17
8. Data management: allows the user to access pulse parameters and results for the
last 100 pulses logged by date and time
9. Measurements: allows the user to measure capacitor values in the Gene Pulser
Xcell or CE Module and to measure sample resistance
10. User Preferences: allows the user to adjust the clock, the screen light intensity, and
the LCD sleep function
Right and left arrows (> and <) appear at the bottom of the first screen and at the top of the second
screen, respectively. These arrows indicate that the Right and Left Arrow keys may be used to toggle
between these related screens.
Home Screen
Fig. 3.2. Gene Pulser Xcell Home screens. All menu functions may be readily accessed from
these screens. Return to the Home screen from anywhere in the program by pressing the Home
key. Press the Left and Right Arrow keys to toggle between the two screens.
3.2.3 Help Screens
Help screens are accessible from all screens by pressing the Help key. Each help menu describes the
keystrokes that may be entered to choose the next function or to continue the current operation. Where
there is more than one help screen, use the Right and Left Arrow keys to scroll between them. To
return to the program screen, press the Back key.
11
HOME
<6. Last pulse
7. Optimize
8. Data management
9. Measurements
10. User preferences
HOME
1. Exponential protocol
2. Time constant protocol
3. Square wave protocol
4. Pre-set protocols
>5. User protocols
Page 18

3.3 Manual Operation
3.3.1 Manual Operation (Quick Guide)
• From the Home screen:
• Press Enter to select exponential decay;
• Press 2, then Enter to select exponential decay but specifying a time constant;
• Press 3, then Enter to select square wave.
• Use the Up and Down Arrow keys to scroll through the parameter value spaces on the screen.
When a parameter value is highlighted, use the keypad to enter a value, then press Enter to accept
that value.
• When the necessary parameter values have been entered, the Pulse button on the Gene Pulser
Xcell is active.
• Press the Pulse button to electroporate the sample.
• Press the Back key to return to the Protocol Detail screen and to deliver another pulse.
3.3.2 Electroporation using Exponential Decay Pulses
See Section 4.1 for a discussion of electroporation using exponential decay pulses.
• When the Home screen (Figure 3.2) is selected, the number 1, corresponding to “Exponential
protocol” is highlighted as the default choice. Press Enter to view the Protocol Detail Screen. If the
number 1 on the Home screen is not highlighted, press 1 or use the Up or Down Arrow keys to
highlight “Exponential protocol”, then press Enter to select. The Protocol Detail screen appears
(Figure 3.3).
• The following combination of parameters may be entered:
Capacitance + Voltage
Capacitance + Voltage + Resistance
The three variables may be selected in any order, however, the set voltage will determine whether the
high voltage or the low voltage circuit is to be used and will limit the range of the capacitance as
indicated in Table 3.1. If a value for the capacitance is chosen that outside the range of the system, this
value will default to the closest allowable value.
Specifying a resistance value requires that the PC Module be attached. This is always recommended with
high resistance media (i.e., >600 ohm) such as water, sucrose, glycerol, sorbitol, or polyethylene glycol. The
PC Module places a resistor in parallel with the sample to reduce the resistance of the circuit. In this way,
the time constant of a high-resistance sample may be reduced and controlled.
12
Page 19

When the Resistance value indicates infinity, no resistors from the PC Module are used. The Measurements
function of Gene Pulser Xcell may be used to determine sample resistance (see Section 3.9).
Exponential Decay Protocol Detail Screen
Fig. 3.3. Exponential Decay Protocol Detail Screen. This screen shows the parameters that may
be specified for exponential decay electroporation. Entering the cuvette size is optional and is for
user information only.
Table 3.1. Capacitance and voltage ranges for the high voltage / low capacitance
circuit and the low voltage / high capacitance circuit of the Gene Pulser Xcell for
delivering an exponential decay pulse.
Capacitance (uF) Voltage (V)
R
sample
< 600 R
sample
> 600
HV Circuit 10 – 50 200 – 2500 200 – 3000
LV Circuit 25 – 3275 10 – 500 10 – 500
*Use of the low voltage circuit requires that the CE Module be in place.
• Use the Up and Down Arrow keys to scroll through and highlight the parameter value spaces for
capacitance, voltage, or resistance. Use the alpha-numeric keypad to input the desired value, or
use the Right and Left Arrow keys to increase or decrease the parameter value by the smallest
allowable increment. Use the Delete or Clear keys to change the value. When the desired value has
been specified, press Enter. If a value outside the limits of the Gene Pulser Xcell is selected, the
value in the field will default to the closest permitted value. Entering a value for cuvette size is
optional and is for user information only.
• When the necessary parameter values have been specified, a flashing “P” appears in the character
space in the lower right corner of the LCD display indicating that the pulse button on the Gene
Pulser Xcell is active and that a pulse may be delivered.
• Press the Pulse button to deliver a pulse. When the Pulse button is depressed, the LCD display
will blank then show “Pulsing”. Upon completion, a tone will sound and the pulse measurements
will be displayed on the Protocol Results screen (see Figure 3.6, Section 3.3.5).
• Use the Left and Right Arrow keys to toggle between the Protocol Results screen and the last
Protocol Detail screen.
13
PROTOCOL DETAIL: EXPONENTIAL
Voltage (V) XXXX
Capacitance (uF) XXXX
Resistance (ohm) XXXX
Cuvette (mm) X
Page 20

• With the Protocol Detail screen on the LCD display another pulse can be delivered using the same
pulse parameters. To change the pulse conditions, press Enter; the cursor appears in the voltage
parameter value. The parameters may be changed as described above. To save the pulse parameters,
see Section 3.3.6.
• To review previously delivered pulses, see Section 3.8.
3.3.3 Electroporation Specifying Time Constant
See Section 4.1 for a discussion of electroporation using exponential decay. Use the Gene Pulser Xcell
to deliver an exponential decay pulse with a specific time constant as follows.
• From the Home screen (Figure 3.2), press 2 or use the Up or Down Arrow keys to highlight “Time
constant protocol”, then press Enter. The Time Constant Protocol Detail screen appears
(Figure 3.4).
• A sample must be in place to use Gene Pulser Xcell in this mode. The following parameters must
be entered:
Time constant + Voltage
In time constant mode, Gene Pulser Xcell will maximize the parallel resistance (and minimize the capacitance)
in order to maximize the energy delivered to the sample. Therefore, the PC Module is required to deliver pulses
using the high voltage circuit, while both the CE Module and PC Module are needed to deliver pulses using
the low voltage circuit. The set voltage will determine whether the high voltage or the low voltage circuit is to
be used and will limit the range of the capacitance as indicated in Table 3.2. If a voltage is chosen that spans
both the low voltage and high voltage range, the Gene Pulser Xcell will preferentially use the low voltage circuit.
As indicated in Table 3.2, the acceptable values for the time constant are a function of the sample resistance
and the voltage circuit used.
Immediately before delivering a pulse, Gene Pulser Xcell will determine sample resistance to determine
whether a pulse can be delivered using the set parameters of voltage and time constant. If the set time
constant is outside the range by more than 20%, the Gene Pulser Xcell will show a message indicating
the approximate time constant that can be delivered, and ask that you press the pulse button again to
deliver the pulse. To estimate sample resistance using Gene Pulser Xcell, see Section 3.9.
Table 3.2. Ranges of time constants that may be achieved using Gene Pulser Xcell
at several sample resistances.
Sample resistance Time constant range (msec)
L
V Cir
cuit* HV Circuit**
20 ohms 0.5 – 65.5 0.2 – 1.0
200 ohms 5.0 – 655 2.0 – 10
>1000 ohms 25 – 3275 10 – 50
* LV Circuit range: 10 – 500 V
** HV Circuit range: 200 – 3000 V
14
Page 21
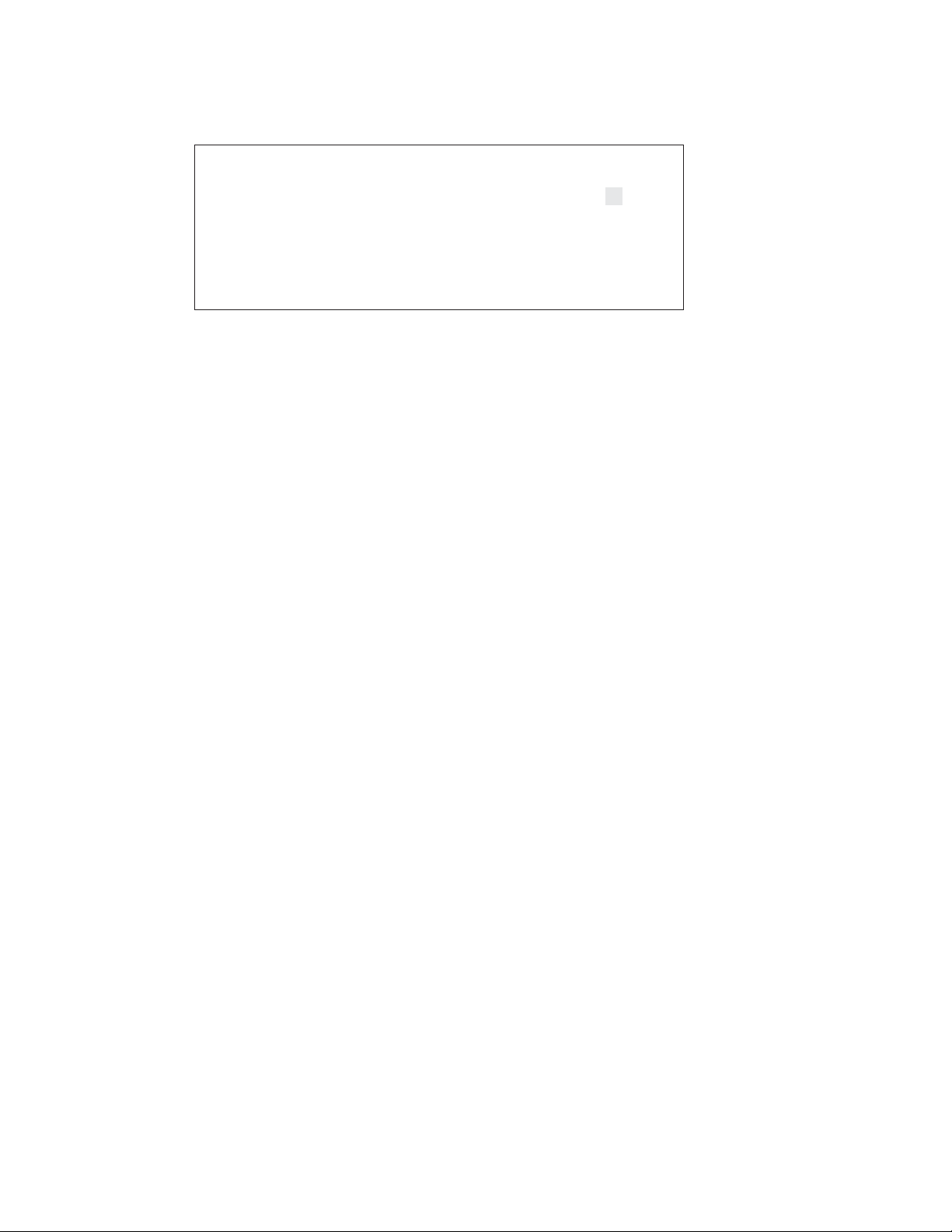
Time Constant Protocol Detail screen
Fig. 3.4. Time Constant Protocol Detail Screen. This screen shows the parameters that may be
specified for exponential decay electroporation when the time constant is designated. Entering
the cuvette size is optional and is for user information only.
• Use the Up and Down Arrow keys to highlight the parameter value space for voltage or time
constant. Use the alpha-numeric keypad to input the desired value, or use the Right and Left Arrow
keys to increase or decrease the parameter value by the smallest allowable increment. Use the
Delete or Clear keys to correct entries. When the desired value has been specified, press Enter. If a
value outside the limits of the Gene Pulser Xcell is selected, the value in the field will default to the
closest permitted value. Entering a value for cuvette size is optional and is for user information only.
• When the necessary parameter values have been specified, a flashing “P” appears in the character
space in the lower right corner of the LCD display indicating that the pulse button on the Gene
Pulser Xcell is active and that a pulse may be delivered.
• Press the Pulse button to deliver a pulse. When the Pulse button is depressed, the LCD display will
blank then show “Pulsing”. Upon completion, a tone will sound and the pulse measurements will be
displayed on the Protocol Results screen (see Figure 3.7, Section 3.3.5).
• Use the Left and Right Arrow keys to toggle between the Protocol Results screen and the last
Protocol Detail screen.
• With the Protocol Detail screen on the LCD display another pulse can be delivered using the same
pulse parameters. To change the pulse conditions, press Enter; the cursor appears in the voltage
parameter value. The parameters may be changed as described above.
• To save the pulse parameters, see Section 3.3.6.
• To review previously delivered pulses, see Section 3.8.
3.3.4 Electroporation using Square Wave Pulses
See Section 4.2 for a discussion of electroporation using square wave pulses. Use Gene Pulser Xcell to
deliver square wave pulses as follows.
• From the Home screen (Figure 3.2), press 3 or use the Up or Down Arrow keys to highlight
“Square wave protocol”, then press Enter. The Square Wave Protocol Detail screen appears
(Figure 3.5).
15
PROTOCOL DETAIL: TIME CONSTANT
Voltage (V) XXXX
Time constant (msec) XXXX.X
Cuvette (mm) X
Page 22

• The following combination of parameters may be entered:
Pulse length + Voltage
Pulse length + Voltage + Number of pulses + Pulse interval
Square Wave Protocol Detail screen
Fig. 3.5. Square Wave Protocol Detail Screen. This screen shows the parameters that may be specified for square wave electroporation. Entering the cuvette size is optional and is for user information only.
Electroporation using square waves may be used to deliver a series of multiple pulses to the cells. This
is specified by the parameter “Number of pulses.” The time between each pulse is specified as the
“Pulse interval.” The default values for the number of pulses and pulse interval are 1 and 0, respectively;
it is only necessary to enter other values if more than one pulse is to be delivered. See Section 4.2 for a
discussion of pulse parameters.
The pulse parameters may be selected in any order. However, the set voltage will determine whether
the high voltage or the low voltage circuit is to be used. Consequently, the set voltage will limit the
ranges of the pulse length and pulse interval as indicated in Table 3.3. If values for the second and third
parameters are chosen that are outside the range of the system, the values will default to the closest
allowable values.
Table 3.3. Pulse length and pulse interval ranges for the high voltage / low
capacitance circuit and the low voltage / high capacitance circuit of the Gene
Pulser Xcell for delivering a square wave pulse.
Voltage (V) Pulse Length Pulse Interval No. Pulses (max)
LV Circuit* 10–500 0.05–100 ms 0.1–10 s 10
HV Circuit 200–3000 0.05–5 msec 5–30 s 2
*Use of the low voltage circuit requires that the CE Module be in place.
• Use the Up and Down Arrow keys to scroll through and highlight the parameter value space for
voltage, pulse length, number of pulses, or pulse interval. Use the alpha-numeric keypad to input
the desired value, or use the Right and Left Arrow keys to increase or decrease the parameter
value by the smallest allowable increment. Use the Delete or Clear keys to correct entries. When
the desired value has been specified, press Enter. If a value outside the limits of the Gene Pulser
Xcell is selected, the value in the field will default to the closest permitted value. Entering a value for
cuvette size is optional and is for user information only.
PROTOCOL DETAIL: SQUARE WAVE
Voltage (V) XXXX
Pulse length (msec) XXX.XX
Number of pulses 1
Pulse interval (sec) 00.0
Cuvette (mm) X
16
Page 23

• When the necessary parameter values have been specified, a flashing “P” appears in the character
space in the lower right corner of the LCD display indicating that the pulse button on the Gene
Pulser Xcell is active and that a pulse may be delivered.
• Press the Pulse button to deliver a pulse. When the Pulse button is depressed, the LCD display will
blank then show “Pulsing”. Upon completion, a tone will sound and the pulse measurements will be
displayed on the Protocol Results screen (see Figure 3.8, Section 3.3.5).
• Use the Left and Right Arrow keys to toggle between the Protocol Results screen and the last
Protocol Detail screen.
• With the Protocol Detail screen on the LCD display another pulse can be delivered using the same
pulse parameters. To change the pulse conditions, press Enter; the cursor appears in the voltage
parameter value. The parameters may be changed as described above.
• To save the pulse parameters, see Section 3.3.6.
• To review previously delivered pulses, see Section 3.8.
3.3.5 Results Screens
After delivering a pulse, the LCD displays the results on a Protocol Results screen. This screen shows
the results in both graphic and tabular form. Figures 3.6, 3.7, and 3.8 show examples of the results
from an exponential decay pulse, an exponential decay pulse in which the time constant was specified,
and a square wave pulse, respectively.
Results of the last 100 pulses as well as of the pulse parameters are stored in Gene Pulser Xcell
memory and are accessible from the Data Management program (Section 3.8).
17
Page 24
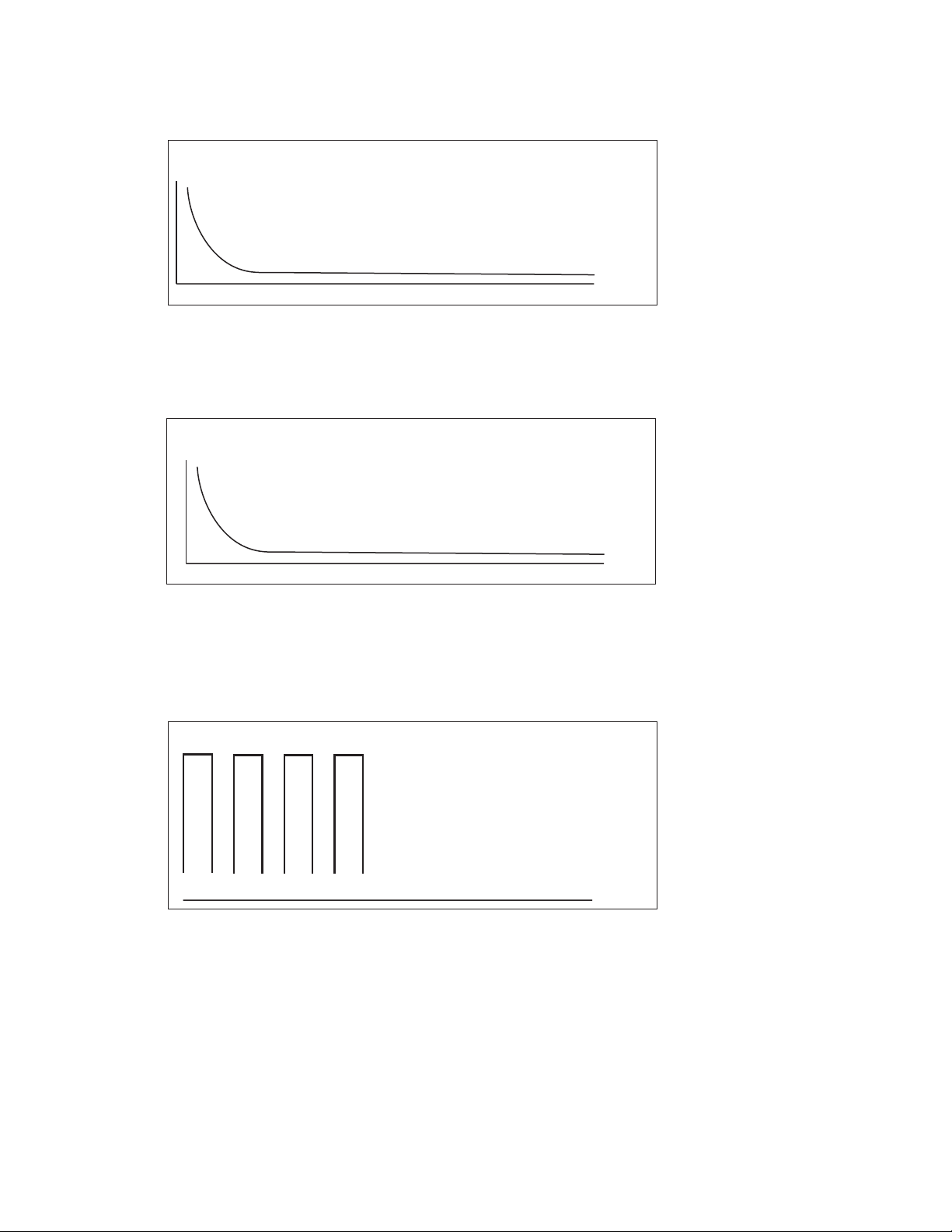
18
Exponential Decay: Results Screen
Fig. 3.6. Exponential Decay Protocol Results screen. The graph shows the exponential decay
pulse. The table gives the calculated time constant (TC) and the delivered voltage (Voltage).
Time Constant: Results screen
Fig. 3.7. Time Constant Protocol Results screen. The graph shows the exponential decay pulse.
The table gives the delivered time constant (TC) and voltage (Voltage) as well as the selected
capacitor (Capacitance) and resistor (Resistance).
Square Wave: Results screen
Fig. 3.8. Square Wave Protocol Results screen. The graph shows the square wave pulse; note
that the x-axis may not be to scale. The table gives the actual pulse length (Pulse Length), the
delivered voltage (Voltage), the calculated droop (% Droop), and, when multiple pulses are given,
the pulse interval (Interval) and the number of pulses (Number).
Results: Exponential Decay
TC(msec) XXXX.X
Voltage (V) XXXX
Results: Time Constant
TC(msec) XXXX.X
Voltage (V) XXXX
Capacitance (uF) XXXX
Resistance (ohms) XXXX
Results: Square Wave
Voltage XXXX
Pulse Length
(msec) XXX.XX
Interval XX.X
Number XX
%Droop XX
Page 25
3.3.6 Saving a program from Manual Operation
A protocol created in manual mode may be saved as a User Protocol as follows.
• Create a protocol as described in Section 3.3.2, 3.3.3 or 3.3.4.
• With the Protocol Detail screen open, press Save.
• The first User Directory screen will appear (Figure 3.9); the second line will read “Choose location
for protocol”.
• Use the Right and Left Arrow keys to toggle between the two User Directory screens. Press 1–12
or use the Up and Down Arrow keys to highlight the User Name under which to store the protocol.
Press Enter to select the User Name. The User Protocols screen will appear (Figure 3.10); the
second line will read “Choose location for protocol”. If it is necessary to create a new User Name,
see Section 3.5.2.
• Use the Right and Left Arrow keys to toggle between the two User Protocols screens. Press 1–12
or use the Up and Down Arrow keys to highlight a location for the new protocol. A protocol may
be stored in a position without an entry (see Section 3.3.6A) or in a position with an entry (see
Section 3.3.6B). If necessary, delete a User Protocol as described in Section 3.5.5.
• To use the saved protocol, press Enter to view the Protocol Detail screen. Press the Pulse button
to electroporate the sample.
User Directory screen
Fig. 3.9. User Directory screen. This is an example of the first User Directory screen. Toggle
between this screen and the second screen with Users 7–12 using the Left and Right Arrow keys.
When purchased, the initial User Directory screen will have only numbers without any names.
19
USER DIRECTORY
Choose location for protocol
1. User1
2. Labguy
3. Malcom
4. Smith
5. Wesson
> 6.
Page 26

