Page 1

®
INSTRUCTIONS
AUTOMATED IMAGE AND DATA
ANALYSIS SOFTWARE
This instruction manual describes the Olympus Automated Image and Data
Analysis Software for Life Science. To ensure safety, obtain optimum performance
and familiarize yourself fully with the use of these products, we recommend that you
study this manual thoroughly before operation. Together with this manual, please also
read the
well as the manuals of all other devices controlled by that software in order to understand general operation methods. Retain this manual in an easily accessible place
near a system for future reference.
Automated Image Acquisition Software and Hardware manuals as
Page 2

OLYMPUS SOFT IMAGING SOLUTIONS GMBH
Rupert-Mayer-Strasse 44
D-81379 München
Tel: +49 89 - 89 55 805 660
Fax +49 89 - 89 55 805 6606
Email: info@olympus-sis.com
www.olympus-sis.com
Page 3

Imaging Excellence
We at Olympus Soft Imaging Solutions GmbH have tried to make the information in this manual as
accurate and reliable as possible. Nevertheless, Olympus Soft Imaging Solutions GmbH disclaims any
warranty of any kind, whether expressed or implied, as to any matter whatsoever relating to this manual, including without limitation the merchantability or fitness for any particular purpose. Olympus Soft
Imaging Solutions GmbH will from time to time revise the product described in this manual and reserves the right to make such changes without obligation to notify the purchaser. In no event shall
Olympus Soft Imaging Solutions GmbH be liable for any indirect, special, incidental, or consequential
damages arising out of purchase or use of this manual or the information contained therein.
No part of this document may be reproduced or transmitted in any form or by any means, electronic or
mechanical, for any purpose, without the prior permission of Olympus Soft Imaging Solutions GmbH.
© 2006 – 2008 by Olympus Soft Imaging Solutions GmbH. All rights reserved.
Manual version: 2.1, December 2008
Page 4

LICENSE AGREEMENT
Preamble
OLYMPUS SOFT IMAGING SOLUTIONS GMBH
scan^R software and drivers/interfacing software to Olympus Soft
Imaging Solutions manufactured hardware. These components are
referred to as OSIS scan^R SOFTWARE PRODUCT.
LICENSE AGREEMENT between END USER and OLYMPUS SOFT
IMAGING SOLUTIONS regarding the OSIS scan^R SOFTWARE
PRODUCT.
IMPORTANT-READ CAREFULLY: Below you will find the contractual
agreements governing the use of the OSIS scan^R SOFTWARE
PRODUCT. These conditions apply to you, the user, and to OLYMPUS
SOFT IMAGING SOLUTIONS. With any of the following actions you
explicitly agree to be bound by the conditions of this contract: purchasing the software, opening the package, breaking of one of the seals or
using the software.
In case you do not agree with any of the conditions of this contract,
please return all parts of the product including manuals without delay.
Remove all software installations of the product from any computer you
might have installed it on. Return all electronic media of the product or
completely destroy all electronic media of the product and send proof
that this has been accomplished. For a refund, please return everything
to where you purchased the product.
§ 1. Scope
(1) This License agreement explicitly covers only the software diskettes
or other media you received with the purchase and the software stored
on these media, the manuals, as far as they were developed and produced by OLYMPUS SOFT IMAGING SOLUTIONS.
§ 2. User rights
(1) OLYMPUS SOFT IMAGING SOLUTIONS permits the User, for the
duration of this contract, to use the software on a single computer and a
single terminal on that computer. This license is explicitly non-exclusive,
i.e., the User does not have an exclusive right to use the software. As a
licensed user you can copy the software from one computer to another
by using a computer network or other storage devices, as long as it is
assured, that the software can only be used on a single computer or
terminal at any time and that the conditions set forth under § 4 are
observed.
(2) The User has the right to produce a copy of the software only for
backup purposes.
§ 3. Additional user rights
Only if OLYMPUS SOFT IMAGING SOLUTIONS provides the User with
permission in written form the User can incorporate parts of the software
into other software developed by the User. A distribution of the software
can only be made in compiled form as part of the software developed by
the User under strict observation of the conditions set forth in the written
permission to the User. The User must include the OSIS scan^R
SOFTWARE PRODUCT copyright notification with the User's software.
The User has to make sure, that OLYMPUS SOFT IMAGING
SOLUTIONS cannot be held liable for any damages or injuries resulting
from the use of the User's software, that include parts of the OSIS
scan^R SOFTWARE PRODUCT.
§ 4. Copyright
(1) OLYMPUS SOFT IMAGING SOLUTIONS or its subsidiaries remain
owners of the software and it's documentation. With the purchase, the
User obtains ownership of the diskettes or other physical storage de-
is manufacturer of the
vices (excluding the software and other data contained thereon), and the
manuals.
(2) OLYMPUS SOFT IMAGING SOLUTIONS reserves the right to all
publications, duplication, editing, and marketing of the software and the
software documentation.
Without prior written permission the User may not:
– change, translate, de-compile or de-assemble the software,
– copy any of the written or printed documentation of the software,
– rent, lease, or license the software to a third party,
(3) The license, property, and user rights to the OLYMPUS SOFT
IMAGING SOLUTIONS software, disks, and manuals may only be sold or
transferred to a third party on a permanent basis, if the third party agrees
to abide by the conditions in this contract.
(4) OLYMPUS SOFT IMAGING SOLUTIONS is the legal owner of all
copyrights and trademarks of the OSIS scan^R SOFTWARE PRODUCT
and documentation. National and international law protect copyrights
and trademarks. OLYMPUS SOFT IMAGING SOLUTIONS reserves all
rights, which are not explicitly expressed in written form.
§ 5. Warranty
(1) OLYMPUS SOFT IMAGING SOLUTIONS guarantees for the period of
12 months after the date of purchase, that the software works in all
major aspects according to the descriptions in the manuals. OLYMPUS
SOFT IMAGING SOLUTIONS, as the producer of the software, provides
this warranty. It does not replace or restrict other warranties or liabilities
provided to the User by local or other sales people or organizations.
