Frame-Seal™ Incubation Chambers
for Sealing Reactions on Slides
Frame-Seal incubation chambers provide vapor-tight sealing for FISH, Polony, in situ PCR,
and PRINS applications. The adhesive frames are placed over the specimen area and
fastened to the slide. Reaction mix is then added and sealed in place with a flexible plastic
coverslip. The gas-tight seal withstands temperatures up to 97°C. After cycling, the entire
chamber can easily be removed from the slide, and the reaction mix can be retrieved.
Frame-Seal chambers can be UV-treated and autoclaved (with a weight on top to keep
them flat), and are compatible with MJ-line and other horizontal-format slide thermal cyclers.
Directions for Use
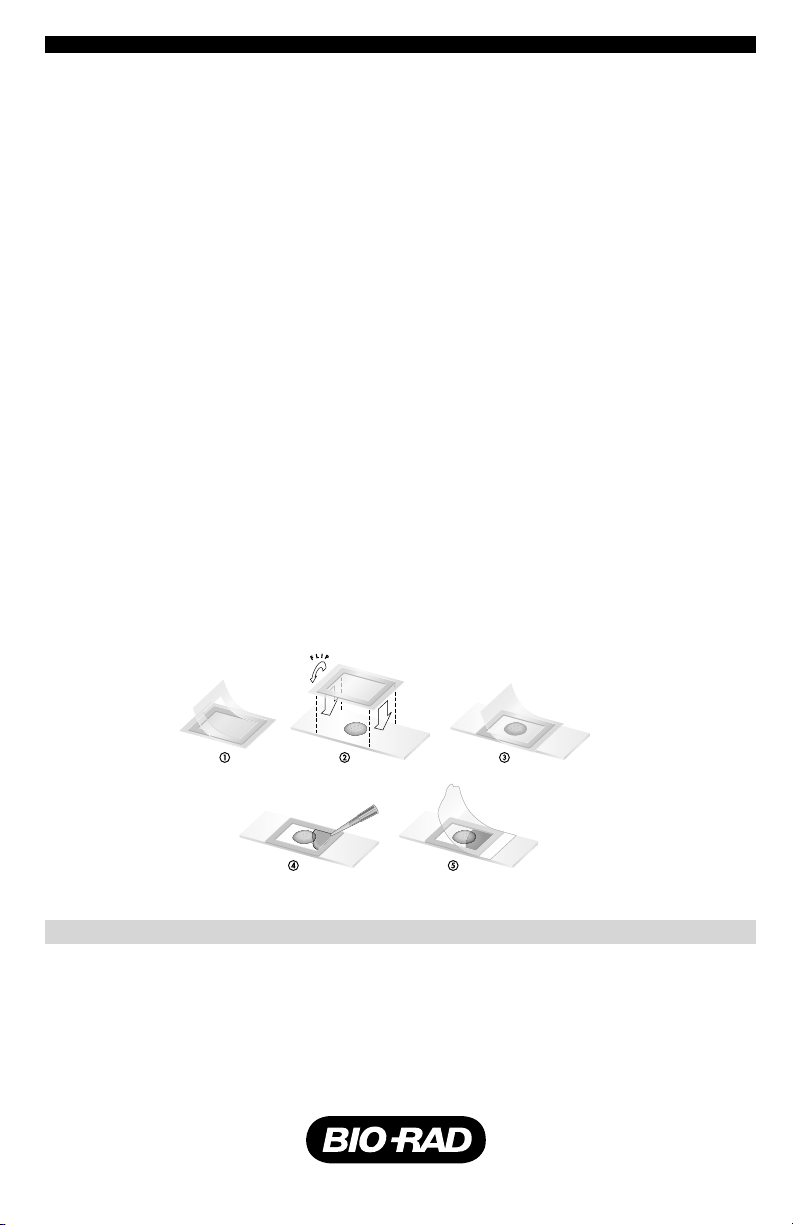
1. Each frame comes sandwiched between two plastic liners — one thin rectangular liner
and one thicker liner with a rectangular hole in it. Lined frames are packaged as strips
with perforations between frames. Separate one frame from a strip and remove the
protective plastic liner that has the rectangular hole, leaving the frame adhered to the
thinner liner (Fig. 1).