User Protocols screen
Fig. 3.10. User Protocols screen. This is an example of the first User Protocols screen. Toggle
between this screen and the second screen with Protocols 7–12 using the Left and Right Arrow
keys. When purchased, the initial User Directory screen will have only numbers without any
names.
3.3.6A Saving a protocol in a location without a named User Protocol entry
• With a User Protocols screen displayed on the LCD, press 1–12 or use the Up, Down, Left, and
Right Arrows to highlight a User Protocol without an entry (e.g., positions 4, 5, or 6 on the screen
in Figure 3.10).
• Press Enter to select the position.
• Type in the name using the alpha-numeric keypad (alphabetic entries are the default; press Shift to
toggle between alphabetic and numeric entries; the maximum length is 10 characters; to use the
same key twice, advance the cursor using the Right Arrow Key, spaces are not allowed). Press
Save or Enter. The protocol is saved in the location named on the User Protocols screen.
• To use the saved protocol, press Enter to view the Protocol Detail screen. Press the Pulse button
to deliver a pulse.
3.3.6B Saving a protocol in a location with a named User Protocol entry
• With a User Protocols screen displayed on the LCD, press 1–12 or use the Up, Down, Left, and
Right Arrows to highlight a User Protocol with a named entry (e.g., positions 1, 2, or 3 on the
screen in Figure 3.10).
• Press Enter to select the position.
• A Warning screen will appear asking whether you want to overwrite the selected User Protocol
(Figure 3.11).
20
USER PROTOCOLS: User1
Choose location for protocol
1.CHO
2.Monkey
3.Human
4.
5.
> 6.
Page 27

Warning screen: overwrite protocol
Fig. 3.11. Warning screen: overwrite protocol.
• The default selection is “No”. Press Enter if you do not want to overwrite the file. The program will
return to the previous User Protocols screen.
• Press the Left Arrow key to select “Yes”, then press Enter to overwrite the file. The User Protocol
will have the same name and will contain all of the parameters specified in the new User Protocol.
The program will return to the User Protocols screen with the cursor highlighting the location (number)
of the selected User Protocol.
• To use the saved protocol, press Enter to view the Protocol Detail screen. Press the Pulse button
to deliver a pulse.
3.4 Pre-set Protocols
The Gene Pulser Xcell is pre-programmed with optimized settings for a number of commonly used
bacterial, fungal, and mammalian cells. The settings are given in Table 3.4. The settings may also be
viewed on the LCD display as described below (see Section 3.4.2). It is possible to change any of the
parameters prior to pulsing a sample. While permanent changes cannot be made to any Pre-set
Protocol, changes may be saved as a User Protocol (see Sections 3.4.4).
3.4.1 Using a Pre-set Protocol (Quick Guide)
• From the Home Screen, press 4, then Enter, to show the Pre-set Protocols screen.
• Press 1–3 to choose between bacterial, fungal, or mammalian cells; press Enter to select the
organism type and to show the list of organisms. For Bacterial and Mammalian Pre-set Protocols,
use the Right and Left Arrow keys to toggle between the two screens.
• Press the number next to the desired protocol to highlight it; press Enter to select and to show the
Protocol Detail screen.
• Press the Pulse button to electroporate the sample.
• Press the Back key to return to the Protocol Detail screen and to deliver another pulse.
21
Do you want to overwrite the
protocol?
YES NO
Press BACK or ENTER to return
to the previous screen.
Press the LEFT arrow then
ENTER to overwrite the name.
Page 28

3.4.2 Electroporation using Pre-set Protocols
There are nine Pre-set Bacterial Protocols, six Pre-set Fungal Protocols, and 12 Pre-set Mammalian
Protocols. These protocols are pre-programmed with the optimal parameters for the given organism.
Use the Pre-set Protocols as follows.
• From the Home screen, press 4 or use the Up and Down Arrow keys to highlight “Pre-set
Protocols”, then press Enter to select and to show the Pre-set Protocols screen (Figure 3.12).
• Press 1–3, or use the Up and Down Arrow keys, to highlight Bacterial, Fungal, or Mammalian
Pre-set Protocols, then press Enter to select.
• Use the alpha-numeric keypad or the Up and Down Arrow keys to scroll through the list of names.
For the Bacterial and Mammalian Pre-set Protocols, use the Right and Left Arrow keys to toggle
between the two screens. When the number corresponding to the desired name is highlighted,
press Enter to select and to view the Protocol Detail Screen showing the electroporation parameters
for that protocol. A flashing “P” in the character space in the lower right corner of the LCD display
indicates that the Pulse button is active.
• For example, from the Pre-set Protocols screen, press 3 to highlight “Mammalian”, then press
Enter to select and to bring up the first Pre-set Mammalian Protocols screen with the names of
six pre-set mammalian protocols (Figure 3.13). Press the Right and Left Arrow keys to toggle
between the two Mammalian Pre-set Protocols screens. Use the alpha-numeric keypad or the
Up and Down Arrow keys to scroll through the list of names. When the desired name on the
Mammalian Pre-set Protocols screen is highlighted, press Enter to select that protocol and to
view the Protocol Detail Screen showing the electroporation parameters for that protocol. For
example, from the Mammalian Pre-set Protocols screen, press 1, then Enter to bring up the
Protocol Detail Screen for CHO cells in a 2 mm cuvette (Figure 3.14).
• Press the Pulse button to deliver a pulse. When the Pulse button is depressed, the LCD display will
blank then show “Pulsing”. Upon completion, a tone will sound and the pulse measurements will be
displayed on the Protocol Results screen (see Section 3.3.5).
• Use the Left and Right Arrow keys to toggle between the Protocol Results screen and the last
Protocol Detail screen.
• With the Protocol Detail screen on the LCD display another pulse can be delivered using
the same pulse parameters. To change the pulse conditions, press Enter; the cursor
appears in the voltage parameter value. The parameters may be changed as described in
Section 3.4.3.
• To review previously delivered pulses, see Section 3.8.
22
Page 29

Pre-Set Protocols Screen
Fig. 3.12. Pre-set Protocols Screen. There are Pre-set protocols for bacteria, fungi, and mammalian cells. Use this screen to make a selection from the list.
Mammalian Pre-set Protocols Screen (screen 1)
Mammalian Pre-Set Protocols Screen (screen 2)
Fig. 3.13. Mammalian Pre-set Protocols Screens. Protocols are provided for 12 mammalian cell
lines. The names of the first six are shown on Screen 1; the names of the second six are shown
on Screen 2.
23
PRE-SET PROTOCOLS: MAMMALIAN
7.A549
8.CV1
9.K562
10.HL60
11.Jurkat
>12.HuT78
PRE-SET PROTOCOLS: MAMMALIAN
1.CHO
2.Cos7
3.M3T3
4.293
5.HeLa
> 6.BHK21
PRE-SET PROTOCOLS
1.Bacterial
2.Fungal
3.Mammalian
Page 30

Pre-set Protocol: CHO
Fig. 3.14. Protocol Detail Screen for CHO cells in the Pre-set Protocols menu.
Table 3.4. Optimized settings found in the Pre-set protocols function of the Gene
Pulser Xcell.
Pre-set
Protocol PL C PC Cuvette Cell vol
Designation Cells Pulse type (msec) (µF) (ohm) V (cm) (µl)
Mammalian 1 CHO Square wave 15 160 0.2 100
Mammalian 2 COS7 Square wave 20 110 0.2 100
Mammalian 3 3T3 Exponential decay 500 ∞ 160 0.2 100
Mammalian 4 293 Square wave 25 110 0.2 100
Mammalian 5 HeLa Exponential decay 500 ∞ 160 0.2 100
Mammalian 6 BHK21 Square wave 25 140 0.2 100
Mammalian 7 A549 Square wave 10 150 0.2 100
Mammalian 8 CV1 Square wave 25 100 0.2 100
Mammalian 9 K562 Exponential decay 1000 ∞ 155 0.2 100
Mammalian 10 HL60 Square wave 25 140 0.2 100
Mammalian 11 Jurkat Exponential decay 1000 ∞ 140 0.2 100
Mammalian 12 HuT78 Square wave 25 130 0.2 100
Bacterial 1 E coli Exponential decay 25 200 1800 0.1 20
Bacterial 2 25 200 2500 0.2 20–40
Bacterial 3 25 200 3000 0.2 20–40
Bacterial 4 A tumefaciens Exponential decay 25 200 2400 0.1 20
Bacterial 5 P aeruginosa Exponential decay 25 200 2500 0.2 100
Bacterial 6 S aureus Exponential decay 25 100 2900 0.2 50
Bacterial 7 B cereus Exponential decay 50 200 1000 0.2 100
Bacterial 8 S pyogenes Exponential decay 25 200 2100 0.2 200
Bacterial 9 L plantarum Exponential decay 25 400 2000 0.2 40
Fungal 1 S cerevisiae Exponential decay 25 200 1500 0.2 40
Fungal 2 25 200 2500 0.4 80
Fungal 3 S pombe Exponential decay 25 200 2300 0.2 40
Fungal 4 C albicans Exponential decay 25 200 1500 0.2 40
Fungal 5 P pastoris Exponential decay 25 200 2000 0.2 40
Fungal 6 D discoideum Square wave 1.0 10 1000 0.4 800 2 pulses
24
PROTOCOL DETAIL: CHO
Voltage (V) 160
Pulse length (msec) 15.00
Number of pulses 1
Pulse interval (sec) 00.0
Cuvette (mm) 2
P
Page 31

3.4.3 Modifying Pre-set Protocol Parameters
The parameters for a Pre-set protocol may be changed as follows.
• From the Protocol Detail screen, press the Up or Down Arrow keys to highlight the value for one of
the parameter settings (voltage, capacitance, or resistance for exponential decay pulses; voltage or
time constant for time constant mode; pulse length, voltage, number of pulses, or pulse interval for
square wave pulses). (Note: the waveform cannot be changed in the Pre-set Protocols Mode.)
When the desired parameter is selected, use the alpha-numeric keypad to input the new value.
Alternatively, use the Right and Left Arrow keys to incrementally increase or decrease, respectively,
the parameter value. Use the Delete or Clear keys to correct entries. When the correct value has
been specified, press Enter. If a value outside the limits of the Gene Pulser Xcell is selected, the
value in the field will default to the closest permitted value. Use the Up and Down Arrow keys to
select other parameter values to be changed, then use the alpha-numeric keypad or the Left and
Right Arrow keys to enter the desired value.
• A pulse may be delivered when appropriate parameters have been entered in the Protocol Detail
screen and the character space at the lower right of the LCD display is flashing “P”.
• To return to the last Protocol Detail screen, press the Back key or the Left Arrow key. Another
pulse may be delivered using the same parameters shown on the LCD display. To return to the
Protocol Results Screen, press the Right Arrow key. (Note: Returning to the Protocol Detail Screen
returns to the modified parameters. To return to the Pre-set Protocol, press the Back key again to
return to the Pre-set Protocols screen. This will remove any changes made.)
• To change the pulse conditions, with the Protocol Detail screen on the LCD display, press Enter;
the cursor appears in the voltage parameter value. The parameters may be changed as described
above.
• To review previously delivered pulses, see Section 3.8.
3.4.4 Saving Changes to Pre-set Protocols
Changes to a Pre-set Protocol may be saved as a User Protocol as follows:
• Change the Pre-set Protocol as described in Section 3.4.3.
• With the Protocol Detail screen open, press Save.
• The first User Directory screen will appear (Figure 3.9); the second line will read “Choose
location for protocol”.
• Use the Right and Left Arrow keys to toggle between the two User Directory screens. Press 1–12 or
use the Up and Down Arrow keys to highlight the User Name under which to store the protocol.
Press Enter to select the User Name. The User Protocols screen will appear (Figure 3.10); the second
line will read “Choose location for protocol”. If it is necessary to create a new User Name, see
Section 3.5.2.
• Use the Right and Left Arrow keys to toggle between the two User Protocols screens. Press 1–12 or
use the Up and Down Arrow keys to highlight a location for the new protocol. A protocol may be
stored in a position without an entry (see Section 3.3.6A) or in a position with an entry (see
Section 3.3.6B). If necessary, delete a User Protocol as described in Section 3.5.5.
• To use the saved protocol, press Enter to view the Protocol Detail screen. Press the Pulse button
to deliver a pulse.
25
Page 32

3.5 User Protocols
User Protocols enable users to store their own protocols for the Gene Pulser Xcell. Up to 144
electroporation protocols may be programmed and stored. (12 users (User Names in the User
Directory) each with 12 protocols per user (User Protocols).)
A new User Protocol may be created either by:
• Creating a new protocol by entering all new parameters in manual mode (Sections 3.3.2, 3.3.3,
and 3.3.4).
• Modifying (editing) a Pre-set protocol (Section 3.4.3).
• Creating a new protocol as a User Protocol (Section 3.5.3).
• Modifying (editing) an existing User Protocol (Section 3.5.4).
Protocols created or modified in the User Protocols menu may be used without being saved (Section 3.5.4).
3.5.1 Using a User Protocol (Quick Guide)
• From the Home screen, press 5, then Enter to open the User Protocols menu and to show the first
User Directory screen. Use the Right and Left Arrow keys to toggle between the two screens.
• Press 1–12 to highlight the desired name; press Enter to select that name and to show the first
User Protocols screen. Use the Right and Left Arrow keys to toggle between the two screens.
• Press 1–12 to highlight the desired protocol name; press Enter to select that protocol and to show
the Protocol Detail screen. The Pulse button on the Gene Pulser Xcell is now active.
• Press the Pulse button to electroporate the sample.
• Press the Back key to return to the Protocol Detail screen and to deliver another pulse.
3.5.2 Creating a New User Name
A new User Name can be created when the space adjacent to that number on the User Directory
screen is clear (Figure 3.15). If it is necessary to delete a User Name or a User Protocol, see Section
3.5.5.
Create a new User Name in the User Name screen as follows.
• From the Home screen (Figure 3.2), press 5 or use the Up and Down Arrow keys to highlight “User
Protocols”, then press Enter to view the User Directory screen (Figure 3.15).
• Use the Right and Left Arrow keys to toggle between the two User Directory screens. Press 1–12
or use the Up or Down Arrow keys to highlight a position without an entry. Press Enter to select
the position.
• Use the alpha-numeric keypad to type in the desired User Name. A name can have up to ten
characters. Press Save when complete. The new User Name is saved.
3.5.3 Creating a New User Protocol
This section describes how to create a new User Protocol from the User Protocols screen and save the
protocol. A new User Protocol can be created when the space adjacent to that number on the User
Protocol screen is clear (Figure 3.16). Section 3.5.4 describes how to edit (change) an existing User
Protocol and save the changes.
26
Page 33

27
• To enter the User Protocols screen from the Home screen (Figure 3.2):
• Press 5 or use the Up and Down Arrow keys to highlight “User Protocols”, then press Enter
to view the first User Directory screen (Figure 3.15).
• Use the Right and Left Arrow keys to toggle between the two User Directory screens. Press
1–12 or use the Up and Down Arrow keys to highlight the number corresponding to the
desired User Name.
• Press Enter to select the name and to view the User Protocols Screen (Figure 3.16).
• Press 1–12 or use the Up, Down, Left, and Right Arrow keys to highlight a User Protocol without
an entry. If necessary, delete a User Protocol as described in Section 3.5.5. Press Enter to view the
Select Method screen (Figure 3.17).
• Press 1–3 or use the Up and Down Arrow keys to highlight the number corresponding to delivering
an exponential decay pulse (Exponential protocol), an exponential decay pulse specifying the time
constant (Time constant protocol), or a square wave pulse (Square wave protocol). Press Enter to
make a selection and to view the appropriate Protocol Detail screen.
Page 34

User Protocols – User Directory Screen (screen 1)
User Protocols – User Directory Screen (screen 2)
Fig. 3.15. User Directory screen.
User Protocols Screen
Fig 3.16. User Protocols screen.
28
USER DIRECTORY
<7.
8.
9.
10.
11.
12.
USER PROTOCOLS: User1
1.CHO
2.Monkey
3.Human
4.
5.
>6.
USER DIRECTORY
1.User1
2.Labguy
3.Malcom
4.Smith
5.Wesson
>6.
Page 35