OLYMPUS SOFT IMAGING SOLUTIONS does not guarantee, that the
software is defect free, that the software fulfills the specific requirements
of the User, or that the OBS scan SOFTWARE PRODUCT works wit h
other software provided by the User.
(2) OLYMPUS SOFT IMAGING SOLUTIONS further guarantees, that the
software storage devices (floppy disks, CD-ROMs, etc.) and the manuals
are free of material defects. Defective storage devices or manuals will be
replace free of charge, if they are returned to OLYMPUS SOFT IMAGING
SOLUTIONS within 90 days of purchase and accompanied by a proof of
purchase.
§ 6. Liability
(1) OLYMPUS SOFT IMAGING SOLUTIONS or their sales organizations
cannot be held liable for damages or injuries resulting from the use of the
software or the lack of capabilities of the software, unless the User can
show gross negligence on the part of OLYMPUS SOFT IMAGING
SOLUTIONS. This applies, without exceptions, also to losses of productivity or profit, interruptions in the flow of business or manufacture, loss
of information, and other financial losses. Without exceptions the possible liability of OLYMPUS SOFT IMAGING SOLUTIONS is limited to the
amount that the User paid for the product. These limitations on the
liability do not influence claims for reasons of product liability.
§ 7. Contract duration, legal consequences of violating the license
(1) The contract is deemed to be in force for an unspecified period. The
User rights are automatically terminated if one of the conditions of the
contracts has been violated.
(2) In case of a contract violation the User has to return the original
storage devices and all copies thereof including all modified copies, and
all printed and written documentation to OLYMPUS SOFT IMAGING
SOLUTIONS, or the User has to destroy these items.
(3) In addition OLYMPUS SOFT IMAGING SOLUTIONS reserves the right
to file a lawsuit to claim reparations for damages, non-compliance, or
removal of the software in case of license violations. The following laws
and/or conditions are in effect: the conditions of this contract, copyright
laws, and the laws of the civil code.
Page 5

Analysis Software Manual
Contents
1 Introduction............................................................................................ 1
1.1 Abstract.............................................................................................. 2
1.2 Technical Support.............................................................................. 2
2 Main User Interface................................................................................ 3
2.1 General.............................................................................................. 4
2.2 General Settings................................................................................ 6
2.3 The scan^R Data Structure................................................................ 7
2.4 Managing Histograms and Scatter Plots............................................ 7
2.4.1 The Histogram Context Menu.......................................................... 9
2.4.2 The Region Context Menu............................................................. 10
2.5 Using the Image Viewer................................................................... 11
2.6 Adjusting the Image Displays........................................................... 12
2.7 Selecting Wells for analysis............................................................. 13
3 Assays.................................................................................................. 15
3.1 General............................................................................................ 16
3.2 Object Finder: Detecting Main Objects............................................ 16
3.3 Sub-object Finder: Detecting Sub-objects ....................................... 18
3.4 Object Finder Modules..................................................................... 20
3.4.1 Entire Image................................................................................... 20
3.4.2 Intensity Threshold......................................................................... 20
3.4.3 Edge Detection............................................................................... 21
3.5 Measurement Parameters ............................................................... 25
3.6 Derived Parameters......................................................................... 26
3.7 Image Processing............................................................................ 27
3.7.1 Background Correction .................................................................. 28
3.7.2 XY Shift.......................................................................................... 29
3.7.3 Inversion......................................................................................... 29
3.7.4 Cut Image....................................................................................... 30
3.8 Virtual Channels............................................................................... 31
3.8.1 Simple Math................................................................................... 31
3.8.2 Spectral Unmixing.......................................................................... 33
3.9 Tracking – Analyzing Time-Lapse Data........................................... 35
3.9.1 Tracking Configuration................................................................... 37
3.9.2 Track Analysis Parameters............................................................ 38
3.9.3 Trace Viewer.................................................................................. 43
4 Analysis Results.................................................................................. 45
4.1 Running an Analysis........................................................................ 46
4.2 Managing Gates............................................................................... 47
4.2.1 Gates and Regions. ....................................................................... 47
4.2.2 The Gate Manager......................................................................... 47
Page 6

Analysis Software Manual
4.2.3 Color Gating................................................................................... 49
4.3 Well Results..................................................................................... 52
4.3.1 Measurement Results.................................................................... 52
4.3.2 Well Results: Populations.............................................................. 53
4.3.3 Export Definitions........................................................................... 54
4.3.4 Export results of individual objects ................................................ 56
5 Example Assay – Step by Step........................................................... 57
5.1 Setting-up and Executing an Assay................................................. 58
5.2 Analyzing the Data .......................................................................... 61
5.3 Time-Lapse Analysis....................................................................... 64
6 Appendix.............................................................................................. 71
6.1 File Conversion................................................................................ 72
6.1.1 Images List .................................................................................... 72
6.1.2 Image Channel Definition .............................................................. 73
6.1.3 Scan Settings................................................................................. 74
6.1.4 View Conversion Results............................................................... 74
6.2 FCS Export Functionality................................................................. 75
6.3 Libraries........................................................................................... 77
6.3.1 Object Analyzers Library (OAL)..................................................... 77
6.3.2 Object Finders Library (OFL)......................................................... 78
6.3.3 Image Processing Library.............................................................. 78
6.3.4 Virtual Channel Library.................................................................. 79
6.4 Valid functions for derived parameters............................................ 80
Page 7

Analysis Software Manual Chapter 1 – Introduction 1
1 Introduction
Page 8
2 Chapter 1 – Introduction
Thank you very much for purchasing Olympus’ Screening Station for Life Science and for your confidence in our products and services.
The
the Olympus
tended for the use in biomedical research.
Analysis Software is designed for the automated analysis of images that were acquired by
Screening Station and with the Acquisition Software. The software is in-
The
ponents of the Olympus Screening Station for Life Sciences are for research use
only.