2. Position the frame adhesive side down over the desired area of the slide (Fig. 2).
3. Remove the top liner (Fig. 3).
4. Pipet the appropriate volume of reagent into the frame at one end (Fig. 4).
5. The polyester covers are packaged in a separate bag. Remove one cover from the bag
and roll it over the frame, beginning at the end where the reagent was added (Fig. 5).
6. Rub all four edges with a blunt, hard object, such as a pen barrel, to ensure good
adhesion. The assembly is now ready to use.
Frame-Seal Chambers 100 frames and coverslips per package
SLF-0201 9 x 9 mm chambers, 25 µl capacity
SLF-0601 15 x 15 mm chambers, 65 µl capacity
SLF-1201 17 x 28 mm chambers, 125 µl capacity
SLF-3001 19 x 60 mm chambers, 300 µl capacity

05-0459 0905 Sig 1204Part# 03526 US/EG Rev C
Tips for Best Results
• Ensure that the slide surface that will contact the frame is clean and dry
• Use forceps when handling the frame to reduce contact with the exposed adhesive
• With a hard object, firmly press on the frame around its perimeter, eliminating any air
bubbles trapped between the adhesive and the glass slide
• Allow the adhesive to set by applying the frame the day before use or by incubating at
95°C for 5 min
• Remove top liner just before adding the reaction mix
• Combine reaction components in a tube, creating excess to avoid trapping air bubbles;
for example, use 70 µl of reaction mix for the 65 µl capacity frame. Place the open tube in
a vacuum chamber for 5 minutes to de-gas the mixture before pipetting it into the frame
• After the coverslip has been applied, press firmly along the frame perimeter with a hard
object. The coverslip will bind tightly to the frame, regardless of any reagent that has
spilled out
• The frame is easily removed post-incubation; a single-edge razor blade will help break the
seal between the frame and the glass slide
Protocol Considerations
• Optimize the reaction conditions, especially annealing temperature, in a tube before
optimizing in the slide format. Use optimized tube conditions as the starting point for
further optimizing conditions in the slide format. Optimize without tissue present first,
and then with tissue
• To prevent binding of proteins and nucleic acids to the glass slide, add 0.05–0.1%
BSA or other carrier protein to the reaction mix
• When a tissue is first used, optimize the protease digestion steps to achieve sufficient
reagent penetration without damaging tissue morphology. A simple hybridization assay
to an abundant target (e.g., a human Alu-repeat sequence) can be used for this purpose.
Optimize the protease concentration and digestion time for each new tissue type and
each new lot of protease
• To minimize tissue damage, minimize the total number of heating cycles performed
(try doing 15–20 cycles) and the total time spent above 90°C. The amplification steps
only need to increase the amount of target to a few hundred or a few thousand copies
— enough to be detected by probe hybridization
• Don’t use one-step methods that incorporate biotin–dNTPs into the amplification mix.
These often lead to nonspecific background. Instead, amplify in one step, wash, then
hybridize with a biotinylated probe in a separate step
Alternative Slide-Sealing Products
SLH-2001 20 x 35 mm Hyb-Seal™ chambers, 165 µl capacity
SLH-4001 20 x 54 mm Hyb-Seal chambers, 355 µl capacity
SLH-6001 20 x 70 mm Hyb-Seal chambers, 465 µl capacity
SLR-0101 Self-Seal™ reagent, 2x; 5 x 1 ml, sufficient for 200 x 50 µl reactions or 600 x 15 µl reactions
Note: Frame-Seal chambers should not be used with Self-Seal reagent
Practice of the patented polymerase chain reaction (PCR) process requires a license. Bio-Rad and MJ brand thermal cyclers and
systems include an Authorized Thermal Cycler and may be used with PCR licenses available from Applied Biosystems. Their use
with Authorized Reagents also provides a limited PCR license in accordance with the label rights accompanying such reagents.
Some applications may also require licenses from other third parties.
 Loading...
Loading...