Select Method Screen
Fig. 3.17. Select Method screen.
• When “Exponential protocol” is selected, the Exponential Decay Protocol Detail screen appears
(Figure 3.18) with the User Name on the first line.
Exponential Decay Protocol Detail Screen
Fig. 3.18. Exponential Decay Protocol Detail screen.
• When “Time constant protocol” is selected, the Time Constant Protocol Detail screen appears
(Figure 3.19) with the User Name on the first line.
Time Constant Protocol Detail Screen
Fig. 3.19. Time Constant Protocol Detail screen.
29
Labguy: Protocol
Voltage (V) XXXX
Time constant (msec) XXXX.X
Cuvette (mm) X
SELECT METHOD
1.Exponential protocol
2.Time constant protocol
3.Square wave protocol
Labguy: Protocol:
Voltage (V) XXXX
Capacitance (uF) XXXX
Resistance (ohm) XXXX
Cuvette (mm) X
Page 36

• When “Square wave protocol” is selected, the Square Wave Protocol Detail screen appears
(Figure 3.20) with the User Name on the first line.
Square Wave Protocol Detail Screen
Fig. 3.20. Square Wave Protocol Detail screen
• Use the Up and Down Arrow keys to scroll through the parameter value spaces on the
appropriate Protocol Detail screen. Use the alpha-numeric keypad to input the desired value, or
use the Right and Left Arrow keys to incrementally increase or decrease the parameter value. Use
the Delete or Clear keys to correct entries. When the desired value has been specified, press Enter.
If a value outside the limits of the Gene Pulser Xcell is selected, the value in the field will default to
the closest permitted value (see Tables 3.1, 3.2, and 3.3). Entering a value for cuvette size is
optional and is for user information only.
• When the necessary parameter values have been specified, a flashing “P” appears in the character
space in the lower right corner of the LCD display indicating that the pulse button on the Gene
Pulser Xcell is active and that a pulse may be delivered.
• Press the Pulse button to deliver a pulse. When the Pulse button is depressed, the LCD display will
blank then show “Pulsing”. Upon completion, a tone will sound and the pulse measurements will
be displayed on the Protocol Results screen (see Section 3.3.5).
• To save the pulse parameters, with the Protocol Detail screen on the LCD, press Save.
• The User Protocols screen will appear with the cursor flashing on the first character space of the
selected position. Type in the name using the alpha-numeric keypad (alphabetic entries are the
default; press Shift to toggle between alphabetic and numeric entries). Press Save or Enter when
complete. The protocol is saved in the location named in the User Protocols screen.
3.5.4 Modifying a User Protocol
A User Protocol may be modified (changed) as follows.
• To enter the User Protocols screen from the Home screen (Figure 3.2):
• Press 5 or use the Up and Down Arrow keys to highlight “User Protocols”, then press Enter
to view the first User Directory screen (Figure 3.15).
• Use the Right and Left Arrow keys to toggle between the two User Directory screens. Press
1–12 or use the Up and Down Arrow keys to highlight the desired User Name. Press Enter to
select the name and to view the first User Protocols screen (Figure 3.16).
• Press 1–12 or use the Up, Down, Left, and Right Arrow keys to highlight the desired User Protocol
to be edited. Press Enter to view the Protocol Detail screen.
Labguy: Protocol:
Voltage (V) XXXX
Pulse length (msec) XXX.XX
Number of pulses 1
Pulse interval (sec) 00.0
Cuvette (mm) X
30
Page 37

• Use the Up and Down Arrow keys to highlight the value of the first parameter to be changed. Use
the alpha-numeric keypad to enter a new value. Alternatively, use the Right and Left Arrow keys to
incrementally increase or decrease, respectively, the parameter value. Use the Delete or Clear keys
to correct entries.
• When the desired value has been specified, press Enter. If a value outside the limits of the Gene
Pulser Xcell is entered, the value in the field will default to the closest permitted value.
• Use the Up and Down Arrow keys to highlight the parameter values to be changed and repeat the
previous two steps. The Pulse button becomes active when the necessary parameters have been
entered into the Protocol Detail screen. The edited protocol may be used without saving.
• To save the edited protocol, with the Protocol Detail screen on the LCD, press Save. A warning
screen will appear asking if you want to overwrite the protocol (Figure 3.21). The default selection is
“No”.
• If you do not want to overwrite protocol name press the Back key to return to the Protocol
Detail screen. Press Enter to return to the User Directory and to choose a new location
(Figure 3.9). See Section 3.3.6 for instructions on selecting a location for the new protocol.
• To overwrite the protocol, press the Left Arrow key to select “Yes”, then press Enter to
overwrite the file. The User Protocol will have the same name as the old file and will contain
the parameters specified in the new protocol. The LCD will display the User Protocols screen
with the cursor highlighting the location (number) of the protocol.
Warning screen: overwrite protocol
Fig. 3.21. Warning screen: overwrite protocol.
3.5.5 Deleting a User Name and a User Protocol
Before a User Name may be deleted, it is necessary to first delete all of the User Protocols under that
name.
Delete a User Protocol as follows.
• To enter the User Protocols screen from the Home screen (Fig 3.2):
• Press 5 or use the Up and Down Arrow keys to highlight “User Protocols”, then press Enter
to view the first User Directory screen (Figure 3.15). Use the Right and Left Arrow keys to
toggle between the two User Directory screens.
• Enter the number corresponding to the desired User Name or use the Up and Down Arrow
keys to highlight the desired User Name.
• Press Enter to select the name and to view the User Protocols screen.
31
Do you want to overwrite the
protocol?
YES NO
Press BACK or ENTER to return
to the previous screen.
Press the LEFT arrow then
ENTER to overwrite the name.
Page 38

• Use the alpha-numeric keypad or the Up and Down Arrow keys to select the position with the
protocol to be deleted.
• Press the Delete key. A warning screen will appear asking whether you want to delete the selected
User Protocol (Figure 3.22). The default selection is “No”.
• If you do not want to delete the Protocol, press Back or Enter. The program will return to the
previous User Protocols screen.
• To delete the Protocol, press the Left Arrow key to select “Yes”, then press Enter. The User
Protocol name as well as all of the parameters under that User Protocol is deleted. The
program will return to the User Protocols screen with the selected User Protocol deleted.
Warning screen: delete protocol
Fig. 3.22. Warning screen: delete protocol.
Delete a User Name as follows.
• With the User Directory screen on the LCD, press the Up or Down Arrow keys or use the alphanumeric keypad to select the name to be deleted. Use the Left and Right Arrow keys to toggle
between the two User Directory screens.
• Press the Delete key. If there are no protocols in the User Name file, a warning screen will appear
asking whether you want to delete the selected User Name (Figure 3.23). The default selection is
“No”.
• If you do not want to delete the file, press Enter. The program will return to the User Directory
screen.
• To delete the User Name, press the Left Arrow key to select “Yes”, then press Enter. The
program will return to the User Directory with the selected User Name deleted.
• If there are protocols in the User Name file, the following screen will appear: “All User Protocols
under this name must be deleted before this user name can be deleted.” Follow the steps above
for deleting the User Protocols.
32
Do you want to delete the
protocol?
YES NO
Press BACK or ENTER to return
to the previous screen.
Press the LEFT arrow then
ENTER to delete the name.
Page 39

Warning screen: delete name
Fig. 3.23. Warning screen: delete name.
3.5.6 Renaming a User Name or a User Protocol name
Rename a User Name or a User Protocol name as follows.
• Choose the User Name or User Protocol to rename from the User Directory screen or the User
Protocols screen, respectively.
• Press the Clear key. A warning screen will appear asking whether you want to change the selected
name (Figure 3.24). The default selection is “No”.
• If you do not want to change the name, press Enter. The program will return to the
corresponding User Directory or User Protocols screen.
• To change the name, press the Left Arrow key to select “Yes”, then press Enter. The previous
screen will appear but with the selected name cleared and with the cursor on the field for the
new name. Use the alpha-numeric keypad to type in the desired new name. A name can have
up to ten characters. Use the Shift key to toggle between alphabetic and numeric characters.
Press Save when complete. The new name is saved.
Warning screen: change name
Fig. 3.24. Warning screen: name change.
33
Do you want to delete the user
protocol?
YES NO
Press BACK or ENTER to return
to the previous screen.
Press the LEFT arrow then
ENTER to delete the name.
Do you want to change the
name?
YES NO
Press BACK or ENTER to return
to the previous screen.
Press the LEFT arrow then
ENTER to change the name.
Page 40

3.6 Last Pulse
From the Home screen (Figure 3.2), press 6 or use the Up or Down Arrow keys to select “Last pulse”,
then press Enter to display the Protocol Detail screen showing the last pulse delivered. This is also
accessible after the instrument has been turned off and restarted.
3.7 Optimize Operation
The Optimize mode allows the user to program a voltage that incrementally increases or decreases
when the Right and Left Arrow keys are pressed. This mode operates similarly to Manual Protocols
(Section 3.3.2, 3.3.3, and 3.3.4) but requires that a voltage increment be specified.
• Access this screen from the Home screen (Figure 3.2) by pressing 7 or use the Up or Down Arrow
keys to highlight “Optimize” then press Enter. The Select Method screen appears (Figure 3.17).
• Press 1–3 or use the Up and Down Arrow keys to highlight the number corresponding to optimizing
voltage by delivering (i) an exponential decay pulse (“Exponential protocol”), (ii) an exponential decay
pulse specifying the time constant (“Time constant protocol”), or (iii) a square wave pulse (“Square
wave protocol”). Press Enter to make a selection and to view the appropriate Optimize screen.
Select Method screen
Fig. 3.17. Select Method screen.
• When “Exponential protocol” is selected, the Exponential Decay Protocol Optimize screen appears
(Figure 3.25).
• When “Time constant protocol” is selected, the Time Constant Protocol Optimize screen appears
(Figure 3.26).
• When “Square wave protocol” is selected, the Square Wave Protocol Optimize screen appears
(Figure 3.27).
34
SELECT METHOD
1.Exponential protocol
2.Time constant protocol
3.Square wave protocol
Page 41

Exponential Decay Protocol Optimize Screen
Fig. 3.25. Exponential Decay Protocol Optimize screen.
Time Constant Protocol Optimize Screen
Fig. 3.26. Time Constant Protocol Optimize screen.
Square Wave Protocol Optimize Screen
Fig. 3.27. Square Wave Protocol Optimize screen.
• Use the Up and Down Arrow keys to scroll through the parameter value spaces on the screen.
When the desired parameter space is highlighted, use the alpha-numeric keypad to input the
desired value (only numeric entries are permitted). Alternatively, use the Right and Left Arrow keys
to incrementally increase or decrease the parameter value. Use the Delete or Clear keys to change
the value. When the desired value has been specified, press Enter. If a value outside the limits of
the Gene Pulser Xcell is selected, the value in the field will default to the closest permitted value.
35
OPTIMIZE: EXPONENTIAL DECAY
Starting voltage (V) XXXX
Increment (V) XXX
Capacitance (uF) XXXX
Resistance (OHM) XXXX
Cuvette (mm) X
OPTIMIZE: SQUARE WAVE
Starting voltage (V) XXXX
Increment (V) XXX
Pulse length (msec) XXX.XX
Number of pulses 1
Pulse interval (sec) XX.X
Cuvette (mm) X
OPTIMIZE: TIME CONSTANT
Starting voltage (V) XXXX
Increment (V) XXX
Time const (msec) XXXX.X
Cuvette (mm) X
Page 42

• When the necessary parameter values have been specified, a flashing “P” appears in the character
space in the lower right corner of the LCD display indicating that the pulse button on the Gene
Pulser Xcell is active and that a pulse may be delivered.
• To use the program:
• Press Pulse to deliver the pulse at the specified voltage.
• After the pulse, the Protocol Results screen is displayed; use the Left and Right Arrow keys to
toggle between the Protocol Results screen and the last Protocol Detail screen.
• From the Protocol Detail screen, another pulse at the same voltage may be delivered by
pressing Pulse. Press Enter to bring the cursor on the screen; the cursor will appear at the
right character space of the first parameter (the voltage). To increase the voltage by the
specified increment value, press the Right Arrow key; to decrease the voltage by the
specified value, press the Left Arrow key. Press Pulse to deliver another pulse.
3.8 Data Management
This section of the program allows the user to view the parameter values and output of the last 100 pulses.
Data for each pulse is presented on a single screen; examples are given below. Each data set includes the
date and time of the pulse, an identifier for how the pulse was programmed, the set parameters, and the
delivered pulse. The pulses are displayed in chronological order or in reverse chronological order and are
logged by date and time. (Check that the date and time are correct under User Preferences (Section 3.10,
Setting the Clock).) Selecting chronological order displays the earliest pulse in the data bank or, if a specific
date was selected, the earliest pulse from the specified date. Selecting reverse chronological order displays
the latest pulse in the data bank or, if a specific date was selected, the last pulse from the specific date.
This is a read-only file. No changes are permitted to this file.
• From the Home screen (Figure 3.2) press 8 or use the Up and Down Arrow keys to highlight “Data
Management”, then press Enter to display the Data Management screen (Figure 3.28).
• Press 1–4, then Enter to select the data to view. Selecting 3 or 4 brings up a Date screen
(Figure 3.29). Use the keypad to enter a date, then press Enter to select that date and to view the
data. If no data was collected on that date, a screen will appear indicating that no data is present
for that date.
Data Management Screen
Fig. 3.28. Data Management screen.
36
What data do you want to view?
All data
1.Reverse chronological order
2.Chronological order
Data from a specific date
3.Chronological order
4.Reverse chronological order
Page 43

Date Screen
Note: this is the default screen with the default clock preference set as MM/DD/YY; if the user
changes the clock preference to DD/MM/YY, the order on the Date screen is Date, Month, Year.
Fig. 3.29. Date screen.
• Press the Left and Right Arrow keys to view earlier and later pulse data, respectively. Press the
Back key to return to the Data Management screen.
Data screens contain the following information:
Line 1: Date and Time
Line 2: Identification of how pulse was programmed (e.g., Manual, Pre-set, User)
Line 3: Type of pulse
Lines 4–8: Pulse conditions
37
From what date do you want
to view data?
Month MM
Date DD
Year YY
Page 44

The following are examples of data screens. It is not possible to edit a Data screen.
Data screen (User Protocol, Square Wave)
Data screen (Manual Mode, Exponential Decay)
Data screen (Manual Mode, Time Constant)
Data screen (Standard Protocol, S. pombe setting)
Fig. 3.30. Examples of data screens.
38
Data output 12-12-01 20:20
User 11 Protocol 11
Settings Output
V 1230 1230
PL (msec) 123.00 123.00
#P 12 12
PI (sec) 12.3 12.3
%droop 12 12
Data output 12-12-01 20:21
Manual exponential
Settings Output
V 1230 1230
C(uF) 1230 1230
R(OHM) 1000 1000
TC(msec) 111.1
Cuvette(mm) 1
Data output 12-12-01 20:22
Manual time constant
Settings Output
V 1230 1230
TC (msec) 1234.5 1234.5
Cuvette(mm) 1
Data output 12-12-01 20:23
Std Protocol: S.pombe
Settings Output
V 2000 2047
C(uF) 25 25
R(ohm) 200 200
TC (msec) 5.1
Cuvette(mm) 2 2
Page 45

3.9 Measurements
This program allows the user to measure the resistance of a sample prior to pulsing in the Gene Pulser
Xcell and to measure the values of the capacitors in a Capacitance Extender attached to the Gene
Pulser Xcell.
• From the Home screen, press 9 or use the Up and Down Arrow keys to highlight “Measurements”,
then press Enter to view the Measurements Screen (Figure 3.31).
Measurements Screen
Fig. 3.31. Measurements screen.
3.9.1 Sample Resistance Measurements
Placing a cuvette containing a sample in the shocking chamber will allow the Gene Pulser Xcell to
measure the sample resistance. The resistance determination is accurate for samples having a resistance
between 5 and 1100 ohms. Samples with a resistance less than 5 ohms will show “5 ohms”. Samples
with a resistance greater than 1100 ohms will show “>1100 ohms”.
• When the Measurements screen (Figure 3.31) is selected, the number 1, corresponding to “Sample
Resistance” is highlighted as the default choice. Press Enter to view the Sample Resistance screen.
If the number 1 on the Measurements screen is not highlighted, press 1 or use the Up or Down
Arrow keys to highlight, then press Enter to select. The Sample Resistance screen appears
(Figure 3.32).
Sample Resistance Screen
Fig. 3.32. Sample Resistance screen.
• Place a sample between the electrodes to allow the Gene Pulser Xcell to monitor sample resistance.
Close the top of the ShockPod and press Enter. The sample resistance is displayed on the LCD
screen. The display range is 5–1100 ohms.
39
Sample resistance
Resistance (ohms) xxxx
Place sample between
electrodes.
Measurements
1.Sample Resistance
2.Capacitor
Page 46

3.9.2 Calibration and Measurement of Capacitors in the CE Module
Because electrolytic capacitors change value slowly over time, to keep the instrument within its original
specifications, the capacitors should be calibrated approximately every one to three months or whenever
there is concern about the accuracy of the capacitors. The Capacitor Measurement function gives the
user a more accurate determination of the capacitor values in the unit by making a measurement of the
capacitors in the units. Capacitor calibration and measurement may be performed as follows.
• Unplug the ShockPod cables from the Gene Pulser Xcell. Connect the CE Module to the Gene
Pulser Xcell.
• From the Measurements screen, press 2 or use the Up and Down Arrow keys to highlight
“Capacitor”, then press Enter to display the Capacitor Measurement screen (Figure 3.33).
• To calibrate the capacitors, press the decimal point (.) on the alpha-numeric keypad. The Gene
Pulser Xcell main unit will begin calibrating the capacitors in both the CE Module and the Gene
Pulser Xcell main unit. Calibration will take about two minutes. At the completion of the test,
12 numbers will be displayed at the bottom of the screen. The numbers on the top row and the first
three on the bottom row are the actual values of the capacitors in the CE Module. These capacitors
are used in combination to set the capacitance in the exponential decay and time constant modes.
When pulsing using the low voltage circuit in square wave mode, all of the capacitors from the CE
Module are used together; when pulsing using the high voltage circuit in square wave mode, the
50 uF capacitor in the Gene Pulser Xcell main unit is used.
• To measure a specific capacitance, with the Capacitor Measurement screen on the LCD, use the
alpha-numeric keypad to type in a capacitance value to be measured. Alternatively, use the Right
and Left Arrow keys to increase or decrease, respectively, the capacitance values. Press Enter,
then press Pulse. The actual capacitance value appears on the Measured Capacitance line. (No
sample should be in place and no pulse occurs during this process.)
Capacitor Measurement Screen
Fig. 3.33. Capacitor Measurement screen.
40
Capacitor measurement
Capacitance (uF) xxxx
Measured capacitance (uF) xxxx
Page 47

3.10 User Preferences
• From the Home screen, press 10 or use the Up or Down Arrow keys to highlight “User preferences”,
then press Enter to display the User Preferences Screen (Figure 3.34).
User Preferences Screen
Fig. 3.34. User Preferences screen.
3.10.1 Setting the Clock
This section of the program allows the user to select how the date is presented, to enter a correct
date, and to enter the correct time. The default setting for the date is Month/Date/Year; this may be
changed to Date/Month/Year. The clock is a 24-hour clock.
• When the User Preferences screen (Figure 3.34) is selected, the number 1, corresponding to
“Clock” is highlighted as the default choice. Press Enter to view the Set Clock screen. If the
number 1 on the User Preferences screen is not highlighted, press 1 or use the Up or Down
Arrow keys to highlight, then press Enter to select. The Set Clock screen appears (Figure 3.35).
Set Clock Screen
Note: the default setting is MM/DD/YY
Fig. 3.35. Set Clock screen.
41
Set clock
PREFERENCE MM/DD/YY DD/MM/YY
DATE XX/XX/XX XX/XX/XX
TIME XX:XX
User Preferences
1.Clock
2.Screen intensity
3.Sleep function
Page 48