Analysis Software, the Acquisition Software as well as the hardware com-
1.1 Abstract
This user manual will guide you through the usage of the analysis software of the Olympus Screening
Station scan^R. It will assist you in setting up efficient and reliable assays from scratch. This scan^R
module is intended to be used for the analysis, quantification and navigation through your results. The
analysis module of scan^R allows you to run the analysis during acquisition or in “offline mode” afterwards.
Special care has been taken to guarantee correct and accurate information within this documentation,
although this is subject to changes due to further development of the Screening System. Thus, the
manufacturer cannot assume liability for any possible errors. We would appreciate reports of any mistakes as well as suggestions or criticism.
1.2 Technical Support
If you find any information missing in this manual or you need additional support, please contact
Olympus directly.
Page 9

Analysis Software Manual Chapter 2 – Main User Interface 3
2 Main User Interface
This chapter explains the features of the image displays and briefly introduces the different menu
points and buttons accessible from the main user interface.
2.1
General.........................................................................................................4
2.2 General Settings...........................................................................................6
2.3 The scan^R Data Structure........................................................................... 7
2.4 Managing Histograms and Scatter Plots....................................................... 7
2.4.1 The Histogram Context Menu.....................................................................9
2.4.2 The Region Context Menu........................................................................10
2.5 Using the Image Viewer..............................................................................11
2.6 Adjusting the Image Displays...................................................................... 12
2.7 Selecting Wells for analysis........................................................................13
Page 10
4 Chapter 2 – Main User Interface
2.1 General
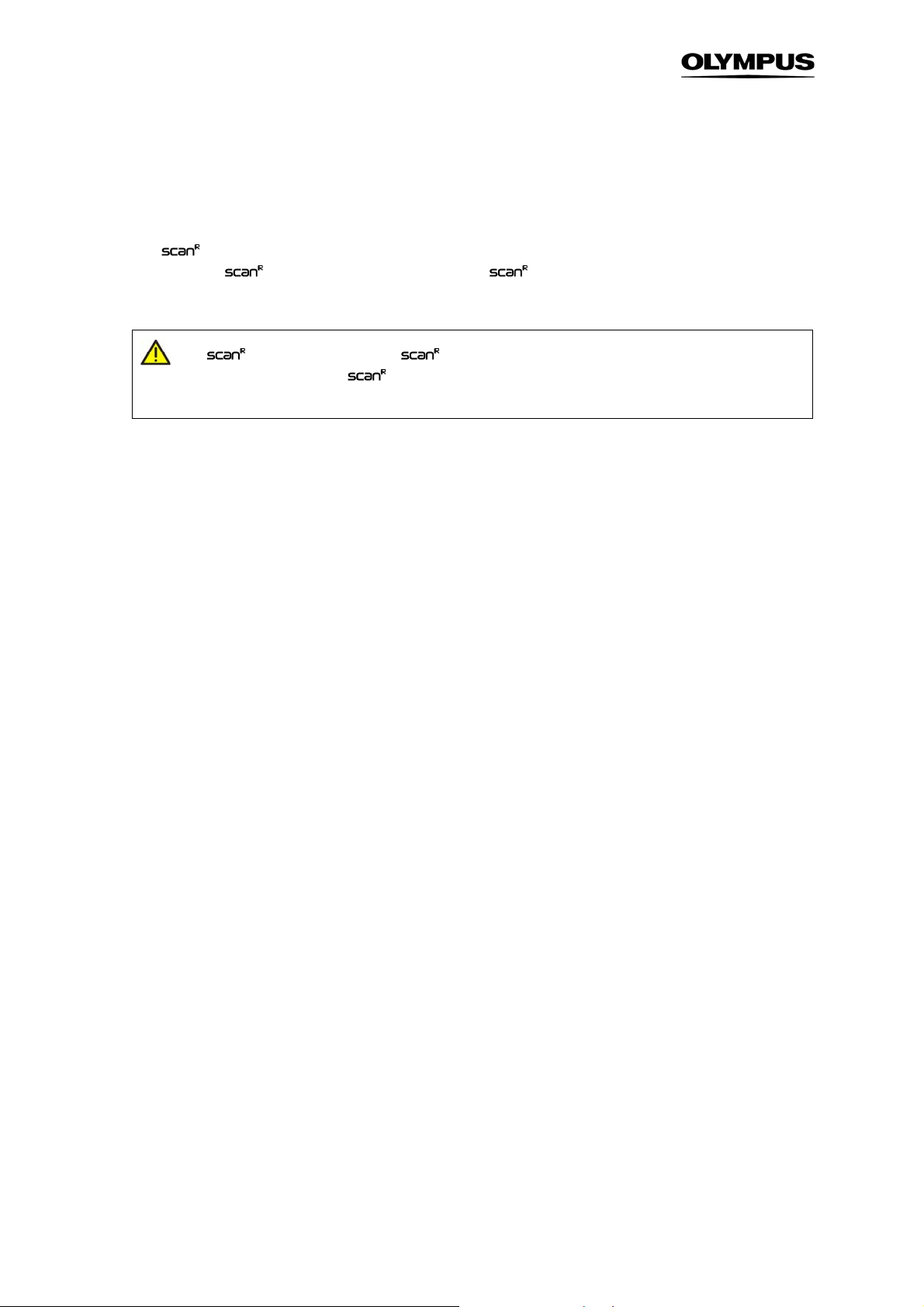
The scan^R Analysis main graphical user interface contains four histograms and an image viewer window. The functions of these histograms are explained in Chapter 2.4,
The image viewer window shows the image with the object corresponding to a data point selected in a
histogram. A description of the image viewer functions is given in Chapter 2.5,
Navigation through the images of a scan is possible with the tools in the Image box. Color channel
selection is done with respective pull-down shortlists in the Display box.
An assay can be started and followed online with the tools and displays in the field at the lower right of
the main window.
The menu bar at the top contains several pull-down menus and commands. They are listed in the following overview:
Managing Histograms
Using the Image Viewer
.
.