• Press the Left or Right Arrow keys to toggle between MM/DD/YY and DD/MM/YY. Press Enter to
make a selection and to bring the cursor to the field below the choice where entries can be made.
• If no change in the date is needed, press Enter again. Use the keypad to type in the date or time
(only numeric entries are permitted). “0” must be typed to specify numbers 1–9 in all fields (i.e.,
01–09). Use the Delete and Clear keys to correct entries. Press Enter when complete. The cursor
will move to the field for entering a new time (XX:XX).
• If no change in the time is needed, press Save. Otherwise, use the alpha-numeric keypad to enter
a new time. The clock is a 24-hour clock. Press Save when complete and return to the User
Preferences screen.
3.10.2 Adjusting the Screen Intensity
• From the User Preferences screen press 2 or use the Up and Down Arrow keys to select “Screen
intensity”, then press Enter. The LCD Brightness screen appears (Figure 3.36).
LCD Brightness Screen
Fig. 3.36. LCD Brightness screen.
• Press the Up and Down Arrow keys to increase or decrease the screen brightness, respectively.
• Press Enter to select. The LCD display returns to the User Preferences screen.
3.10.3 Sleep Function Setting
The sleep function is an energy-saving mode of operation, designed to maximize the life of the battery
and the LCD screen. The default setting causes the LCD display to turn off after 30 min of inactivity.
Pressing any alpha-numeric key will restore the last screen displayed and return the Gene Pulser Xcell
to its former operating status. The user may change the time before the
instrument enters sleep mode using the Sleep Function Setting.
42
LCD BRIGHTNESS
Press UP/DOWN arrow keys
to change the brightness of
the LCD display.
Press ENTER to select.
Page 49

• From the User Preferences screen press 3 or use the Up or Down Arrow keys to highlight “Sleep
function”, then press Enter to select and to view the Sleep Function screen (Figure 3.37).
Sleep Function Screen
Fig. 3.37. Sleep Function screen.
• Use the Right and Left Arrow keys to change the time before the LCD enters sleep mode.
• Press Enter to select. The LCD returns to the User Preferences screen.
3.11 The Pulse Trac™System
The Gene Pulser Xcell uses the Pulse Trac waveform delivery system to generate the most accurate exponential
decay pulses possible for optimal cell transformation. Pulse Trac accurately calculates the time constant and
amplitude of each pulse based on what is actually delivered to the sample. However, more importantly, the
Pulse Trac system compensates for amplitude changes caused by varying sample resistances and improves
the accuracy of the low voltage capacitors. This provides:
• Sample conductivity measurement integrated with pulse output for true waveform delivery for samples
with resistance in the 10–1000 ohm range;
• Internal calibration and circuit monitoring program for accurate pulse delivery throughout the lifetime
of the unit;
• Time constant and voltage amplitude measurements to allow pulse delivery verification.
3.11.1 Pulse Trac System Description
The Pulse Trac system monitors and adjusts for the internal system resistance used to limit current
and the sample resistance in the cuvette. Sample resistance depends on its conductivity, the
distance between the electrodes in the cuvette, and the volume of media within the cuvette. Pulse
Trac circuitry monitors the resistance of the sample and delivers the desired voltage regardless of
sample volume or conductivity. When you are optimizing electroporation with the Pulse Trac system, the electrical variables are controlled with exacting precision so that your results reflect only
the biological variables in your experimental design. This Pulse Trac diagnostic algorithm examines
the complete electrical circuit and electronically selects the right combination of capacitors to
deliver the most accurate and reproducible pulse for optimal and consistent electroporation over
the lifetime of the unit.
43
SLEEP FUNCTION
Time before entering sleep
mode (min):
5 10 20 30 60 OFF
Page 50

3.11.2 Pulse Trac Diagnostic Algorithm
This test algorithm tests and selects the optimal capacitor circuit of the Gene Pulser Xcell unit and the
CE Module module in the range of 25–3,275 µF. This is the key bank of capacitors used in low
voltage/high capacitance precision pulse delivery. The test algorithm tightens the already rigorous
capacitor tolerance from ±20% to ±10% (other unit designs can have a capacitor variance as high as
±40%) in the 200–1075 µF range. The high voltage capacitors in the Gene Pulser Xcell unit are not part
of this system since they are pre-selected to the same 10% tolerance that this test algorithm provides.
The Pulse Trac test algorithm is activated upon startup of the Gene Pulser Xcell when the Capacitor
Extender module is installed.
Section 4
Overview of Electroporation Theory
Electroporation is a physical process in which cells are exposed to a high-voltage electric field resulting
in a temporary rearrangement of the cell membrane. As a result, the cells become permeable and may
take up solutes from their surrounding environment, including nucleic acids, proteins, carbohydrates,
and small molecules. While much work has been done to determine how cells become permeabilized
during the process of electroporation, the membrane changes that occur are still largely hypothetical
(see Chang, et al., 1992).
The Gene Pulser Xcell is the only electroporation instrument capable of delivering both exponential decay
and square wave pulses (Figure 4.1). The system consists of a pulse generator system (the main unit with
either or both the CE Module and PC Module), a shocking chamber, and a cuvette with incorporated
electrodes (see Section 2). Activating the pulse button on the Gene Pulser Xcell charges the capacitors in
the unit and, if attached, in the CE Module, to a high voltage; the instrument then discharges the current
in the capacitor into the sample in the cuvette. Discharging the charged capacitor generates either the
exponential decay or the square wave pulse.
There are two instrument parameters that describe the changes that the cells experience upon
electroporation. The first of these, the electric field strength, E, measured in volts/cm, describes the
electrical environment in the electroporation chamber. Standard electrodes used in electroporation
consist of two parallel plates separated by a distance, d (cm); therefore,
E = V / d,
where V is the applied voltage and d is the distance between the electrodes. In practical terms, the field
strength is manipulated by altering the voltage of the instrument or by changing the distance between
the electrodes. Because the electric conductance of the cell cytoplasm is much higher than that of the
cell membrane, placing the cell in an electric field creates a voltage potential across the cell membrane.
As the field strength increases, the transmembrane voltage experienced by the cell increases as does
the likelihood that a pore will form in the membrane due to breakdown of the lipid bilayer, allowing
molecules to enter the cell from the outside (Hui, 1995; Neumann, et al, 2000).
The second parameter that affects the cell is the length of time that it is exposed to the electric field.
For exponential decay pulses, this is controlled by the capacitance of the instrument and the resistance
within the circuit. For square wave pulses the pulse length is controlled directly by setting the time that
the cells are exposed to the electric field. These are discussed for each pulse type below.
44
Page 51

4.1 Exponential Decay Pulses
The exponential decay circuit of the Gene Pulser Xcell generates an electrical pulse by discharging a capacitor.
When a capacitor is discharged into the sample, the voltage across the electrodes rises rapidly to the peak
voltage then declines over time, t, with an exponential decay waveform (Figure 4.1A) according to the
following equation:
V
t
= V0[e
-(t/RC)
],
where V
0
is the initial voltage in the capacitor, Vtis the voltage at time = t (msec) after the pulse, e is the base
of the natural logarithm, R is the resistance of the circuit (expressed in ohms), and C is the capacitance
(expressed in microfarads). The time required for the initial voltage to drop to V
0
/e is referred to as the time
constant, τ, a convenient expression of the pulse length (expressed in msec). When t = τ = R x C, the
voltage has declined to 1/e (~37%) of the initial value, V
0(Vτ
= V0/ e).
By changing the capacitor of the instrument or by changing the resistance of the circuit, the time
constant may be readily changed. When high-resistance media is used (e.g., low ionic-strength media
used for most bacteria and yeast), the resistance of the circuit may be controlled using the PC Module
which places a resistor in parallel with the sample. For resistors connected in parallel, the total resistance
of the circuit is given by the equation:
R
T
= (R
sample
* RPC) / (R
sample
+ RPC).
When the sample resistance is much greater than the resistor in the PC Module (R
sample
>> RPC), the
latter is the primary determinant of the resistance of the circuit and R
T
~ RPC. Therefore, the PC Module
reduces the resistance of the circuit thereby reducing the time constant of the circuit.
When low-resistance media is used (e.g., high ionic-strength media such as PBS or growth media
used for most mammalian cells), the time constant is most easily manipulated by selecting the proper
capacitor for the circuit using the CE Module. Additionally, changing the volume of low -resistance
media in the cuvette will alter the resistance of the circuit (resistance is inversely proportional to volume).
4.2 Square Wave Pulses
Truncating the pulse from a capacitor after discharging it into the sample generates square wave pulses.
The ideal square wave pulse has the same voltage at the end as at the beginning of the pulse (Figure
4.1B). However, using a charged capacitor to produce this waveform (as is done in all commercially
available electroporation instruments), the voltage at the end of the pulse, V
t
, is always less than the
voltage at the beginning of the pulse, V
0
. This is because when the switch is closed across a charged
capacitor, maximum current instantaneously flows through the circuit and gradually falls to zero. To
produce a square wave, the pulse is terminated at some time, t, following discharge of the capacitor.
This time (t) is termed the pulse length. The longer the pulse length, the greater is the difference in voltage
between the beginning and the end of the pulse. This voltage decay may be determined from the
following equation:
ln (V0/ Vt) = t / (R C).
45
Page 52

The decrease in voltage that occurs with a square wave pulse is inversely related to both the capacitance of the instrument and the resistance of the sample. The fractional decrease in voltage at the end
of the pulse as a function of the initial voltage, V0, is termed droop and is given by the following equa-
tion:
Droop = (V0– Vt) / V0.
Combining the two equations:
ln [1 / (1 – Droop)] = t / (R C).
In order for the pulse to most closely approximate a true square wave, droop must be minimized (i.e.,
V
t
= V0and V0- Vt= 0). Experimentally, this is achieved by choosing the highest values for R and C.
For any given sample, R may be considered a constant. The Gene Pulser Xcell uses a 50 uF capacitor for
the high-voltage circuit and a 3275 uF capacitor for the low-voltage circuit. For each of these circuits,
C may also be considered a constant. Therefore, for the same sample, as pulse length increases,
droop also increases. However, increasing sample resistance reduces the droop at any given pulse
length. Increasing the sample resistance may be accomplished by (1) reducing the temperature of the
sample; (2) reducing the ionic concentration of the solution; and (3) reducing the volume of liquid in the
electroporation cuvette in the case of low resistance media. Never use the PC Module when performing
square wave pulsing because attaching a resistor from the PC Module in parallel with the sample will
result in increasing the droop of the pulse. Table 4.1 gives the pulse lengths corresponding to a given
droop as a function of resistance in the high voltage and low voltage ranges. For example, pulsing into
a 200-ohm load, the pulse length will be 0.510 msec with a 5% droop in the high voltage range and
33.4 msec with a 5% droop in the low voltage range.
46
Page 53

Table 4.1 Droop associated with pulse length at various sample resistances for the
high-voltage and low-voltage ranges for the Gene Pulser Xcell.
High voltage circuit Low voltage circuit
Droop % 10 20 5 10 20
Sample
Resistance Pulse length (msec) Pulse length (msec)
(ohms)
20 0.051 0.109 0.223 3.34 7.14 14.6
200 0.510 1.09 2.23 33.4 71.4 146
1000 2.55 5.45 11.2 167 357 730
3500 8.93 19.1 39.0 585 1249 2556
.
Fig. 4.1. (A) Exponential decay pulse from a capacitance discharge system. When a capacitor,
charged to an initial voltage V
o
, is discharged into cells, the voltage applied to the cells decreases
over time in an exponential surve such that the voltage “V” at any given time “t” is given by
V = V
o
e
–(t/RC)
. In the special case where t = CR then Vo/e. The value CR is known as the time con-
stant of the voltage decay. The shorter the time constant the faster the decay. (B) Square wave
pulse from a capacitance discharge system. The pulse length is the time the cells are subjected
to the discharge. During the pulse the voltage again decreases in an exponential decay so that at
the end of the pulse the voltage is lower than at the beginning. We call this drop in voltage the
pulse droop and measure it as a percentage of the initial voltage.
47
Time
V
0
V
0
t
V
t
ττ
V
0
e
Time
Voltage
(V)
Voltage
(V)
Page 54

Section 5
Factors Affecting Electroporation: Optimizing Electroporation
The electrical conditions for the electroporation of cells have been verified through years of research (see
Shigekawa & Dower, 1988, Change, et al., 1992, and Nickoloff, 1995a,b, for overviews as well as for
protocols on electroporation of numerous species). For suggestions on optimizing electroporation
parameters, see the following references: Calvin & Hanawalt, 1988, Dower, et al., 1988 (for bacterial
cells); Shillito, et al., 1985, Fromm, et al., 1987, Dekeyser, et al., 1990, Joersbo & Brunstedt, 1991 (for
plant cells); Chu, et al., 1987, Knutson & Yee, 1987, Andreason & Evans, 1989, Anderson, et al., 1989,
and Heiser, 1999 (for mammalian cells). For microorganisms, optimum electrotransformation occurs
using exponential decay pulses under electrical conditions similar to those used for E. coli and S. cerevisiae,
two species that are most commonly used in research today. The pulse conditions consist of a low
capacitance (short pulse duration) at high voltage. For E. coli, conditions that result in a field strength of
~18 kV/cm and a time constant of ~5 msec are optimal. For many bacterial species, including
Salmonella, Borrelia, Lactococcus, and Enterococcus, the conditions for electroporation are generally
identical to those used for E. coli. For many other bacterial species, altering the field strength will often
result in higher electrotransformation. For S. cerevisiae, conditions that result in a field strength of
~7.5 kV/cm and a time constant of ~5 msec are near optimal. Similar optimal conditions are also found
with other species of yeast.
Successful electroporation of plant protoplasts has been obtained using exponential decay pulses at
both low-capacitance and high-voltage settings or high-capacitance (long pulse duration) and low-voltage
settings. Typical conditions are time constants of 10–100 µsec and field strengths of 2.5 –5 kV/cm or
time constants of 2–5 msec and a field strength of about 500 V/cm.
For mammalian cells, optimum electrotransformation conditions have been reported using both
exponential decay and square wave pulses. Typical conditions for both types of pulses employ
pulse lengths or time constants of 10–40 msec and field strengths of 400–900 V/cm. Generally
there is an inverse relationship between field strength and pulse length or time constant such that,
over a limited range, one variable may be increased and the other decreased in order to maintain
the optimum electroporation conditions. Additionally, as cell size increases, the field strength at
which optimum electrotransformation occurs generally decreases. Other conditions that are
important for maximizing electroporation efficiency are discussed in the following sections.
5.1 Cell growth
The optimal portion of the growth phase to harvest cells is generally dependent on the cell type. When
preparing competent cells of a new species it is generally best to start with conditions employed for use within
the same genus. Suggestions for factors to consider and general methods for producing electrocompetent
cells are discussed in the articles by Dower, et al. (1992) and Trevors, et al. (1992). For most bacterial species,
the highest transformation efficiencies are obtained when cells are harvested in early to mid-log growth. With
E.coli, as the cells reach stationary phase, the transformation efficiency declines precipitously (Dower, 1990). In
contrast, most yeast species are generally harvested in mid- to late-log growth. For S. cerevisiae, the
transformation efficiency increases as much as 60-fold from early to late-log cultures (Becker and Guarente,
1991). For mammalian cells, the highest expression following electroporation is obtained when cells are in
mid-log phase growth (Anderson, et al., 1991).
48
Page 55

5.2 DNA
While the majority of electroporation applications involve delivery of plasmid DNA to cells, nearly any type
of molecule can be introduced into cells by electroporation, including RNA, proteins, carbohydrates, and
small molecules, such as nucleoside triphosphates and fluorescent dyes. With few exceptions, when
delivering autonomously replicating plasmids, the highest transformation efficiencies are obtained when
electroporating supercoiled plasmid; this is the case for microorganisms, plant, and animal cells.
However, integration of electroporated plasmid into the host genome is usually most efficient using linearized
plasmid, such as when isolating stable transformants of mammalian cells (Barsoum, 1995), or studying
homologous recombination in Candida (Thompson, et al., 1998), Pichia (Cregg & Russell, 1998), and
Tetrahymena (Gaertig & Gorovsky, 1995). Addition of carrier, such as salmon sperm DNA or plasmid has
also been shown to increase gene expression in some cell lines (Chu, et al., 1987; Showe, et al., 1990).
Although transformation of most cell types has been accomplished using plasmid DNA isolated by a variety
of methods, the plasmid purity has an effect on transformation efficiency. Significantly lower transformation
efficiencies are generated with unpurified miniprep plasmid DNA than with plasmid DNA that has been purified. Plasmid produced using the Bio-Rad Aurum matrix is as efficient as CsCl-purified plasmid for
transformation of both microorganisms and mammalian cells. However, as long as plasmids used for
are all prepared in the same manner, changes in expression levels are due to differences in transcription or
translation of the gene of interest.
Finally, when electroporating ligation mixtures into E. coli, components in the reactions have been shown
to interfere with transformation (Dower, 1990). Heating the ligation mixture (65°C for 15 min) followed
either by dilution with water (Willson and Gough, 1988), dialysis (Heery and Dunican, 1989; Jacobs et al.,
1990), or ethanol precipitation (Böttger, 1988; Zabarovsky and Winberg, 1990) significantly increases the
transformation efficiency of the DNA.
5.3 Electroporation Media
Many types of microorganisms are most efficiently electroporated in high resistance media
(resistance >3000 ohms). For this reason, when preparing electrocompetent cells, it is important to wash cells thoroughly to remove all traces of ions present in the growth media. Failure
to thoroughly remove the growth media from the cells may result in the sample arcing during
electroporation at high field strengths. Cells should be washed at least three times with water
or with non-ionic solutions, such as glucose, glycerol, sucrose, or sorbitol. For many microorganisms,
10–15% glycerol is a convenient electroporation medium, since it is recommended as a cryoprotectant for storage of cell cultures.
In some bacterial species, including low (~1 mM) amounts of MgCl
2
in the electrophoresis buffer increases
the transformation efficiency. At least in some bacteria, Mg
++
ions probably maintain the structural integrity
of the cell membrane. For example, Pseudomonas aeruginosa is very sensitive to divalent cation chelators;
incubation of cells in the presence of EDTA destabilizes the outer membrane of the cells (Haque & Russell,
1974). In the absence of Mg
++
ions, the membrane may not be able to re-form following electroporation.
Figs 5.1A and B show the effect of concentration of several biologically important ionic solutions on
sample resistance. Note that: (1) the volume of liquid in the cuvette has a significant effect on sample
resistance—for ionic solutions, sample resistance is inversely proportional to the volume of solution in
the cuvette; (2) the resistance of a solution containing divalent ions is lower than a solution containing
the same concentration of monovalent ions; and (3) the resistance of a buffered solution is affected by
its pH.
49
Page 56

The addition of even small concentrations of ionic compounds significantly reduces the resistance of
high resistance samples and may cause arcing. Residual salt from ethanol precipitation of DNA should
be removed by washing the DNA pellet prior to dissolving it in either water or Tris-EDTA. Table 5.1
shows that adding a solution of plasmid in 10 mM Tris, pH 8.0 –1 mM EDTA to water does reduce the
sample resistance. This should be considered when electroporating bacterial samples in high-resistance
media. DNA may be used directly from enzyme reactions for transformation, but the final salt concentration
in the electroporation sample should be kept below ~5 meq for high-voltage operation.
5.4 Temperature
The temperature at which cells are maintained during electroporation would be expected to have a role in
the efficiency of the electroporation process for several reasons. First, since passing an electric pulse
through the cells results in heating, keeping the cells at a low temperature during the pulse might reduce
the heating and therefore increase the cell viability. Second, since electroporation involves the transient formation of pores in the cell membrane, keeping cells at a low temperature after the pulse might allow the
pores to remain open longer giving the DNA in the medium more time to enter the cells. Alternatively, a
higher temperature may speed pore closure and increase cell viability. Third, changing the temperature of
a solution changes its conductivity. The conductivity of the media increases with increasing temperature,
resulting in a decrease in the media resistance and a decrease in the time constant, τ. Finally, diffusion
rate is highly dependent on temperature, and keeping cells at a low temperature would be expected to
reduce the diffusion of molecules across the cell membrane. In practice, the most efficient temperature at
which to electroporate cells must be determined empirically. Most bacterial cells are most electrocompetent
when maintained at <4°C from the time of harvest until pulsing. The transformation efficiency of E. coli
decreases by ~100-fold if pulsed at 20°C (Shigekawa & Dower, 1988); however, electrocompetent S.
aureus should be incubated at room temperature prior to electroporation (Lee, 1995). For most mammalian
cells, electroporation is most efficient when cells are maintained at room temperature both before and
after the pulse (Chu, et al., 1987), although some cell types are more efficiently transformed at low
temperature (Potter, et al., 1984).
50
Page 57

Fig. 5.1. Resistance of solutions (A) NaCl and MgCl2and of (B) buffers of NaPO4at pH 6.1 and 7.3
and HEPES at pH 7.5. Resistance was measured in 0.2 cm cuvettes containing either 40 µl or 200
µl of solution at room temperature.
51
Concentration (mM)
Concentration (mM)
0246810
0246810
4000
3500
3000
2500
2000
1500
1000
500
0
3500
3000
2500
2000
1500
1000
500
0
B
A
Sample resistance (ohms)
Sample resistance (ohms)
NaCI (40 µl)
NaCI (200 µl)
MgCI
2
(40 µl)
MgCI
2
(200 µl)
PO
4
6.1, 40 µl
PO46.1, 200 µl
PO
4
7.3, 40 µl
PO
4
7.3, 200 µl
HEPES, 7.5, 40 µl
HEPES, 7.5, 200 µl
Page 58

Table 5.1. Resistance of Water in 0.2 cm Cuvettes to which TE has been added
1
SAMPLE R
sample
(ohms) R
sample
(ohms)
(40 µl volume) (200 µl volume)
Water > 600,000 > 600,000
Water + 1 µl TE > 600,000 35,000
Water + 5 µl TE 11,200 8,700
Water + 10 µl TE 4,850 4,700
1
Sample resistance in 0.2 cm cuvettes containing either 40 or 200 µl of water and the indicated volume
of TE (10 mM Tris, pH 8.0, 1 mM EDTA) measured at 1000 V.
Section 6
Electroporation of Bacterial Cells
For most bacterial species, electroporation provides a rapid and simple method for transforming cells.
Particularly for gram negative bacteria, but also for some gram positive species, this method provides
higher transformation efficiencies than conjugation or chemical methods. We are interested in hearing of
additional species and strains transformed by electroporation and including this information in subsequent
versions in our Electroprotocols manual. Please contact your local Bio-Rad representative, access our
web site at www.bio-rad.com, or, in the U.S., call our Technical Services at (800) 424-6723 with any
comments or questions.
6.1 E. coli
6.1.1 Preparation of Electrocompetent Cells
See Ausubel, et al. (1987) and Miller and Nickoloff (1995) for additional information.
Gene Pulser Xcell conditions: C = 25 µF; PC = 200 ohm; V = 1.8 kV (in 0.1 cm cuvettes);
C = 25 µF; PC = 200 ohm; V = 2.5 kV, or
C = 25 µF; PC = 200 ohm; V = 3.0 kV (in 0.2 cm cuvettes).
This procedure requires a Gene Pulser Xcell main unit and PC Module.
1. Inoculate 5 ml of a fresh overnight E. coli culture into 500 ml of L-broth in a 2.8 L Fernbach flask.
2. Grow the cells at 37°C shaking at 300 rpm to an OD
600
of approximately 0.5–0.7. The best results
are obtained with cells that are harvested at early- to mid-log phase; the appropriate cell density
depends on the strain and growth conditions but should be about 4–5 x 10
7
cells/ml.
3. Chill the cells on ice for ~20 min. For all subsequent steps, keep the cells as close to 0°C as possible (in an ice/water bath) and chill all containers in ice before adding cells. Transfer the cells to a
sterile, cold 500 ml centrifuge bottle and centrifuge at 4000 x g for 15 minutes at 4°C.
4. Carefully pour off and discard the supernatant. It is better to sacrifice yield by pouring off a few
cells than to leave any supernatant behind.
5. Gently resuspend the pellet in 500 ml of ice-cold 10% glycerol. Centrifuge at 4000 x g for
15 minutes at 4°C; carefully pour off and discard the supernatant.
52
Page 59

53
6. Resuspend the pellet in 250 ml of ice-cold 10% glycerol. Centrifuge at 4000 x g for 15 minutes at
4°C; carefully pour off and discard the supernatant.
7. Resuspend the pellet in ~20 ml of ice-cold 10% glycerol. Transfer to a 30 ml sterile Oakridge tube.
Centrifuge at 4000 x g for 15 minutes at 4°C; carefully pour off and discard the supernatant.
8. Resuspend the cell pellet in a final volume of 1–2 ml of ice-cold 10% glycerol. The cell
concentration should be about 1–3 x 10
10
cells/ml.
9. This suspension may be frozen in aliquots on dry ice and stored at -70°C. The cells are stable for
at least 6 months under these conditions.
6.1.2 Electroporation
1. Thaw the cells on ice. For each sample to be electroporated: place a 1.5 ml microfuge tube on
ice, place either a 0.1 or 0.2 cm electroporation cuvette on ice, and place a 17 x 100 mm tube
with 1 ml of SOC at room temperature.
2. To a cold, 1.5 ml polypropylene microfuge tube, add 20 µl of cell suspension if electroporating in 0.1 cm
cuvettes, or 20–40 µl of cell suspension if electroporating in 0.2 cm cuvettes. Add 1 to 2 µl of DNA
(DNA should be in a low ionic strength buffer such as water or TE). Mix well and incubate on ice for
~1 minute. (Note: it is best to mix the plasmids and cells in a microfuge tube since the narrow gap of
the cuvettes prevents uniform mixing.)
3. From the Home screen on Gene Pulser Xcell open the Pre-set Protocols screen, then the Bacterial
Protocol screen (press 4, then Enter twice). When using the 0.1 cm cuvettes, press Enter to open
E. coli, 1mm cuvette Protocol Detail screen. When using the 0.2 cm cuvettes, press 2 then Enter, or
3 then Enter, to select the Protocol Detail screens for E. coli to pulse at 2.5 or 3.0 kV, respectively.
See Section 3.4 for operating instructions.
4. Transfer the mixture of cells and DNA to a cold electroporation cuvette and tap the suspension to
the bottom. Place the cuvette in the ShockPod. Push the chamber lid down to close.
5. Pulse once.
6. Remove the cuvette from the chamber and immediately add 1 ml of SOC medium to the cuvette.
Quickly but gently resuspend the cells with a Pasteur pipette. (The period between applying the
pulse and transferring the cells to outgrowth medium is crucial for recovering E. coli transformants
(Dower et al., 1988). Delaying this transfer by even 1 minute causes a 3-fold drop in transformation.
This decline continues to a 20-fold drop by 10 minutes.)
7. Transfer the cell suspension to a 17 x 100 mm polypropylene tube and incubate at 37°C for 1 hour,
shaking at 225 rpm.
8. Check and record the pulse parameters. The time constant should be close to 5 milliseconds. The
field strength can be calculated as actual volts (kV) / cuvette gap (cm).
9. Plate on LB plates with antibiotic.
6.1.3 Solutions and Reagents
1. L-Broth: 10 g Tryptone peptone, 5 g Yeast extract, 5 g NaCl; dissolve in 1.0 L water.
Autoclave.
2. LB agar plates with selective antibiotic: prepare L broth as above, adding 15 g of agar per
liter. Autoclave. Cool to 55–60°C and add antibiotic. Pour 12–15 ml per 100 mm plate.
3. 10% (v/v) Glycerol: 12.6 g glycerol (density = 1.26 g/cc) in 90 ml of water. Autoclave or filter
sterilize.
4. TE: 10 mM Tris-HCl pH 8.0, 1 mM EDTA.
Page 60

5. SOB: 2.0 g Tryptone peptone, 0.5 g Yeast extract, 0.2 ml 5 M NaCl, 0.25 ml 1 M KCl; dissolve
in 90 ml water. Adjust pH to 7.0. Bring volume to 100 ml. Autoclave. Add 1.0 ml sterile
1 M MgCl2and 1.0 ml sterile 1 M MgSO4.
6. SOC: to 100 ml SOB, add 2.0 ml sterile 1 M glucose (sterilize by filtration).
6.2 Staphylococcus aureus
6.2.1 Preparation of Electrocompetent Cells
See Lee (1995) for additional information.
Using this method, we have obtained transformation efficiencies of >1 x 10
6
transformants/µg
electroporating S. aureus RN4220 with pLI50.
Gene Pulser Xcell conditions: C = 25 µF; PC = 100 ohm; V = 2.9 kV.
This procedure requires a Gene Pulser Xcell main unit and PC Module.
1. Inoculate a colony of S. aureus from a TSA plate into 3 ml of B2 broth in a 17 x 100 mm tube.
2. Incubate at 37°C overnight, shaking at 250 rpm.
3. Inoculate 1.5 ml of the overnight culture into 150 ml of B2 broth in a 1 liter flask. Incubate at 37°C,
shaking at 250 rpm, to 3–5 x 10
8
cells/ml (OD
600
~ 0.8–0.85). The doubling time of S. aureus is
about 30 min at 37°C; growth will take about 4 hrs.
4. Chill the cells in an ice water bath for 15 min to stop growth. Decant the cells into a sterile 500 ml
centrifuge bottle. Harvest the cells by centrifugation at 12,000 x g for 15 min at 4°C.
5. Carefully pipette off the supernatant, keeping the cell pellet on ice.
6. Resuspend the cell pellet in 500 ml of sterile, ice-cold water. Pellet the cells by centrifugation at
12,000 x g for 15 min at 4°C; carefully remove the supernatant. Wash the cells 2 more times in
500 ml of sterile, ice-cold water.
7. Resuspend the cell pellet in 25 ml of sterile, ice-cold 10% glycerol. Transfer to a 30 ml sterile
Oakridge tube. Pellet the cells by centrifugation at 12,000 x g for 15 min at 4°C; carefully remove
the supernatant.
8. Resuspend the cells in 20 ml of 10% glycerol. Incubate at 20°C for 15 min. Pellet the cells by
centrifugation at 12,000 x g for 15 min at 4°C; carefully remove the supernatant.
9. Resuspend the cell pellet in 2 ml of 10% glycerol; the final cell concentration should be
~1 x 10
10
cells/ml.
10. Dispense 250 µl aliquots of the electrocompetent cells into sterile 1.5 ml microfuge tubes; freeze
the cells in an isopropanol-dry ice bath, then store at -70°C. The cells are stable for several months
under these conditions.
6.2.2 Electroporation
1. Pipette the DNA samples (5 ng - 2 µg in no more than 3 µl) to be electroporated into sterile 1.5 ml
microfuge tubes.
2. Thaw the competent cells at room temperature for several minutes. Add 50 µl of cells to each DNA
sample; gently pipette up and down to mix. Incubate the samples at room temperature for 30 min.
3. For each sample to be electroporated, add 1 ml of SMMP medium containing a subinhibitory concentration of antibiotic to a 17 x 100 mm tube at room temperature and place a 0.2 cm cuvette at
room temperature.
54
Page 61

4. From the Home screen on Gene Pulser Xcell open the Pre-set Protocols screen, then the Bacterial
Protocol screen (press 4, then Enter twice). To select the Protocol Detail screen for S. aureus, press
6 or the Down Arrow key to highlight S. aureus, then press Enter. See Section 3.4 for operating
instructions.
5. Transfer the mixture of cells and DNA to a 0.2 cm electroporation cuvette at room temperature and
tap the suspension to the bottom. Place the cuvette in the ShockPod. Push the chamber lid down
to close.
6. Pulse once.
7. Remove the cuvette from the chamber and immediately add 1 ml of SMMP medium containing a
subinhibitory concentration of antibiotic; gently transfer the cells to a sterile 17 x 100 mm tube using
a Pasteur pipette. Incubate 1 hr at 37°C, shaking at 250 rpm.
8. Check and record the pulse parameters. The time constant should be close to 2.5 milliseconds. The
voltage should be approximately 2.9 kV.
9. Plate aliquots of the electroporated cells on trypticase soy agar containing selective antibiotic.
Incubate plates for 36–48 hrs at 37°C.
6.2.3 Solutions and Reagents
1. Trypticase soy agar (TSA): dissolve 40 g trypticase soy agar (Becton Dickinson, Sparks, MD) in 1 L
of water. Autoclave.
2. B2 medium: 10 g casein hydrolysate (casamino acids), 25 g yeast extract, 5 g glucose, 25 g
NaCl, 1 g K2HPO4; dissolve in 900 ml of water and adjust pH to 7.5; bring volume to 1.0 L.
Autoclave.
3. 0.2 M sodium hydrogen maleate: either 11.6 g maleic anhydride and 4 g NaOH dissolved in 500
ml of water, or 13.8 g maleic acid, 4 g NaOH dissolved in 500 ml water. Autoclave.
4. 2X SMM: 25 ml of 0.2 M sodium hydrogen maleate, 40 ml of 0.1 N NaOH; adjust the pH to 6.5. Add
5 ml of 1 M MgCl
2
, 42.7 g sucrose; dissolve and bring volume to 125 ml. Filter sterilize.
5. 4X Penassay broth: 17.5 g Antibiotic Medium 3 (Becton Dickinson) dissolved in 250 ml of water.
Autoclave.
6. SMMP: 55 ml of 2X SMM, 40 ml of 4X Penassay broth, 5 ml of 10% (w/v) bovine albumin; adjust
pH to 7.0; filter sterilize. For selection of antibiotic-resistant transformants, add a sub-inhibitory
concentration of antibiotic.
7. 10% (v/v) Glycerol: 12.6 g glycerol (density = 1.26 g/cc) in 90 ml of water. Autoclave or filter sterilize.
55
Page 62

6.3 Agrobacterium tumefaciens
6.3.1 Preparation of Electrocompetent Cells
See Lin (1995) for additional information.
Gene Pulser Xcell conditions: C = 25 µF; PC = 200 ohm; V = 2.4 kV.
This procedure requires a Gene Pulser Xcell main unit and PC Module.
1. Inoculate an aliquot from log phase culture of A. tumefaciens into 1.5 L of YM broth in a 2.8 L
Fernbach flask.
2. Incubate at 30°C overnight, shaking at 300 rpm to a density of 5–10 x 10
7
cells/ml (OD550 ~ 1.0).
3. Decant the cells into sterile 500 ml centrifuge bottles and pellet the cells by centrifugation at 3000 x g
for 10 min at 4°C.
4. Carefully pour off and discard the supernatant; place the centrifuge bottles with the cell pellets on
ice.
5. Add ~50 ml of sterile, ice-cold 10% glycerol to each of the bottles and vortex to resuspend the cell
pellets; bring the volume in each of the centrifuge bottles to 500 ml with sterile, ice-cold 10% glycerol.
Pellet the cells by centrifugation at 3000 x g for 10 min at 4°C; pour off and discard the supernatant.
6. Combine the cell pellets and wash the cells in one bottle as in step 5.
7. Resuspend the cell pellets in 25 ml of sterile, ice-cold 10% glycerol and transfer to a chilled 30 ml
Oakridge tube. Pellet the cells by centrifugation at 3000 x g for 5 min at 4°C; pour off and discard
the supernatant.
8. Resuspend the cell pellet in 0.5 ml of sterile, ice-cold 10% glycerol; the final cell volume should be
~1.5 ml and the cell concentration should be ~ 5 x 10
10
cells/ml. The cells may be used immediately
or frozen. To freeze, dispense 200 µl aliquots of the electrocompetent cells in sterile 1.5 ml
microfuge tubes; freeze the cells in an isopropanol-dry ice bath, then store at -70°C. The cells are
stable for about 6 months under these conditions.
6.3.2 Electroporation
1. Pipette the DNA samples (up to 100 ng in no more than 5 µl of TE) to be electroporated into sterile
1.5 ml microfuge tubes. Place tubes on ice.
2. Thaw the electrocompetent A. tumefaciens cells on ice. For each DNA sample to be electroporated,
add 20 µl of electrocompetent cells to each DNA sample; gently tap the tubes to mix.
3. For each DNA sample to be electroporated, add 1 ml of YM broth to a 17 x 100 tube at room
temperature and place a 0.1 cm electroporation cuvette on ice.
4. From the Home screen on Gene Pulser Xcell open the Pre-set Protocols screen, then the Bacterial
Protocol screen (press 4, then Enter twice). To select the Protocol Detail screen for A. tumefaciens,
press 4 or the Down Arrow key to highlight A. tumefaciens, then press Enter. See Section 3.4 for
operating instructions.
5. Transfer the DNA - cell samples to the electroporation cuvettes and tap the suspension to the bottom. Place the cuvette in the ShockPod. Push the chamber lid down to close. Pulse once.
6. Remove the cuvette from the chamber and immediately use the 1 ml of YM broth in the
17 x 100 mm tube to transfer the cells from the cuvette to the tube.
7. Check and record the pulse parameters. The time constant should be about 5 milliseconds. The
voltage should be about 2.4 kV.
56
Page 63

8. Incubate the cells 3 hr at 30°C, shaking at 250 rpm. Plate aliquots of the electroporated cells on
YM agar plates containing the appropriate selective media. Incubate plates for 48 hrs at 30°C.
6.3.3 Solutions and reagents
1. YM broth: 0.4 g yeast extract, 10 g mannitol, 0.1 g NaCl, 0.1 g MgSO4, 0.5 g K2HPO4.3H20,
dissolve in 1.0 L of water and adjust to pH 7.0. Autoclave. For YM plates, add 15 g agar/1 L of YM
broth.
2. 10% (v/v) Glycerol: 252 g glycerol (density = 1.26 g/cc) in 1800 ml water. Autoclave or filter sterilize.
6.4 Bacillus cereus
6.4.1 Preparation of Electrocompetent Cells
Slight modification of the method of Silo-Suh, et al. (1994).
Using this method, we have obtained transformation efficiencies of 2 x 10
6
transformants/µg
electroporating B. cereus UW85 with pAD123.
Gene Pulser Xcell conditions: C = 50 µF; PC = 200 ohm; V = 1.0 kV.
This procedure requires a Gene Pulser Xcell main unit and PC Module.
1. Inoculate a colony of B. cereus into 10 ml of LB in a 125 ml flask; incubate overnight at 28°C
shaking at 300 RPM.
2. Inoculate 2 ml of the overnight culture into 500 ml of BHI in a 2.8 L Fernbach flask; incubate at
28°C shaking at 300 RPM.
3. Monitor the culture until OD
600
= 0.3 (cell density is ~1 x 107cells/ml).
4. Chill the culture on ice for 10 min.
5. Transfer the cells into 2 chilled, sterile 250 ml centrifuge bottles.
6. Pellet the cells by centrifugation at 4°C for 10 min at 10,000 x g in a chilled rotor.
7. Discard the supernatant. Resuspend each pellet in 50 ml of sterile, ice cold EP buffer. Pellet the
cells as above.
8. Discard the supernatant. Resuspend each pellet in 15 ml of sterile, ice cold EP buffer. Transfer to a
chilled 30 ml Oakridge tube.
9. Pellet the cells by centrifugation at 4°C for 10 min at 10,000 x g in a chilled rotor.
10. Discard the supernatant. Resuspend the cell pellet in 0.5 ml of EP buffer. Keep the cells on ice and
use as soon as possible for electroporation. Cells cannot be frozen and used for electroporation.
6.4.2 Electroporation
1. For each sample to be electroporated, pipette the DNA (0.05 - 1 ug in up to 10 µl of either water
or TE) into a sterile 1.5 ml microfuge tube. Place tubes on ice. Add 100 µl electrocompetent cells
to each plasmid sample; mix, and incubate on ice for 5–10 min.
2. Transfer 100 µl of plasmid/cell suspension to chilled 0.2 cm cuvettes on ice; tap the solution to the
bottom of the cuvette.
3. For each DNA sample to be electroporated, prepare a 17 x 100 tube containing 2.0 ml of LB at
room temperature.
57
Page 64