AnalysisRun: starts the analysis with the current settings
AnalysisBatch Run: starts multiple analysis with the current settings
Page 11

Analysis Software Manual Chapter 2 – Main User Interface 5
AnalysisOpen: opens a previous analysis (*.sca-file)
AnalysisSave/Save as: saves the current analysis (*.sca-file)
AnalysisExport Table: exports the values determined for each detected object to a spread sheet. The
values exported depend on the active view. When also sub-objects are detected not only one file is
exported but for every sub-object a separate list is exported. The values that are exported depend on
the active view: population view / trace view (see Chapter4.3.4)
AnalysisEdit Assay: opens the “Assay settings” menu
AnalysisLoad Assay: loads an existing assay (*.say-file). In contrast to the *sca-file the *.say-file contains only the analysis, i.e. the operations to perform on a data set but not the results of a specific
analysis.
AnalysisSave Assay: saves the current assay (*.say-file)
AnalysisAssay Gating: opens the gate manager (see Chapter 4.2.2)
AnalysisExit: exits the analysis
ScanOpen: opens a scan and assigns it to the current assay. The file types that can be opened are
the scan^R experiment descriptor files (*.xml-format) and dotslide images (*.wtp-, and *.ets-format)
ScanRelink images: re-links acquired images to an analysis. To do so, navigate to the folder where
the images are stored and select Current Folder.
ScanCustom conversion: converts a third party data set into the scan^R format (see Chapter 6.1)
ScanSelect wells: selects a set of wells for analysis and data navigation; allows also to display a well
overview, i.e. an overview of the images that were acquired in a well. (see Chapter 2.7)
ScanScan Info: displays the path to the image data
ScanSettings: displays the settings to a scan
TrackingTrace View: toggles between the Population and Trace View modes.
TrackingConfigure Tracer: opens the Trace Configuration window to select how the object tracking is
to be performed.
TrackingDefine Parameters: opens the Trace Parameters window to select trace analysis operations.
TrackingShow traces: opens the Trace Viewer that visualizes the trace graphs.
ViewLayout: display properties for the RGB display (affects only the displays).
ViewParameter View: lists all parameters for a selected object, that were determined during analysis
ModulesObject Finders: list and configure available Object Finder Modules (Chapter 6.3, Libraries)
ModulesObject Analysers: list and configure available Object Analyzer Modules (Chapter 6.3, Libraries)
ModulesImage Processors: list and configure available Image Processing Modules (Chapter 6.3,
Libraries)
ModulesVC Processors: list and configure available Virtual Channel Modules (Chapter 6.3, Libraries)
RepositioningInteractive: the scan^R screening system moves to the position of the selected object
Settings: Gives access to some preferences including galleries, directories, data export and settings for
repositioning an reclassification.
Page 12
6 Chapter 2 – Main User Interface
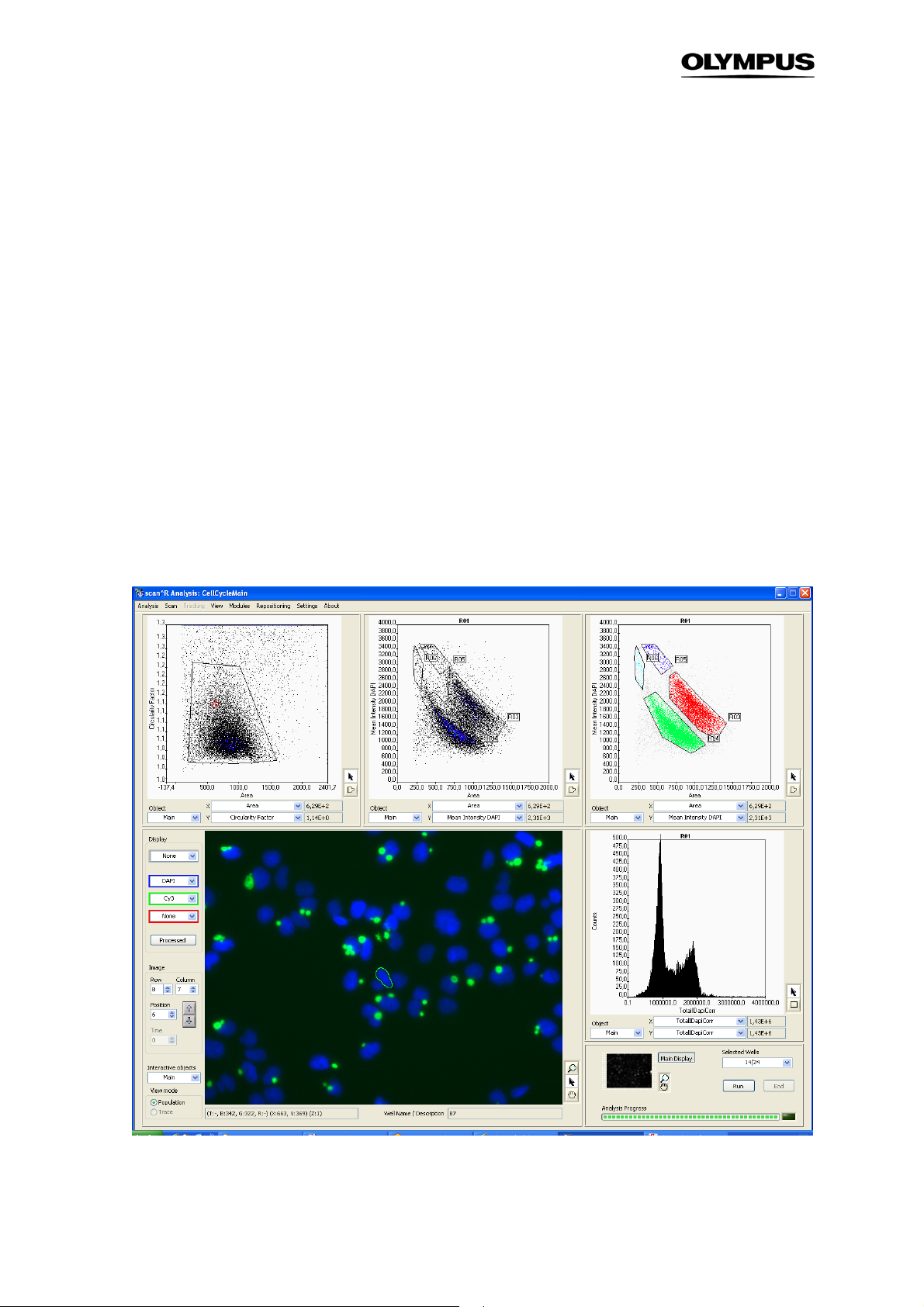
2.2 General Settings
The general preferences can be defined in the Settings menu.
Max Gallery Objects. Sets the number of objects that are displayed in a gallery.
Sort mode. The options are Typical, Random, yx and –yx. When Typical is set, the galleries display the
objects which are closest to the center of gravity of the selected region or histogram in the order of
distance to that center. Random displays randomly selected objects within the selected region. The
options yx and –yx allow to create a gallery that is ordered by one parameter.
Default Data Directory. Enter the directory where the data are read by default (used for “open scan”)
Result Export Directory. Enter the directory where the results are to be stored. When this field is empty
the results will be stored in the scan directory\Population Results. The results of a tracking analysis will
bestored in the scan directory\Trace Results. (Note that in earlier versions the results are stored in the
scan directory\Results folder.
Page 13

Analysis Software Manual Chapter 2 – Main User Interface 7
Export2Txt. The results can be exported as .txt files or as .fcs files. (For export to .fcs format, see
Chapter 6.2)
Repositioning & Reclassification. Set the Port and the Address for communication with the scan^R
screening system for experiments with repositioning.
2.3 The scan^R Data Structure
scan^R analysis data can be separated into the acquired images and the assay being applied on them.
The acquired images as a whole are called a scan; it includes the individual images and their acquisition settings like color channels, integration time, plate information etc. An assay describes the processing and analysis steps applied to extract data out of the images.
This separation between analysis settings and acquired images allows the reuse of once adapted
analysis settings for different scans.
The images acquired during a scan^R scan are stored as 16-bit *.tif files in a Data subfolder in the experiment scan storage folder.
Additionally, the scan settings are stored in an Experiment_descriptor.xml and the stage positions in
the Acquisitionlog.dat file.
The scan^R analysis software serves for the analysis of the scans. The instructions (assays) for these
analyses are stored in the scan^R Analysis/Assays folder as *.say files. These files can be loaded via
AnalysisLoad Assay… to then apply the assay on a scan data set.
Once an assay has been performed on a data set, a *.sca file is generated and can be stored in the
experiment storage folder. These files contain all analysis data including histograms and scatter plots
etc. *.sca files can be loaded via AnalysisOpen… to revisit the analysis results.
To clarify this: if you open a Scan via ScanOpen… the experimental settings of a scan will be loaded
by reading in the Experiment_descriptor.xml file. Thus, you get access to the raw image data. In contrast to this, if you open an Analysis (*.sca) you will get the results in addition to the images.
2.4 Managing Histograms and Scatter Plots
Histograms are used in scan^R for data representation, classification and navigation. scan^R uses 2-D
and 1-D histograms following the common representation in cytometry.
A 1-D histogram shows the frequency distribution of only one parameter. A 1-D histogram is created
when the same parameter for X and Y is selected. On the X-axis of the histogram the selected parameter and on the Y-axis the number of counts is plotted.
A 2-D histogram (scatter plot) is a plot of one parameter of a number of objects against a second parameter. Color-coding is used as "third dimension" to represent the frequency of occurrences.
The data displayed in a histogram are assigned to an object type (main-/ sub-object). The detection of
the objects is defined by the assay. The object type can be chosen from the Object pull down menu
Page 14
8 Chapter 2 – Main User Interface
underneath the histogram. The X and Y pull down menus are then automatically updated to correspond
to the measured parameters of the chosen object type (see assay definition). The X and Y pull down
menus are used to change the parameters displayed in a histogram. The axes of abscissa and ordinate
are labeled with the chosen parameter.
Two buttons are located in the lower right corner of each histogram. They allow toggling between the
Navigation and the Region-Selection modes.
Navigation
The Navigation button with the pointer symbol allows navigating within the data. Each data point within
a histogram is directly linked to the object from which it is derived. A selected data point is highlighted
by a red circle in all histograms in navigation mode as long as the data point is within the displayed
area. The corresponding object is displayed in the Image Viewer. The X and Y values of the data point
are displayed next to the X and Y pull-down menus. Holding and dragging the mouse using this tool
allows to virtually following the objects changes within the parameter set. The navigation tool also allows dragging and modifying existing regions.
Region
The Region tool is used to draw polygons into a 2-D histogram and to set a range in 1-D histograms.
Regions define bi-dimensional intervals within the parameter range and thus subpopulations of data
points. Double-click in order to close a region in a 2-D histogram.
Page 15

Analysis Software Manual Chapter 2 – Main User Interface 9
2.4.1 The Histogram Context Menu
The histogram can be managed through the context menu accessible via right-click into the histogram.
(Note that you will get the region context menu, if you right-click on a region border as described below). The histogram context menu contains the following commands:
• Set Gate. Apply a gate on the selected histogram by choosing a region from the list that appears.
• Show region. Select a region from the list that appears to generate a zoomed-in view of it with the
X/Y parameters that were used to define this region.
• Clone from. Select a histogram from the list that appears to duplicate the histogram.
• Gallery. This command generates an image gallery of objects in the current histogram. The number
of images and the selection criteria is set according to the gallery preferences given in the Settings
menu.
• Zoom out. This command zooms out of the current view by a factor of 2 (in case of linear axis scal-
ing).
• Color Gating. This command causes the population of each gate to be displayed in a different color
in the histogram.
• Settings… This command opens the Histogram Properties dialog with the following settings.
Axis attributes. This field offers different choices to set the axis scaling.