4. From the Home screen on Gene Pulser Xcell open the Pre-set Protocols screen, then the Bacterial
Protocol screen (press 4, then Enter twice). To select B. cereus, press 7 or the Down Arrow to
highlight B. cereus, then press Enter to open the B. cereus Protocol Detail screen. See Section 3.4
for operating instructions.
5. Transfer the DNA - cell samples to the electroporation cuvettes and tap the suspension to the
bottom. Place the cuvette in the ShockPod. Push the chamber lid down to close. Pulse once.
6. Remove the cuvette from the chamber and immediately use a portion of the L broth in the
17 x 100 mm tube to transfer the cells from the cuvette to the tube.
7. Check and record the pulse parameters. The time constant should be about 8.6 msec and the
voltage about 1.0 kV. The field strength can be calculated as actual volts (kV) / cuvette gap (cm).
8. Incubate the cells for 1–1.5 hr at 37°C, shaking at 250 rpm.
9. Plate aliquots of the electroporated cells on LB agar plates containing the appropriate selective
media. Incubate plates overnight at 28°C.
6.4.3 Solutions and Reagents
1. EP buffer (0.5 mM K
2
HPO4–KH2PO4, 0.5 mM MgCl2, 272 mM Sucrose): 54.4 ml 1M Sucrose,
100 µl 1 M MgCl
2
, 190 µl 0.1 M KH2PO4, 810 µl 0.1 M K2HPO4. Bring volume to 200 ml with
Milli-Q water (the pH should be ~7.4). Filter sterilize. Store at 4°C.
2. Brain Heart Infusion (BHI): dissolve 37 g BHI in 1000 ml water; autoclave.
3. L-Broth: 10 g Tryptone peptone, 5 g Yeast extract, 5 g NaCl; dissolve in 1.0 L water. Autoclave.
6.5 Pseudomonas aeruginosa
6.5.1 Preparation of Electrocompetent Cells
From Dennis & Sokol (1995) and Smith & Iglewski (1989).
Using this method, electroporating P. aeruginosa PAO1 with pUCP19, we have obtained transformation
efficiencies >1 x 10
7
transformants/µg.
Gene Pulser Xcell conditions: C = 25 µF; PC = 200 ohm; V = 2.5 kV.
This procedure requires a Gene Pulser Xcell main unit and PC Module.
1. Inoculate a colony of P. aeruginosa into 25 ml of LB in a 125 ml flask; incubate overnight at 37°C
shaking at 300 RPM.
2. Inoculate 10 ml of the overnight culture into a 2.8 L Fernbach flask with 500 ml of LB; incubate at
37°C shaking at 300 RPM.
3. Monitor the culture until OD
600
= 0.5; the cell density should be ~3 x 108cells/ml.
4. Chill the flask on ice for 10 min.
5. Transfer the cells into 2 chilled, sterile 250 ml centrifuge bottles.
6. Pellet the cells by centrifugation at 4°C for 10 min at 2300 x g in a chilled rotor.
7. Discard the supernatant. Resuspend each pellet in 250 ml of sterile, ice cold SMEB buffer. Pellet
the cells as above.
8. Discard the supernatant. Resuspend each pellet in 125 ml of sterile, ice cold SMEB buffer. Pellet
the cells as above.
58
Page 65

9. Discard the supernatant. Resuspend each pellet in 12 ml of sterile, ice cold SMEB buffer. Transfer
to a chilled 30 ml Oakridge tube.
10. Pellet the cells by centrifugation at 4°C for 10 min at 2300 x g in a chilled rotor.
11. Discard the supernatant. Resuspend the cell pellet in 2.0 ml of SMEB buffer. Keep the cells on ice
and use as soon as possible for electroporation.
6.5.2 Electroporation
1. For each sample to be electroporated, pipette 0.05–0.5 µg of plasmid into a sterile 1.5 ml microfuge
tube. Place tubes on ice. Add 100 µl electrocompetent P. aeruginosa cells to each plasmid sample;
mix, and incubate on ice for 5–10 min.
2. Transfer 100 µl of plasmid/cell suspension to chilled 0.2 cm cuvettes on ice; tap the solution to the
bottom of the cuvette.
3. For each DNA sample to be electroporated, prepare a 17 x 100 tube containing 1.0 ml of SOC at
room temperature.
4. From the Home screen on Gene Pulser Xcell open the Pre-set Protocols screen, then the Bacterial
Protocol screen (press 4, then Enter twice). To select P. aeruginosa, press 5 or the Down Arrow to
highlight P. aeruginosa, then press Enter to open the P. aeruginosa Protocol Detail screen. See
Section 3.4 for operating instructions.
5. Place the cuvette in the ShockPod. Push the chamber lid down to close. Pulse once.
6. Immediately after the pulse, use the 1 ml of SOC broth in the 17 x 100 mm tube to transfer the
plasmid/cell suspension from the cuvette to the tube. Check and record the pulse parameters. The
time constant should be about 5 msec and the voltage about 2.5 kV. The field strength can be
calculated as volts (kV) / cuvette gap (cm).
7. Incubate the cells for 1 hr at 37°C shaking at 250–300 RPM.
8. Plate onto LB agar plates with selective antibiotic. Incubate overnight at 37°C.
6.5.3 Solutions and Reagents
1. SMEB buffer (1 mM HEPES, pH 7.0, 1 mM MgCl2, 300 mM Sucrose): 2.0 ml 0.5 M HEPES, pH
7.0, 1.0 ml 1.0 M MgCl
2
, 102.7 g Sucrose. Dissolve in water and bring volume to 1000 ml.
Autoclave. Store at 4°C.
2. L-Broth: 10 g Tryptone peptone, 5 g Yeast extract, 5 g NaCI; dissolve in 1 L water. Autoclave.
3. LB agar plates with selective antibiotic: prepare L broth as above, adding 15 g of agar per liter.
Autoclave. Cool to 55–60°C and add antibiotic. Pour 12–15 ml per 100 mm plate.
4. SOB: 2.0 g Tryptone peptone, 0.5 g Yeast extract, 0.2 ml 5 M NaCl, 0.25 ml 1 M KCl; dissolve in
90 ml water. Adjust pH to 7.0. Bring volume to 100 ml. Autoclave. Add 1.0 ml sterile 1 M MgCl
2
and 1.0 ml sterile 1 M MgSO4.
5. SOC: to 100 ml SOB, add 2.0 ml sterile 1 M glucose (sterilize by filtration).
59
Page 66

6.6 Streptococcus pyogenes
6.6.1 Preparation of Electrocompetent Cells
From Simon & Ferretti (1995).
Using this method, we have obtained transformation efficiencies of 1 x 105transformants/µg
electroporating S.pyogenes CS101 with pDC123 (Chaffin & Rubens, 1998).
Gene Pulser Xcell conditions: C = 25 µF; PC = 200 ohm; V = 2.1 kV.
This procedure requires a Gene Pulser Xcell main unit and PC Module.
1. Inoculate a colony of S. pyogenes from a fresh plate into a 25 ml tube containing 20 ml of THY
with 20 mM Glycine; cap the tube tightly. Incubate overnight at 37°C without shaking.
2. Inoculate 18 ml of the overnight culture into a 500 ml flask containing 250 ml of THY media with 20
mM glycine. Swirl gently to mix and continue incubation (about 2 hrs) until the OD
600
is ~0.2 (the
cell density should be about 3 x 10
7
cells/ml).
3. Chill the culture on ice for 10 min.
4. Transfer the cells into a chilled, sterile 500 ml centrifuge bottle.
5. Pellet the cells by centrifugation at 4°C for 10 min at 6000 x g in a chilled rotor.
6. Pipette off and discard the supernatant. Resuspend the cell pellet in 10 ml of ice-cold sterile Elpo
medium, then bring the volume to ~225 ml. Centrifuge the cells at 8000 x g.
7. Pipette off and discard the supernatant. Resuspend the pellet in 5 ml of ice-cold sterile Elpo medium
and transfer to a 30 ml Oakridge tube. Bring the volume to 30 ml, then centrifuge the cells at
12,000 x g.
8. Pipette off and discard the supernatant. Resuspend the cell pellet in about 1.5 ml of ice-cold sterile
Elpo medium. Keep the cells on ice and use as soon as possible for electroporation.
6.6.2 Electroporation
1. Pipette the DNA samples to be electroporated (0.05–0.5 µg) into sterile 1.5 ml microfuge tubes.
Place tubes on ice. Add 200 µl electrocompetent S. pyogenes cells to each plasmid sample; mix,
and incubate on ice for 5–10 min.
2. Transfer 200 µl of plasmid/cell suspension to chilled 0.2 cm cuvettes on ice; tap the solution to the
bottom of the cuvette.
3. For each sample to be electroporated, prepare a sterile 17 x 100 mm tube containing 1.0 ml of
THY at room temperature.
4. From the Home screen on Gene Pulser Xcell open the Pre-set Protocols screen, then the Bacterial
Protocol screen (press 4, then Enter twice). To select S. pyogenes, press 8 or the Down Arrow to
highlight S. pyogenes, then press Enter to open the S. pyogenes Protocol Detail screen. See
Section 3.4 for operating instructions.
5. Place the cuvette in the ShockPod. Push the chamber lid down to close. Pulse once.
6. Immediately after the pulse place the cuvette on ice for ~ 5 min. Check and record the pulse
parameters. The time constant should be about 9.8 msec and the voltage about 2.1 kV. The field
strength can be calculated as volts (kV) / cuvette gap (cm).
7. Add 1.0 ml of THY broth to the cuvette and transfer the plasmid/cell suspension to the
17 x 100 mm tube. Incubate the cells for 2 hr at 37°C without shaking.
60
Page 67

8. Plate onto THY agar plates with selective antibiotic. Incubate 1–2 D at 37°C.
6.6.3 Solutions and reagents
1. THY media (Todd-Hewitt broth with 0.2% yeast extract): 30 g Todd Hewitt broth, 2 g Yeast
extract; dissolve in 1 L water. Autoclave.
2. THY + Glycine: 500 ml THY media, 5 ml 2 M Glycine (15 g/100 ml).
3. THY agar with antibiotic: 30 g Todd-Hewitt broth, 2 g Yeast extract, 15 g Bacto agar; dissolve in
1 L water. Autoclave; cool to 55–60°C and add antibiotic. Pour plates.
4. Elpo medium: 122 ml 20% Glucose, 0.5 ml 1 M MgCl
2
; bring to 500 ml with water and pH to 6.5.
Filter sterilize.
6.7 Lactobacillus plantarum
6.7.1 Preparation of Electrocompetent Cells
From Bringel & Hubert (1990). Using this method, we have obtained transformation efficiencies of 4 x 10
4
transformants/µg electroporating L.plantarum NCIB7220 with pGK12.
Gene Pulser Xcell conditions: C = 25 µF; PC = 400 ohm; V = 2.0 kV.
This procedure requires a Gene Pulser Xcell main unit and PC Module.
1. Inoculate L. plantarum into 50 ml of MRS media in a 500 ml flask; incubate overnight at 30°C shaking
at 300 RPM. The cells should be in exponential growth (OD
600
< 1.5).
2. Inoculate a sample of the overnight culture into 500 ml of MRS/SG in a 2.8 L Fernback flask to an
OD
600
= 0.05. Incubate at 30°C shaking at 80 RPM.
3. Monitor the culture until OD
600
= 0.3 (usually ~ 4 hrs).
4. Transfer the cells into a sterile 500 ml centrifuge bottle at room temperature.
5. Pellet the cells by centrifugation at 18°C for 10 min at 3000 x g.
6. Discard the supernatant. Resuspend the cell pellet in 250 ml of sterile water at room temperature.
Centrifuge the cells as above but increase the speed of to 4000 x g.
7. Discard the supernatant. Repeat the water wash as above. Pellet the cells as above.
8. Discard the supernatant. Resuspend the cell pellet in a final volume of 2 ml of 30% PEG. Keep the
cells on ice and use as soon as possible for electroporation.
6.7.2 Electroporation
1. For each sample to be electroporated, pipette 0.1–1.0 µg of plasmid into a sterile 1.5 ml microfuge
tube (<2 µl). Place tubes on ice. Add 40 µl electrocompetent cells to each plasmid sample; mix.
2. Transfer 40 µl of plasmid/cell suspension to chilled 0.2 cm cuvettes on ice; tap the solution to the
bottom of the cuvette.
3. From the Home screen on Gene Pulser Xcell open the Pre-set Protocols screen, then the Bacterial
Protocol screen (press 4, then Enter twice). To select L. plantarum, press 9 or the Down Arrow to
highlight L. plantarum, then press Enter to open the L. plantarum Protocol Detail screen. See
Section 3.4 for operating instructions.
4. Place the cuvette in the ShockPod. Push the chamber lid down to close. Pulse once.
61
Page 68

5. Immediately after the pulse place the cuvette on ice for 30 min. Add 1 ml MRS/SG to the cuvette
and transfer the plasmid/cell suspension to a sterile, 17 x 100 mm tube. Incubate the cells for
3–4 hrs at 30°C without shaking.
6. Plate onto MRS agar plates containing antibiotic.
7. Incubate 2–3 D at 30°C.
6.7.3 Solutions and Reagents
1. 2X MRS (w/o Glucose): 10 g Proteose Peptone No 3 (Difco #211693), 10 g Beef Extract (Difco
#211520), 5 g Yeast Extract (Difco #212750), 1 ml Tween 80, 11.5 ml 1 M K
2
HPO4, 12.3 ml 3 M
Na Acetate, 1.9 g citric acid, 0.8 ml 1 M MgSO
4
, 225 µl 1 M MnSO4. Dissolve in water and bring
the volume to 500 ml. Autoclave
2. 1 X MRS: 500 ml 2 X MRS, 200 ml 10% Glucose (filter sterilized), 300 ml sterile water
3. 1 X MRS Agar with antibiotic: To a 1 L bottle, add 7.5 g Bacto agar and 150 ml water;
autoclave. Cool to 55°C. Warm 250 ml 2 X MRS and 100 ml 10 % Glucose to 55°C; add to the
agar. Add antibiotic. Pour plates.
4. MRS/SG (MRS with 0.75 M Sorbitol + 1% Glycine): 250 ml 2 X MRS, 50 ml 20 % Glucose (sterilized
by filtration), 150 ml sterile 2.5 M Sorbitol, 33 ml sterile 2 M Glycine, 17 ml sterile water.
5. 2 M Glycine: 15.0 g in 100 ml water; filter sterilize.
6. 2.5 M Sorbitol: 227.8 g Sorbitol; dissolve in ~250 ml water then bring volume to 500 ml. Filter
sterilize.
7. 30% PEG 1000: 150 g PEG 1000 (warm to ~50°C to melt; pour into beaker with ~200 ml water).
Bring volume to 500 ml with water. Filter sterilize.
Section 7
Electroporation of Fungal Cells
7.1 Saccharomyces cerevisiae
7.1.1 Preparation of electrocompetent cells
See Becker & Guarante (1991) and Ausubel et al. (1987) for additional information. Using this procedure
we have obtained >5 x 105transformants/µg electroporating S. cerevisiae Sc948 with Yep352.
Gene Pulser Xcell conditions: C = 25 µF; PC = 200 ohm; V = 1.5 kV (in 0.2 cm cuvettes), or
C = 25 µF; PC = 200 ohm; V = 2.5 kV (in 0.4 cm cuvettes).
This procedure requires a Gene Pulser Xcell main unit and PC Module.
1. Inoculate 500 ml of YPD in a 2.8 L Fernbach flask with an aliquot from an overnight culture of
S. cerevisiae. The doubling time of S. cerevisiae is approximately 2 hrs at 30°C.
2. Incubate at 30°C overnight, shaking at 250 rpm, to a density of ~ 1 x 10
8
cells/ml (OD
600
(1:10
dilution) = 0.30–0.35).
3. Chill the cells in an ice water bath for 15 min to stop growth.
4. Decant the cells into a sterile 500 ml centrifuge bottle and pellet the cells by centrifugation at
4000 x g for 5 min at 4°C.
62
Page 69

5. Carefully pour off and discard the supernatant; place the centrifuge bottles with the cell pellets on
ice.
6. Add 80 ml of sterile water to the bottle and vortex to resuspend the cell pellet.
7. Add 10 ml of 10 X TE buffer; swirl to mix. Add 10 ml of Lithium Acetate stock solution; swirl to mix.
Incubate 45 min at 30°C shaking at ~85 RPM.
8. Add 2.5 ml of 1 M DTT; swirl to mix. Incubate 15 min at 30°C shaking at ~85 RPM.
9. Bring the volume to 500 ml with sterile water. Pellet the cells by centrifugation at 4000 x g for 5 min
at 4°C; pour off and discard the supernatant.
10. Add ~50 ml of sterile, ice cold water to the bottle and vortex to resuspend the cell pellet; bring the
volume to 500 ml with sterile, ice-cold water. Pellet the cells by centrifugation at 4000 x g for 5 min
at 4°C; pour off and discard the supernatant.
11. Resuspend the cell pellet in 25–30 ml of sterile, ice-cold 1 M sorbitol and transfer to a chilled 30 ml
Oakridge tube. Pellet the cells by centrifugation at 4000 x g for 5 min at 4°C; pour off and discard
the supernatant.
12. Resuspend the cell pellet in 0.5 ml of sterile, ice-cold 1 M sorbitol; the final cell volume should be
1.3–1.5 ml and the cell concentration should be ~1 x 10
10
cells/ml. Keep the cells on ice and use
as soon as possible for electroporation.
7.1.2 Electroporation
1. For each sample to be electroporated, prepare a 17 x 100 mm sterile tube with 1 ml of 1 M
Sorbitol and place on ice; also, place a 0.2 cm or 0.4 cm cuvette on ice.
2. Pipette the DNA samples (5–100 ng in a volume of 5 µl) to be electroporated into sterile 1.5 ml
microfuge tubes. Place tubes on ice.
3. If 0.2 cm cuvettes are used, add 40 µl of the competent cells to each DNA sample; if 0.4 cm
cuvettes are used, add 80 µl of the competent cells to each DNA sample. Mix gently and incubate
on ice for ~5 min.
4. From the Home screen on Gene Pulser Xcell open the Pre-set Protocols screen, then the Fungal
Protocol screen (press 4, Enter, then 2, Enter). When using 0.2 cm cuvettes, press Enter to open
the S. cerevisiae, 2 mm cuvette Protocol Detail screen. When using 0.4 cm cuvettes, press 2 then
Enter to open the S. cerevisiae, 4 mm Protocol Detail screen. See Section 3.4 for operating
instructions.
5. Transfer the DNA - cell samples to the appropriate electroporation cuvettes that have been chilled
in ice and tap the suspension to the bottom. Place the cuvette in the ShockPod. Push the lid down
to close. Pulse once.
6. Remove the cuvette from the chamber, immediately add 1 ml of ice cold 1 M sorbitol to the
cuvette, then gently transfer the diluted cells into a sterile 17 x 100 mm tube.
7. Check and record the pulse parameters. The time constant should be about 5 milliseconds. The
voltage should be approximately 1.5 kV when pulsing the 0.2 cm cuvettes and approximately 2.5
kV when pulsing the 0.4 cm cuvettes.
63
Page 70