Grid cosmetics. This field offers different choices to set the grid display.
Autoscale. This field offers different choices to set the auto scale of the axes.
BKColor. A click on the colored field opens a window that allows selecting the background color.
Page 16
10 Chapter 2 – Main User Interface
Selector Color. A click on the colored field opens a window that allows selecting the cross-hair color.
Region Color: A click on the colored field opens a window that allows selecting the color of the region
outline.
Color scheme. This color palette defines the coloring of data points (pixels) in the histogram in dependence of the number of counts (events) they represent. For example, if the Color table bin width is set to
three and a data point represents 7, 8 or 9 counts, it will be displayed in the third color from the left.
Color table bin width. It defines the range of counts to be given the same color from the Color scheme.
Binned color table. Click this button to activate the color binning as set in Color table bin width.
Color Gating. This command causes the population of each gate to be displayed in a different color in
the histogram.
Show Legend. This command causes a legend of the colors in the Color Gating mode to be displayed
in the histogram.
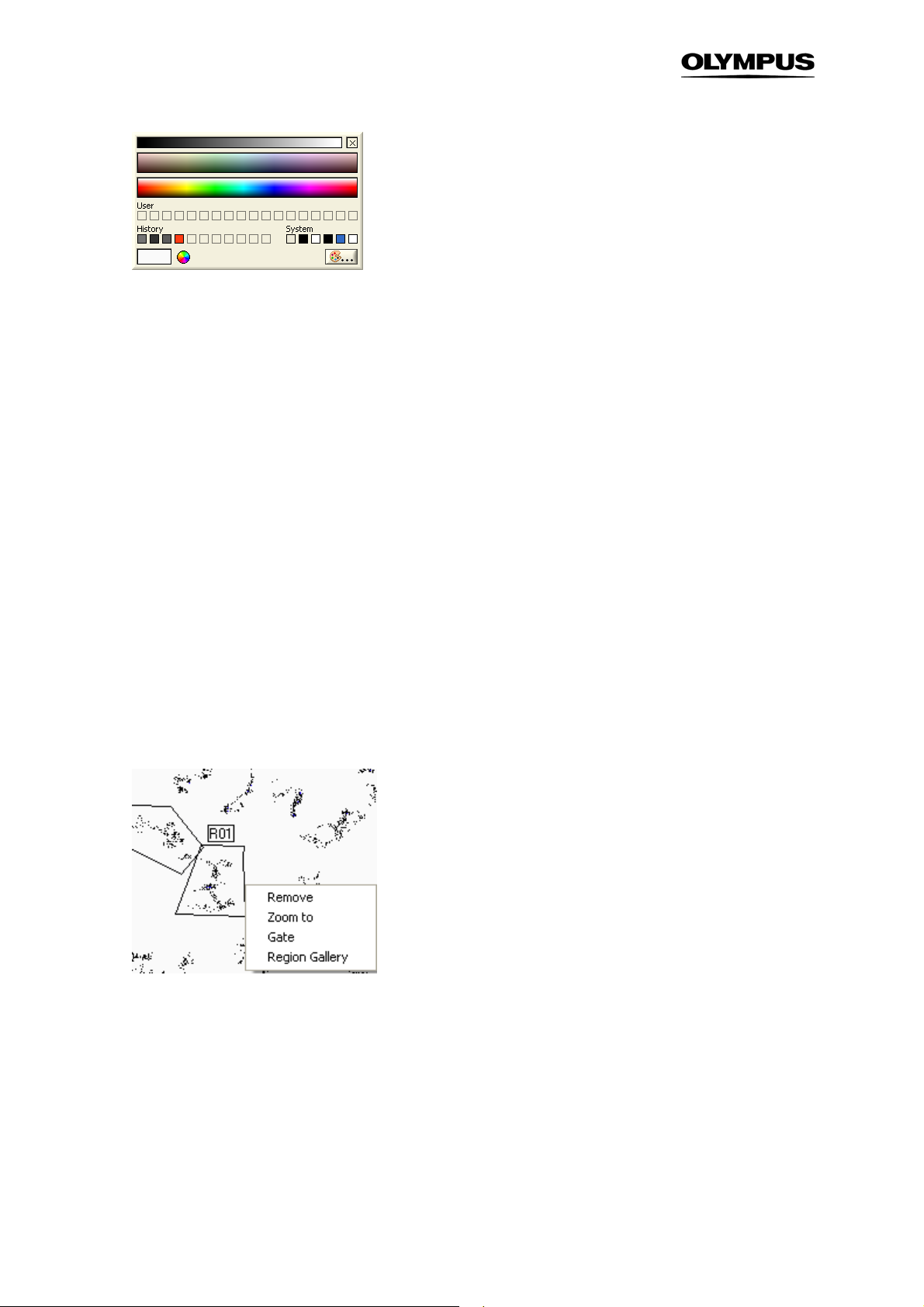
2.4.2 The Region Context Menu
The region context menu will open, if you right-click on a region border
• Remove. Deletes the selected region or gate.
• Zoom to. Gives a zoomed view of the selected region.
• Gate. Converts the region into an AND Gate. (See also Chapter 4.4.2,
The Gate Manager.
) When a
gate is applied to a histogram, only the data points within this gate are displayed. To display all data
points open the histogram context menu and go to Set gatenone.
• Region Gallery. This command generates an image gallery of objects in the selected region. The
number of images and the selection criteria is set according to the gallery preferences given in the
Settings menu. (For more information see Chapter 2.2,
General Settings.
)
Page 17
Analysis Software Manual Chapter 2 – Main User Interface 11
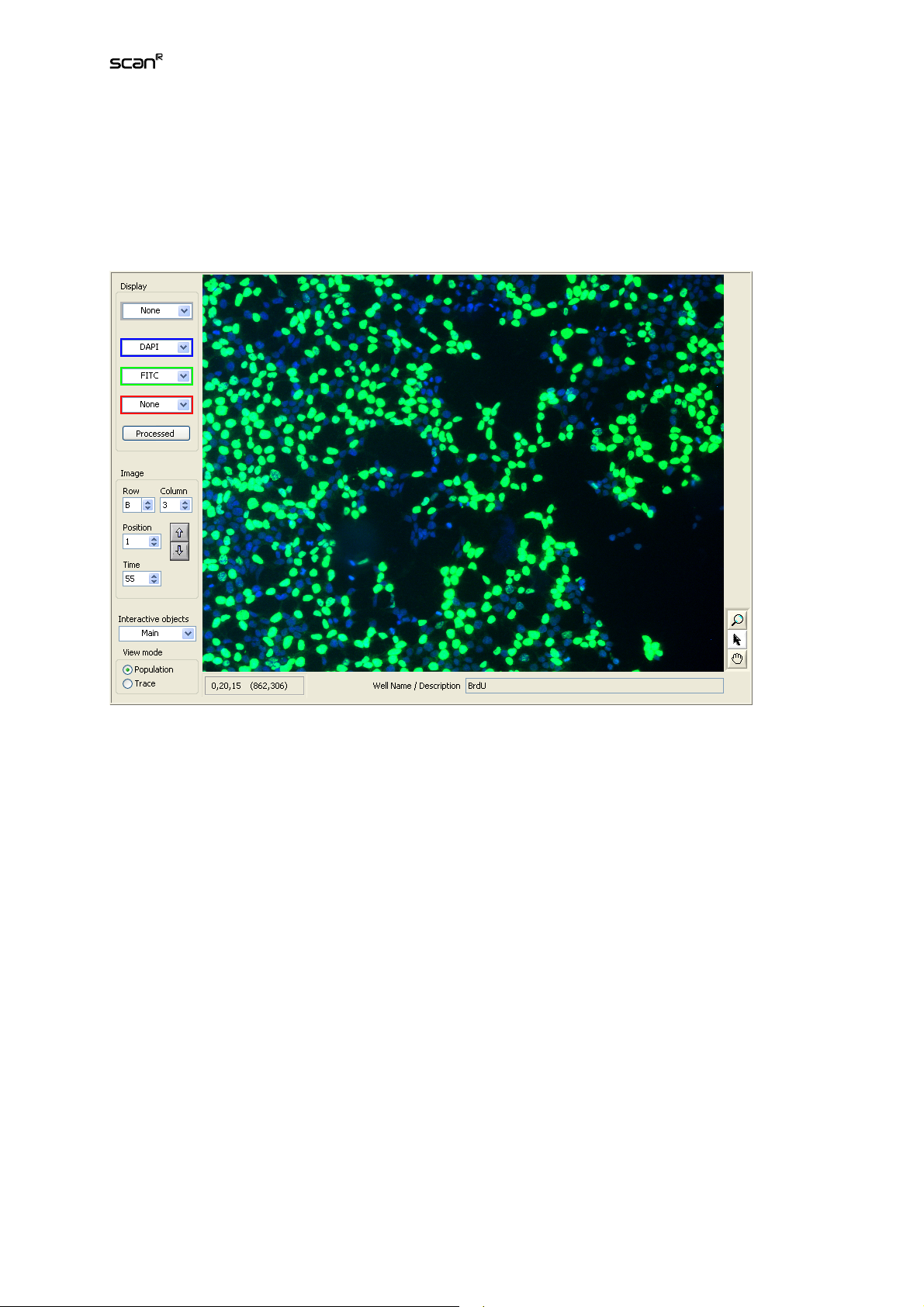
2.5 Using the Image Viewer
Each analysis data point is directly linked to the image object it is generated from. These objects can
be displayed in a close-up view in the image viewer. Browsing in the data set by clicking on data points
with the histograms Navigation tool automatically leads to a corresponding update of the image in the
viewer. Additionally the Well, Position, Time and Well/Name Description fields are updated and give
information about the image origin. The small arrow buttons on the left of these fields can likewise be
used to navigate. Values can be typed in as well.
Display. Multi-color images consist of individual color channels that were recorded with different optical
acquisition settings (e.g. different excitation filters). The red, green and blue pull-down menus serve to
select the input for the three color channels that are displayed in the respective colors. In order to display a channel (e.g., a transmission channel) in grayscale, select it from the gray pull-down menu.
When having both grayscale and any color selection active, a transmission overlay/display will be used
for the grayscale selection.
The clipping of the RGB display, i.e., the scaling of the image display brightness, can be changed via
the menu point ViewLayout that opens the Image Clipping window.
Image: Processed. Click this button to toggle between the original image and the processed image as
defined in AnalysisEdit AssayAssay Settings/Image Processing. Image processing is used to improve the quality of the displayed image but slows down the systems image display. Therefore it is
especially recommended to switch it off for performance when creating galleries . The image processing is described in Chapter 4.9,
Image Processing.
Page 18

12 Chapter 2 – Main User Interface
Row/Column/Position/Time. Use these entries to select a specific image to be displayed.
The up/down arrows allow fast navigation though your image data set by incrementing or decrementing
the Time, Position and/or Well number.
Interactive objects. Select the object type to be outlined in the display when clicking on an object or
data point.
The image viewer is equipped with a tool bar to select different mouse tools:
Zoom
The Zoom mode is used to zoom into the displayed image. A click into the image causes a zoomed-in
view with the cursor position as the center. To zoom out, the Shift key must be pressed simultaneously.
Selection
The Selection mode allows selecting individual objects within the image via mouse click. The object
type to be displayed can be selected in the pull down menu in the bottom right corner of the image
viewer. Main objects are highlighted by a green outline. The data point corresponding to the selected
object is highlighted with a red circle 2-D histograms and a vertical red line in 1-D histograms.
Move
Depending on the zoom factor, only a part of the image will fit into the display. The Move mode allows
moving the visible area via mouse-drag.
The status bar in the lower left corner shows information about the magnification of the displayed image, the current x/y position of the cursor and the pixel value(s) at this position.