8. Plate aliquots of the electroporated cells on selective agar plates containing 1 M sorbitol. Incubate
plates for 48–72 hrs at 30°C.
7.1.3 Solutions and reagents
1. YPD: 10 g yeast extract, 20 g peptone, dissolve in 900 ml water. Autoclave. Add 100 ml sterile
20% glucose.
2. 1M sorbitol: 182.2 g sorbitol, dissolve in 800 ml water. Bring volume to 1.0 L with water.
Autoclave.
3. 20% glucose: 20 g glucose, dissolve in 60 ml water. Adjust volume to 100 ml with water. Sterilize
through a 0.22 µ filter.
7.2 Schizosaccharomyces pombe
7.2.1 Preparation of Electrocompetent Cells
See Prentice (1991) for additional information.
Using this method we have obtained transformation efficiencies up to 3 x 10
5
transformants / µg
electroporating S. pombe CHP408 with pART1 (Apolinario, et al, 1993).
Gene Pulser Xcell conditions: C = 25 µF; PC = 200 ohm; V = 2.3 kV.
This procedure requires a Gene Pulser Xcell main unit and PC Module.
1. Inoculate 200 ml of YCD in a 2.8 L Fernbach flask with an aliquot from an overnight culture of S.
pombe. (An initial OD
600
of 0.1 is usually a good starting point.) The doubling time of S. pombe is
approximately 2 hrs at 30°C.
2. Incubate at 30°C overnight, shaking at 250 rpm, to late log phase (~2 x 10
8
cells/ml; OD
600
(1:20 dil)
= 0.4–0.5).
3. Chill the cells in an ice water bath for 15 min to stop growth.
4. Decant the cells into a sterile 250 ml centrifuge bottle and pellet the cells by centrifugation at 4000
x g for 5 min at 4°C.
5. Carefully pour off and discard the supernatant; keep the cells on ice.
6. Add ~50 ml of sterile, ice-cold 1.2 M sorbitol and vortex to resuspend the cell pellet; bring the
volume in each of the centrifuge bottles to ~250 ml. Pellet the cells by centrifugation at 4000 x g
for 5 min at 4°C; pour off and discard the supernatant.
7. Wash the cells again as in step 6 with a total of ~200 ml sterile, ice-cold 1.2 M sorbitol.
8. Resuspend the cell pellet with ~10 ml of sterile, ice-cold 1.2 M sorbitol and transfer to a chilled 30
ml Oakridge tube; rinse the centrifuge bottle with 15–20 ml of 1.2 M ice cold 1.2 M sorbitol and
transfer to the Oakridge tube. Pellet the cells by centrifugation at 4000 x g for 5 min at 4°C; pour
off and discard the supernatant.
9. Resuspend the cell pellet in 3 ml of sterile, ice-cold 1.2 M sorbitol. Keep the cells on ice and use
as soon as possible for electroporation.
64
Page 71

7.2.2 Electroporation
1. For each sample to be electroporated, prepare a 17 x 100 mm sterile tube with 1 ml of 1.2 M
Sorbitol and place on ice; also place a 0.2 cm cuvette on ice.
2. Pipette the DNA samples (up to 1 µg) to be electroporated into sterile 1.5 ml microfuge tubes.
Place tubes on ice. Add 40 µl of the competent cells to each DNA sample and mix gently.
3. From the Home Screen on Gene Pulser Xcell open the Fungal Protocol Screen (press 4, Enter, 2,
Enter). To select S. pombe, press 3 or the Down Arrow to highlight S. pombe, then press Enter to
open the S. pombe Protocol Detail screen. See Section 3.4 for operating instructions.
4. Transfer the DNA - cell samples to the chilled 0.2 cm electroporation cuvettes and tap the suspension
to the bottom. Place the cuvette in the ShockPod. Push the chamber lid down to close. Pulse
once.
5. Remove the cuvette from the chamber and immediately add 1 ml of ice cold 1.2 M sorbitol to the
cuvette; gently transfer the diluted cells to a sterile 17 x 100 mm tube.
6. Check and record the pulse parameters. The time constant should be close to 5 milliseconds. The
voltage should be approximately 2.3 kV.
7. Incubate the tubes at room temperature for 40–60 min. Plate aliquots of the electroporated cells on
selective agar plates without sorbitol (e.g., EMM). Incubate plates for 6–8 days at 30°C.
7.2.3 Solutions and Reagents
1. YCD media: 10 g yeast extract, 2 g casamino acids, dissolve in 900 ml water. Autoclave. Add
100 ml 20% glucose.
20% Glucose: 100 g glucose, dissolve in 350 ml water; bring volume to 500 ml with water. Filter
sterilize.
2. 1.2 M sorbitol: 218.6 g sorbitol, dissolve in ~700 ml water. Add water to 1.0 L. Autoclave.
3. EMM: 12.35 g EMM – Dextrose (Q-Biogene, Catalog # 4110-122), dissolve in 900 ml water. Pour
450 ml into each of two 1 L bottles and add 10.0 g agar to each bottle. Autoclave 30 min. Cool to
55–60°C and add 50 ml 20% Glucose to each bottle. Pour plates (sufficient for ~60 plates).
7.3 Pichia pastoris
7.3.1 Preparation of Electrocompetent Cells
See Cregg & Russell (1998) for additional information.
Using this method we have obtained transformation efficiencies of ~6 x 104transformants / µg
electroporating P. pastoris X33 with SacI-pPICZA (Invitrogen).
Gene Pulser Xcell conditions: C = 25 uF; PC = 200 ohm; V = 2.0 kV.
This procedure requires a Gene Pulser Xcell main unit and PC Module.
1. Inoculate 500 ml of YPD in a 2.8 L Fernbach flask with an aliquot from a fresh overnight culture of
P. pastoris. The doubling time of P. pastoris is approximately 2 hrs at 30°C.
2. Incubate at 30°C overnight, shaking at 300 rpm, to a density of ~7 x 10
7
cells/ml (OD
600
(1:10 dil)
= 0.24–0.30).
3. Decant the cells into a sterile 500 ml centrifuge bottle and pellet the cells by centrifugation at
3000 x g for 5 min at 4°C.
4. Carefully pour off and discard the supernatant.
65
Page 72

5. Add 100 ml of sterile, YPD/HEPES to each of the bottles and vortex to resuspend the cell pellets;
add 2.5 ml of 1M DTT; mix gently. Incubate the cells for 15 min at 30°C without shaking.
6. Bring the volume to ~500 ml with sterile, ice-cold 1 M sorbitol. Pellet the cells by centrifugation at
3000 x g for 5 min at 4°C; pour off and discard the supernatant.
7. Add ~50 ml of sterile, ice-cold 1 M sorbitol and vortex to resuspend the cell pellets; bring the
volume to 500 ml with sterile, ice-cold 1 M sorbitol. Pellet the cells by centrifugation at 3000 x g for
5 min at 4°C; pour off and discard the supernatant.
8. Resuspend the cell pellet in ~25 ml of sterile, ice-cold 1 M sorbitol and transfer to a chilled
30 ml Oakridge tube. Pellet the cells by centrifugation at 3000 x g for 5 min at 4°C; pour off and
discard the supernatant.
9. Resuspend the cell pellet in 0.5 ml of sterile, ice-cold 1 M sorbitol; the final cell volume should be
~1.3 ml and the cell concentration should be >1 x 10
10
cells/ml. Keep the cells on ice and use as
soon as possible for electroporation.
7.3.2 Electroporation
1. For each sample to be electroporated, prepare a 17 x 100 mm sterile tube with 1.0 ml of 1 M sorbitol
and place on ice; also place a 0.2 cm cuvette on ice.
2. Pipette the DNA samples (up to 0.1 µg) to be electroporated into sterile 1.5 ml microfuge tubes.
Place tubes on ice.
2. Add 40 µl of the competent cells to each DNA sample and mix gently.
3. From the Home Screen on Gene Pulser Xcell open the Fungal Protocol Screen (press 4, Enter, 2,
Enter). To select P. pastoris, press 5 or the Down Arrow to highlight P. pastoris, then press Enter to
open the P. pastoris Protocol Detail screen. See Section 3.4 for operating instructions.
4. Transfer the DNA - cell samples to the 0.2 cm electroporation cuvettes that have been chilled in ice
and tap the suspension to the bottom. Place the cuvette in the ShockPod. Push the chamber lid
down to close. Pulse once.
5. Remove the cuvette from the chamber and immediately add 1.0 ml of ice cold 1.0 M sorbitol to the
cuvette; gently transfer the diluted cells to a sterile 17 x 100 mm tube.
6. Check and record the pulse parameters. The time constant should be close to 5 milliseconds. The
voltage should be approximately 2.0 kV.
7. When selecting for complementation of an auxotrophic mutant, the cells may be plated
immediately onto minimal agar plates lacking the appropriate nutrient. When selecting for antibiotic
resistance, incubate the cells at 30°C for 1 hr without shaking; add 1 ml of 1M sorbitol and
continue incubating for 1 hr; plate aliquots of the electroporated cells on YPD agar plates with the
appropriate antibiotic. Incubate the plates for 72–96 hrs at 30°C.
7.3.3 Solutions and Reagents
1. YPD: 10 g yeast extract, 20 g peptone, dissolve in 900 ml water. Autoclave. Add 100 ml sterile
20% glucose.
2. YPD/HEPES: 100 ml YPD media, 20 ml 1 M HEPES, pH 8.0.
3. 1M sorbitol: 182.2 g sorbitol, dissolve in 800 ml water. Bring volume to 1.0 L with water.
Autoclave.
4. 20% glucose: 20 g glucose, dissolve in 60 ml water. Adjust volume to 100 ml with water. Sterilize
through a 0.22 µ filter.
66
Page 73

5. 1 M HEPES, pH 8.0: 23.8 g HEPES (MW = 238.3), dissolve in ~65 ml water. Adjust pH to 8.0 with
5 N NaOH. Bring volume to 100 ml with water. Sterilize through a 0.22 µ filter.
6. 1M DTT: 3.85 g DTT, dissolve in ~22 ml water. Bring volume to 25 ml with water. Sterilize through
a 0.22 µ filter.
7.4 Candida albicans
7.4.1 Preparation of Electrocompetent Cells
See deBacker, et al. (1999) for additional information.
Using this method we have obtained transformation efficiencies of >400 transformants / µg
electroporating C. albicans CAI4 with NheI-pUX (Thompson, et al., 1998).
Gene Pulser Xcell conditions: C = 25 µF; PC = 200 ohm; V = 1.5 kV.
This procedure requires a Gene Pulser Xcell main unit and PC Module.
1. Inoculate a colony of C. albicans into 5–6 ml of YPD/uridine media in a 17 x 100 mm tube; incubate
overnight at 30°C shaking at 250–300 rpm.
2. Inoculate an aliquot of the overnight culture into 500 ml of YPD/uridine media in a 2.8 L Fernbach
flask; incubate overnight at 30°C shaking at 250–300 rpm to a density of 5–10 x 10
7
cells/ml
(OD
600
of a 1:10 dilution = 0.3–0.45).
3. Chill the culture on ice for 10 min.
4. Transfer the cells into a sterile 500 ml centrifuge bottle.
5. Pellet the cells by centrifugation at 15–20°C for 5 min at 3000 x g.
6. Discard the supernatant. Resuspend the cell pellet in 125 ml of sterile 5 mM LiAc/10 mM DTT;
incubate at 20°C for 1 hr.
7. Pellet the cells at 4°C for 5 min at 3000 x g.
8. Discard the supernatant. Resuspend the pellet in 125 ml of sterile, ice cold water. Pellet the cells
as above.
9. Discard the supernatant. Resuspend the cell pellet in 125 ml of ice cold water. Pellet the cells as
above.
10. Discard the supernatant. Resuspend the cell pellet in 35 ml of ice cold 1 M sorbitol. Transfer the
cells to a chilled Oakridge tube. Pellet the cells at 4°C for 5 min at 3000 x g.
11. Carefully remove the supernatant. Resuspend the pellet in 0.5 ml of ice cold 1 M sorbitol.
12. Keep the cells on ice and use as soon as possible for electroporation.
7.4.2 Electroporation
1. For each sample to be electroporated, prepare a 17 x 100 mm sterile tube with 1.0 ml of 1 M
sorbitol and place on ice; also place a 0.2 cm cuvette on ice.
2. Pipette the linearized DNA samples (0.1–1 µg) to be electroporated into sterile 1.5 ml microfuge
tubes. Place tubes on ice.
3. Add 40 µl of the electrocompetent cells to each DNA sample and mix gently. Incubate the cells on
ice for 5 min.
67
Page 74

4. From the Home Screen on Gene Pulser Xcell open the Fungal Protocol Screen (press 4, Enter, 2,
Enter). To select C. albicans, press 4 or the Down Arrow to highlight C. albicans, then press Enter
to open the C. albicans Protocol Detail screen. See Section 3.4 for operating instructions.
5. Transfer the DNA - cell samples to the 0.2 cm electroporation cuvettes that have been chilled in ice
and tap the suspension to the bottom. Place the cuvette in the ShockPod. Push the chamber lid
down to close. Pulse once.
6. Remove the cuvette from the chamber and immediately add 1.0 ml of ice cold 1.0 M sorbitol to the
cuvette; gently transfer the diluted cells to a sterile 17 x 100 mm tube.
7. Check and record the pulse parameters. The time constant should be close to 5 milliseconds. The
voltage should be approximately 1.5 kV.
8. Immediately plate an aliquot of the cells onto minimal agar plates containing 1 M sorbitol but lacking the appropriate nutrient. Incubate the plates for 4–6 days at 30°C.
7.4.3 Solutions and Reagents
1. YPD / Uridine: 10 g yeast extract, 20 g peptone, dissolve in 900 ml water; add 15 g agar.
Autoclave. Add 100 ml sterile 20% glucose, 1 ml 25 mg/ml uridine.
2. YPD / Uridine agar: 10 g yeast extract, 20 g peptone, dissolve in 900 ml water. Autoclave. Add
100 ml sterile 20% glucose, 1 ml 25 mg/ml uridine.
3. 1M sorbitol: 182.2 g sorbitol, dissolve in 800 ml water. Bring volume to 1.0 L with water.
Autoclave.
4. 20% glucose: 20 g glucose, dissolve in 60 ml water. Adjust volume to 100 ml with water. Sterilize
through a 0.22 µ filter.
5. 5 mM LiAc/10 mM DTT in TE: 1 ml 1 M Lithium Acetate (10.2 g / 100 ml), 20 ml 0.1 M DTT (1.54
g / 100 ml), 2 ml 1 M Tris, pH 7.5, 0.4 ml 0.5 M EDTA, 176.6 ml water. Filter sterilize. Store at
–20°C.
6. 1M sorbitol: 182.2 g sorbitol, dissolve in 800 ml water. Bring volume to 1.0 L with water.
Autoclave.
7.5 Dictyostelium discoideum
7.5.1 Preparation of Electrocompetent Cells
See Howard et al. (1988) and Knecht & Pang (1995) for additional information.
Gene Pulser Xcell conditions: square wave, V = 1.0 kV, 1.0 ms Pulse length, 2 pulses, 5 sec pulse interval;
V = 1.0 kV.
1. Inoculate Dictyostelium cells at a concentration of 5–7 x 10
5
cells/ml into 40 ml of HL5 media in a
500 ml flask. The cells may either be scraped from a plate or transferred from liquid media. The
doubling time of Dictyostelium is approximately 12 hrs at 21°C.
2. Incubate the culture at 21°C for about 24 hrs, shaking at 125 rpm. About 16–20 hrs prior to
preparing the competent cells, dilute the cells to 7 x 10
5
cells/ml with HL5 media. Incubate at 21°C
overnight, shaking at 125 rpm.
3. Transfer 100 ml of the cells into two sterile, disposable, 50 ml centrifuge tubes and incubate on ice
for 15 min to stop growth.
4. Pellet the cells by centrifugation at 400 x g for 5–7 min at room temperature.
68
Page 75

5. Carefully pour off and discard the supernatant; place the centrifuge bottles with the cell pellets on
ice.
6. Pool the cell pellets and resuspend in 50 ml of sterile, ice-cold E buffer. Pellet the cells by centrifugation
at 400 x g for 5–7 min at room temperature.
7. Carefully pour off and discard the supernatant; place the centrifuge bottles with the cell pellets on
ice and resuspend the cells at a concentration of 1 x 10
7
cells/ml. Keep the cells on ice and use as
soon as possible for electroporation.
7.5.2 Electroporation
1. Pipette the DNA samples (up to 50 µg) to be electroporated into sterile 1.5 ml microfuge tubes.
Place tubes on ice.
2. Add 800 µl of the competent cells to each DNA sample and pipette up and down to mix; incubate
on ice ~1 min.
3. From the Home Screen on Gene Pulser Xcell open the Fungal Protocol Screen (press 4, Enter, 2,
Enter). To select D. discoideum, press 6 or the Down Arrow to highlight D. discoideum, then press
Enter to open the D. discoideum Protocol Detail screen. See Section 3.4 for operating instructions.
4. Transfer the DNA - cell samples to 0.4 cm electroporation cuvettes that have been chilled in ice
and tap the suspension to the bottom of the tube. Place the cuvette in the ShockPod. Push the
chamber lid down to close. Pulse once (the program delivers two pulses, each with a ~1 msec
time constant approximately 5 sec apart).
5. Remove the cuvette from the chamber and immediately dilute the cells to 10 ml with the appropriate media.
6. Check and record the pulse parameters. The time constant should be 1 millisecond. The voltage
should be approximately 1.0 kV.
7. When selecting for complementation of an auxotrophic mutant, the cells may be plated
immediately into selective media lacking the appropriate nutrient. When selecting for antibiotic
resistance, incubate the cells overnight at 21°C prior to adding the selective agent.
7.5.3 Solutions and Reagents
1. HL5 media: 17.8 g bacteriological peptone (Oxoid, Ogdensburg, NY), 7.2 g yeast extract, 0.54 g
Na
2
HPO4, 0.4 g KH2PO4, 130 µl B12/Folic acid mix; bring to 1L with water and adjust to pH
6.3–6.5. Autoclave for 25 min on two successive days. Prior to use, add 20 ml of 50% glucose
and 10 ml of 100 X Antibiotic-Antimycotic (Life Technologies, Gaithersburg, MD).
2. B12/Folic acid mix: 5 mg B12, 200 mg folic acid; add 95 ml water, then pH to 6.5–6.8 with 5N
NaOH; bring to 100 ml with water. Filter sterilize and store at -20°C protected from light.
3. E buffer: 10 ml 100 mM NaH2PO4, adjusted to pH 6.1 with KOH, 10 ml 0.5 M sucrose, 80 ml
water; autoclave.
69
Page 76

Section 8
Electroporation of Mammalian Cells
This procedure requires a Gene Pulser Xcell main unit and CE Module.
8.1 Preparation of Electrocompetent Cells
8.1.1 Attached Cells
1. One – two days prior to electroporation, transfer the cells into 75cm2flasks with fresh growth medium
so that they will be 50–70% confluent on the day of the experiment. For most cell lines the cell
density will be 2–10 x 10
6
cells / flask; about 1–10 x 105cells are needed per electroporation.
2. Aspirate the media and rinse the flasks using ~12 ml of phosphate buffered saline (PBS).
3. Aspirate the PBS and add ~0.4 ml of trypsin-EDTA; incubate the cells 2–5 min at room temperature.
Tap the flask to detach the cells from the surface.
4. Neutralize the trypsin by adding 10 ml of growth media with serum.
5. Transfer the cells to a 50 ml sterile centrifuge tube; pellet the cells by centrifugation at 400 x g for
5–7 min at room temperature.
6. Aspirate the media, then resuspend the cell pellets in electroporation buffer (Optimem, growth
media without serum, PBS, Hepes-buffered saline, Phosphate-buffered sucrose, or
Hepes-buffered sucrose) at a density of 1–5 x 10
6
cells/ml. Gently pipette the cells to obtain a sin-
gle-cell suspension.
8.1.2 Suspension Cells
1. One – two days prior to electroporation, dilute the cells into fresh growth medium in a 75cm
2
flask
so that they will be in mid-log phase (generally about 0.5–4 x 10
6
cells/ml) on the day of the
experiment. About 1–10 x 105cells are needed per electroporation.
2. Transfer the cells to a 50 ml sterile centrifuge tube; pellet the cells by centrifugation at 400 x g for
5–7 min at room temperature.
3. Aspirate the media, then resuspend the cell pellets in electroporation buffer (Opti-MEM, growth
media without serum, PBS, Hepes-buffered saline, Phosphate-buffered sucrose, or Hepes-buffered
sucrose) at a density of 1–5 x 10
6
cells/ml. Gently pipette the cells to obtain a single-cell suspension.
For all Pre-set Protocols programmed in Gene Pulser Xcell, we recommend that cells be resuspended
in Opti-MEM or in growth media without serum.
8.2 Electroporation
1. Set the desired pulse conditions on the Gene Pulser Xcell. If a Pre-Set Protocol is used (see Table
3.4), open the Mammalian Pre-set Protocols screen as follows: from the Home screen, press 4,
Enter, 3, Enter; use the alpha-numeric keypad or the Up and Down Arrow keys to select the cell
line, then press Enter to open the appropriate Protocol Detail screen.
70
Page 77

2. Add plasmid DNA(s) to the electroporation cuvettes. The amount of DNA required per sample is
dependent on the cell type. Start with a concentration of 10–50 µg/ml.
3. Add cells to the cuvette and tap the side of the cuvette to mix. See Table 8.1 for suggestions on
cell concentrations and volumes to use.
4. Place the cuvette in the ShockPod. Push down the lid to close. Pulse once.
5. Immediately after the pulse, transfer the cells to a plate using ~0.5 ml of media and a plugged
Pasture pipette.
6. Rock the plates gently to assure even distribution of the cells over the surface of the plate.
Incubate the plates at 37°C in a humidified atmosphere.
7. Assay transient gene expression 24–48 hrs following electroporation.
Table 8.1 Suggested starting cell concentrations and volumes for
electroporating mammalian cells
Cuvette Cell concentration Cell volume Growth conditions following electroporation
(cm) (cells/ml) (µl)
0.2 1 x 10
6
100 48 well plate with 0.5 ml growth media
0.2 5 x 10
6
200 6 well plate with 2 ml growth media
0.4 2.5 x 10
6
400 6 well plate with 2 ml growth media
8.3 Reagents and Solutions for Electroporations
1. Growth medium with 10% FBS.
2. Opti-MEM (Invitrogen, Carlsbad, CA).
3. Trypsin-EDTA: 0.05% trypsin, 0.53 mM EDTA in PBS (Invitrogen).
4. Phosphate-buffered saline: 137 mM NaCl, 2.7 mM KCl, 9.5 mM sodium phosphate, pH 7.3;
autoclave.
5. Hepes-buffered saline: 10 mM Hepes, pH 7.3, 140 mM NaCl; autoclave.
6. Phosphate-buffered sucrose: 272 mM Sucrose, 10 mM sodium phosphate, pH 7.3; filter sterilize.
7. Hepes-buffered sucrose: 272 mM Sucrose, 10 mM Hepes, pH 7.3; filter sterilize.
71
Page 78

Section 9
References
Anderson, M.L.M., Spandidos, D.A., and Coggins, J.R., Electroporation of lymphoid cells: factors
affecting the efficiency of transfection, J. Biochem. Biophys. Meth., 22, 207 (1991)
Andreason, G.L. and Evans, G.A., Optimization of electroporation for transfection of mammalian cell
lines, Anal. Biochem. 180, 279 (1989)
Apolinario, E., Nocero, M., Jin, M., and Hoffman, C.S., Cloning and manipulation of the
Schizosaccharomyces pombe his7+ gene as a new selectable marker for molecular genetic studies, Curr.
Genet. 24, 491 (1993)
Ausubel, F.M., Brent, R., Kingston, R.E., Moore, D.D., Seidman, J.G., Smith, J.A., and Struhl, K. (eds.)
Current Protocols in Molecular Biology, John Wiley & Sons, NY (1987)
Barsoum, J., “Stable integration of vectors at high copy number for high-level expression in animal cells,”
in Methods in Molecular Biology, vol. 48, Nickoloff, J.A. (ed.), Humana Press, Totowa, NJ, 225 (1995)
Becker, D.M. and Guarente, L., High-efficiency transformation of yeast by electroporation, Methods
Enzymol. vol. 194, Academic Press, San Diego, CA, 182 (1991)
Böttger, E. C., High-efficiency generation of plasmid cDNA libraries using electro-transformation,
BioTechniques 6, 878 (1988)
Bringel, F. and Hubert, J.-C., Optimized transformation by electroporation of Lactobacillus
plantarum strains with plasmid vectors, Appl. Microbiol. Biotechnol., 33, 664 (1990)
Calvin, N.M. and Hanawalt, P.C., High-efficiency transformation of bacterial cells by electroporation, J.
Bacteriol. 170, 2796 (1988)
Chaffin, D.O. and Rubens, C.E., Blue/white screening of recombinant plasmids in Gram-positive bacteria
by interruption of alkaline phosphatase gene (phoZ) expression, Gene 219, 91 (1998)
Change, D.C., Chassy, B.M., Saunders, J.A., and Sowers, A.E. (eds.) Guide to Electroporation and
Electrofusion, Academic Press, Inc., San Diego (1992)
Chu, G., Hayakawa, H., and Berg, P., Electroporation for the efficient transfection of mammalian cells
with DNA, Nuc. Acids Res. 15, 1311 (1987)
Cregg, J.M. and Russell, K.A., “Transformation,” in Methods in Molecular Biology, vol. 103, Higgins,
D.R. and Cregg, J.M. (eds.), Humana Press, Totowa, NJ, 27 (1998)
DeBacker, M.D., Maes, D., Vandoninck, S., Logghe, M., Contreras, R., and Luyten, W.H.M.L.,
Transformation of Candida albicans by electroporation, Yeast 15, 1609 (1999)
Dekeyser, R.A., Claes, B., deRycke, R.M.U., Habets, M.E., vanMontagu, M.C., and Caplan, A.B.,
Transient gene expression in intact and organized rice tissues, The Plant Cell 2, 591 (1990)
Dennis, J.J. and Sokol, P.A., “Electrotransformation of Pseudomonas,” in Methods in Molecular Biology,
vol. 47, Nickoloff, J.A. (ed.), Humana Press, Totowa, NJ, 125 (1995)
Dower, W. J., “Electroporation of bacteria: a general approach to genetic transformation,” in Genetic
Engineering—Principles and Methods, vol 12, Plenum Publishing Corp., NY, 275
(1990)
Dower, W. J., Miller, J. F., and Ragsdale, C. W., High efficiency transformation of E. coli by high voltage
electroporation, Nuc. Acids Res. 16, 6127 (1988)
Dower, W.J., Chassy, B.M., Trevors, J.T., and Blaschek, H.P., “Protocols for the transformation of bacteria
by electroporation,” in Guide to Electroporation and Electrofusion, Change, D. C., Chassy, B. M.,
Saunders, J. A., and Sowers, A. E. (eds.), Academic Press Inc., 485, San Diego (1992)
72
Page 79

Fromm, M., Callis, J., Taylor, L.P., and Walbot, V., “Electroporation of DNA and RNA into plant protoplasts”,
in Methods Enzymol., vol. 153, Academic Press Inc., San Diego, 351, (1987)
Gaertig, J. and Gorovsky, M.A., “Gene transfer by electroporation in Tetrahymena,” in Methods in
Molecular Biology, vol. 47, Nickoloff, J.A. (ed.), Humana Press, Totowa, NJ, 331 (1995)
Haque, H. and Russell, A. D., Effect of ethylene-diamine-tetraacetic acid and related chelating agents
on whole cells of gram-negative bacteria. Antimicrob. Agents Chemother., 5, 447 (1974)
Heery, D. M., and Dunican, L. K., Improved efficiency M13 cloning using electroporation, Nuc. Acids
Res. 17, 8006 (1989)
Heiser, W.C., “Optimizing electroporation conditions for the transformation of mammalian cells,” in
Methods in Molecular Biology, vol. 130, Tymms, M.J. (ed.), Humana Press, Totowa, NJ, 117 (1999)
Hui, S.W., “Effects of pulse length and strength on electroporation efficiency,” in Methods in Molecular
Biology, vol. 48, Nickoloff, J.A. (ed.), Humana Press, Totowa, NJ, 195 (1995)
Howard, P.K., Ahern, K.G., and Firtel, R.A., Establishment of a transient expression system for
Dictyostelium discoideum, Nuc. Acids Res. 16, 2613 (1988)
Jacobs, M., Wnendt, S., and Stahl, U., High-efficiency electro-transformation of Escherichia coli with
DNA from ligation mixtures, Nuc. Acids Res. 18, 1653 (1990)
Joersbo, M. and Brunstedt, J., Electroporation: mechanism and transient expression, stable transformation
and biological effects in plant protoplasts, Physiol. Plant 81, 256 (1991)
Knecht, D. and Pang, K.M., “Electroporation of Dictyostelium discoideum,” in Methods in Molecular
Biology, vol. 47, Nickoloff, J.A. (ed.), Humana Press, Totowa, NJ, 321 (1995)
Knutson, J.C. and Yee, D., Electroporation: parameters affecting transfer of DNA into mammalian cells,
Anal. Biochem. 164, 44 (1987)
Lee, J.C., “Electrotransformation of Staphylococci,” in Methods in Molecular Biology, vol. 47, Nickoloff,
J.A. (ed.), Humana Press, Totowa, NJ, 209 (1995)
Lin, J.-J., “Electrotransformation of Agrobacterium,” in Methods in Molecular Biology, vol. 47, Nickoloff,
J.A. (ed.), Humana Press, Totowa, NJ, 171 (1995)
Miller, E.M. and Nickoloff, J.A., “Escherichia coli electrotransformation,” in Methods in Molecular
Biology, vol. 47, Nickoloff, J.A. (ed.), Humana Press, Totowa, NJ, 105 (1995)
Neumann, E., Kakorin, S., and Toensing, K., “Principles of membrane electroporation and transport of
macromolecules,” in Methods in Molecular Medicine vol 37, Jaroszeski, M.J., Heller, R., and Gilbert, R.
(eds.), Humana Press, Totowa, NJ, 1 (2000)
Nickoloff, J.A. (ed.) Methods in Molecular Biology, vol. 47, Humana Press, Totowa, NJ, (1995a)
Nickoloff, J.A. (ed.) Methods in Molecular Biology, vol. 48, Humana Press, Totowa, NJ, (1995b)
Prentice, H.L., High efficiency transformation of Schizosaccharomyces pombe by electroporation, Nuc.
Acids Res., 20, 621 (1991)
Potter, H., Weir, L., and Leder, P., Enhancer-dependent expression of human k immunoglobulin genes
introduced into mouse pre-B lymphocytes by electroporation, Proc. Nat. Acad. Sci. 81, 7161 (1984)
Shigekawa, K. and Dower, W.J., Electroporation of eukaryotes and prokaryotes: A general approach to
the introduction of macromolecules into cells, Biotechniques 6, 742 (1988)
Shillito, R.D., Saul, M.W., Paszkowski, J., Muller, M., and Potrykis, I., High efficiency direct gene transfer
to plants, Bio/Technology 3, 1099 (1985)
Showe, M.K., Williams, D.L., and Showe, L.C., Quantitation of transient gene expression after electroporation,
Nuc. Acids Res. 20, 3153 (1990)
73
Page 80

Silo-Suh, L.A., Lethbridge, B.J., Raffel, S.J., He, H., Clardy, J., and Hanelsman, J., Biological activities
of two fungistatic antibiotics produced by Bacillus cereus UW85, Appl. Environ. Microbiol. 60, 2023
(1994)
Simon, D. and Ferretti, J.J., Electrotransformation of Streptococcus pyogenes with plasmid and linear
DNA, FEMS Micobiol. Lett. 82, 219 (1991)
Smith, A.W. and Iglewski, B.H., Transformation of Pseudomonas aeruginosa by electroporation, Nuc.
Acids Res. 17, 10509 (1989)
Thompson, J.R., Register, E., Curotto, J., Kurtz, M., and Kelly, R., An improved protocol for the preparation
of yeast cells for transformation by electroporation, Yeast 14, 565 (1998)
Trevors, J.T., Chassy, B.M., Dower, W.J., and Blaschek, H.P., “Electrotransformation of bacteria by
plasmid DNA,” in Guide to Electroporation and Electrofusion, Change, D. C., Chassy, B. M., Saunders,
J. A., and Sowers, A. E. (eds.), Academic Press Inc., San Diego, 265, (1992)
Willson, T. A., and Gough, N. M., High voltage E. coli electro-transformation with DNA following ligation,
Nuc. Acids Res. 16, 11820 (1988)
Zabarovsky, E. R., and Winberg, G., High efficiency electroporation of ligated DNA into bacteria, Nuc.
Acids Res. 18, 5912 (1990)
74
Page 81

Section 10
Specifications and Product Information
10.1 System Specifications
165-2660 Gene Pulser Xcell Total System
Includes Main Unit, CE module, PC module
Outputs Waveform: Exponential decay or Square wave.
Voltage: 10–3,000 Volts
Capacitance 10–500 V, 25–3275 mF in 25 mF increments.
500–3000 V, 10, 25, 50 mF.
Resistance (Parallel) 50–1,000 Ω in 50 Ω increments plus infinity.
Sample Resistance 20 Ω minimum at 10–2,500 V
600 Ω minimum at 2,500–3,000 V
Square Wave Timing 10–500 V: 0.05–100 msec pulse length, 1–10 pulses,
0.1–10 sec pulse interval
500–3,000 V: 0.05–5 msec pulse length, 1–2 pulses,
5–30 sec pulse interval
165-2661 Gene Pulser Xcell Eukaryotic System
Includes Main Unit, CE module
Outputs same as Total system without the parallel resistance.
165-2662 Gene Pulser Xcell Microbial System
Includes main unit, PC module
Outputs Waveform: Exponential decay or Square wave.
Voltage: 200–3,000 Volts
Capacitance 10, 25, 50 µF.
Resistance (Parallel) 50–1,000 Ω in 50 Ω increments plus infinity.
Sample Resistance 20 Ω minimum at 200–2,500 V
600 Ω minimum at 2,500–3,000 V
Square Wave Timing 0.05–5 msec pulse length, 1–2 pulses,
5–30 pulse interval
165-2666 Gene Pulser Xcell Main Unit
Outputs Waveform: Exponential decay or Square wave.
Voltage: 200–3,000 Volts
Discharge Capacitance 10, 25, 50 µF.
Sample Resistance 20 Ω minimum at 200–2,500 V
600 Ω minimum at 2,500–3,000 V
Square Wave Timing 0.05–5 msec pulse length, 1–2 pulses,
5–30 sec pulse interval
General
Input Voltage 100–120 VAC or 220–240 VAC, 50/60 Hz.
Power Max 240 W (During short charging periods)
Operating environment Temp. 0–35°C, Humidity 0–95%, (non-condensing).
Regulatory Safety EN 61010, EMC EN61326 Class A
Dimensions, Main Unit: 31 x 30 x 14 cm, weight 6.62 kg.
CE Module: 31 x 28 x 9 cm, weight 3.08 kg.
PC Module: 31 x 28 x 5 cm, weight 1.90 kg.
75
Page 82

10.2 Product Information
Catalog
Number Product Description
165-2660 Gene Pulser Xcell Total System for eukaryotic and microbial cells, 100–240 V, 50/60
Hz, exponential decay and square wave delivery, includes main unit, CE Module,
PC Module, ShockPod, 15 sterile cuvettes (5 each of 0.1, 0.2, and0.4 cm gap),
Instruction Manual
165-2661 Gene Pulser Xcell Eukaryotic system, 100/240 V, 50/60 Hz, exponential decay
(25–3,275 µF range) and square wave delivery, includes main unit, CE Module,
ShockPod, 5 sterile cuvettes (0.4cm gap), Instruction Manual
165-2662 Gene Pulser Xcell Microbial system, 100/240 V, 50/60 Hz, exponential decay and
square wave delivery delivery, includes main unit, PC Module, ShockPod, 10 sterile
cuvettes (5 each of 0.1 and 0.2 cm gap), Instruction Manual
165-2666 Gene Pulser Xcell main unit, 100/240 V, 50/60 Hz
165-2667 Gene Pulser Xcell CE Module, 25–3,275 µF range controlled by main unit, includes
integral leads
165-2668 Gene Pulser Xcell PC Module, 50–1,000 ohm range controlled by main unit,
includes integral leads
165-2669 Gene Pulser Xcell ShockPod shocking chamber, includes integral leads for
connection to Gene Pulser Xcell, Gene Pulser II, or MicroPulser.
165-2095 Gene Pulser Cuvette Rack
165-2094 Gene Pulser Electroprotocols
165-2088 Gene Pulser Cuvettes, 0.4 cm gap, 50, sterile
165-2086 Gene Pulser Cuvettes, 0.2 cm gap, 50, sterile
165-2089 Gene Pulser Cuvettes, 0.1 cm gap, 50, sterile
165-2091 Gene Pulser Cuvettes, 0.4 cm gap, 500, sterile
165-2092 Gene Pulser Cuvettes, 0.2 cm gap, 500, sterile
165-2093 Gene Pulser Cuvettes, 0.1 cm gap, 500, sterile
165-2081 Gene Pulser Cuvettes, 0.4 cm gap, 5, sterile
165-2082 Gene Pulser Cuvettes, 0.2 cm gap, 5, sterile
165-2083 Gene Pulser Cuvettes, 0.1 cm gap, 5, sterile
76
Page 83

4006217 Rev A
S
cience
p
aboratories Ma
ce
a
a
ada
C
a
dFrance
089 318 8
7
, Fx.
089 318 8
g
a
ael
orea
1
oland
g
Swede
8
2
US/EG
A
Bio-R
ad
.
Laboratories, Inc
Life
Grou
ulletin 0000
Web sitewww.bio-rad.com Bio-Rad L
Also in: AustraliaPh. 02 9914 2800, Fx. 02 9914 2889 Austri
zil Ph. 55 21 507 6191 Can
Br
Czech Republic Ph. (420) 2-4141 0532 Fx. (420) 2-4143 1642 Denmark
Finlan
Kong Ph. 852-2789-3300, Fx. 852-2789-1257 Indi
Hon
Isr
Ph. 03 951 4127, Fx. 03 951 4129 Japan Ph. 03-5811-6270, Fx. 03-5811-6272
Ph. 82-2-3473-4460, Fx. 82-2-3472-7003 Latin AmericaPh. 305-894-5950, Fx. 305-894-5960 MexicoPh. 52 5 534 2552 to 54, Fx. 52 5 524 597
K
The NetherlandsPh. 0318-540666, Fx. 0318-542216 New ZealandPh. 64-9-4152280, Fx. 64-9-443 3097 Norway Ph. 47-23-38-41-30, Fx. 47-23-38-41-39
P
Ph. (48) 22-8126 672, Fx. (48) 22-8126 682 Portugal
apore Ph. 65-2729877, Fx. 65-2734835 South Africa 00 27 11 4428508, Fx. 00 27 11 4428525Spain Ph. 34 91 590 5200, Fx. 34 91 590 5211
Sin
n Ph. 46 (0)8-55 51 27 00, Fx. 46 (0)8-55 51 27 80 SwitzerlandPh. 061 717-9555, Fx. 061 717-9550 United Kingdom Ph. 0800-181134, Fx. 01442-25911
Rev
in Offi
hin
Belgium Ph. 09-385 55 11, Fx. 09-385 65 54
4-17
iaPh. 7 095 721 1404, Fx. 7 095 721 1412
00-000 0000 Sig 040
4-123
 Loading...
Loading...