2.6 Adjusting the Image Displays
ViewLayout opens the Image Clipping window that allows adjusting the display brightness. Note that
these image settings affect the front panel display as well as well overviews and galleries A raw image
will always have a certain background intensity. Also, one will avoid to over saturate images and – especially in fluorescence applications – rather use only a fraction of the camera chip capacity. The consequence is that a raw image is usually low in contrast and may even appear entirely black. Clipping is
applied to change the image brightness by defining a range of low pixel counts to be displayed black
as well as a range of high pixel counts to be displayed with maximum brightness.
Clipping Type. This button toggles between Dynamic [%] and Absolute clipping.
Page 19
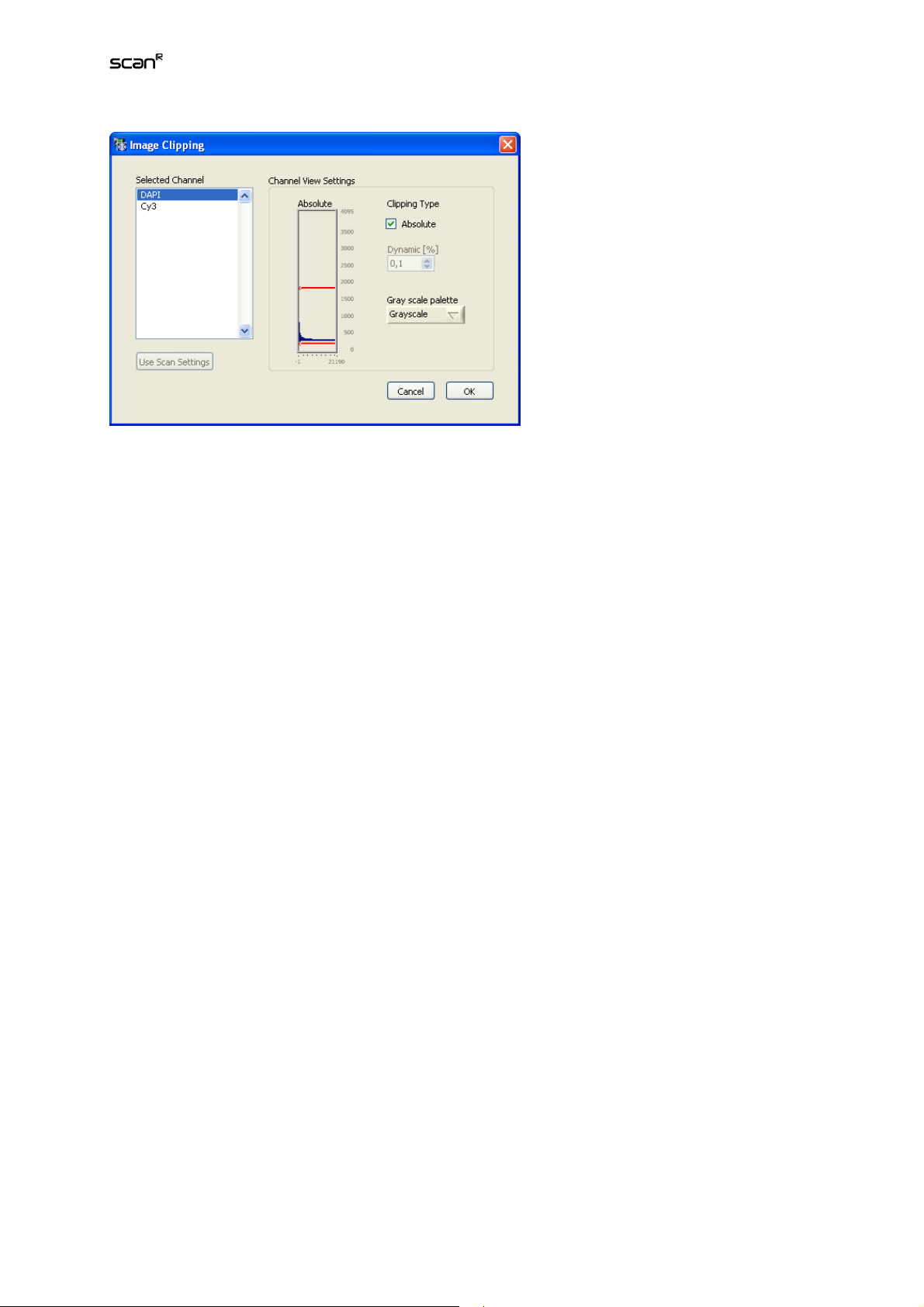
Analysis Software Manual Chapter 2 – Main User Interface 13
Dynamic [%]. Define here how many pixels (as a percentage of the total number of pixels) will be displayed with maximum and minimum brightness. The intensity of the remaining range of pixels will then
be scaled linearly in between. The higher the numbers the stronger the resulting contrasts.
Absolute. Define here the range of pixel counts to be displayed with maximum and minimum brightness
by dragging the red horizontal lines – that represent the maximum and minimum thresholds – with the
mouse. The intensity of the remaining range of pixels will then be scaled linearly in between.
Gray scale palette. The channel selected in the gray pull-down menu in the main GUI can be displayed
in different false-color palettes that can be selected here.
Use scan settings.
2.7 Selecting Wells for analysis
The command ScanSelect Wells opens the Select Wells display. It features a graphical representation of all wells of the well plate or slide.
When the window is opened for the first time, green circles represent wells that were selected for scanning. These wells will be considered in the analysis . To change this selection double-click on the wells.
These wells are also removed from the list on the right. A second double-click reactivates the well.
Dragging a rectangle around a group of wells likewise carries out deactivation and activation.
In the list on the right you can also change the name of the wells and group wells by assigning the
same name.
Restore. This function restores the initial well pattern
Name/Description. In certain cases it may be advantageous to group wells (see also Chapter 4.3,
Results
list. The default entry is the alphanumerical code of the well position. The name can be changed at will.
). Wells of one group need to have an identical entry in the Name/Description column of the well
Well
Page 20
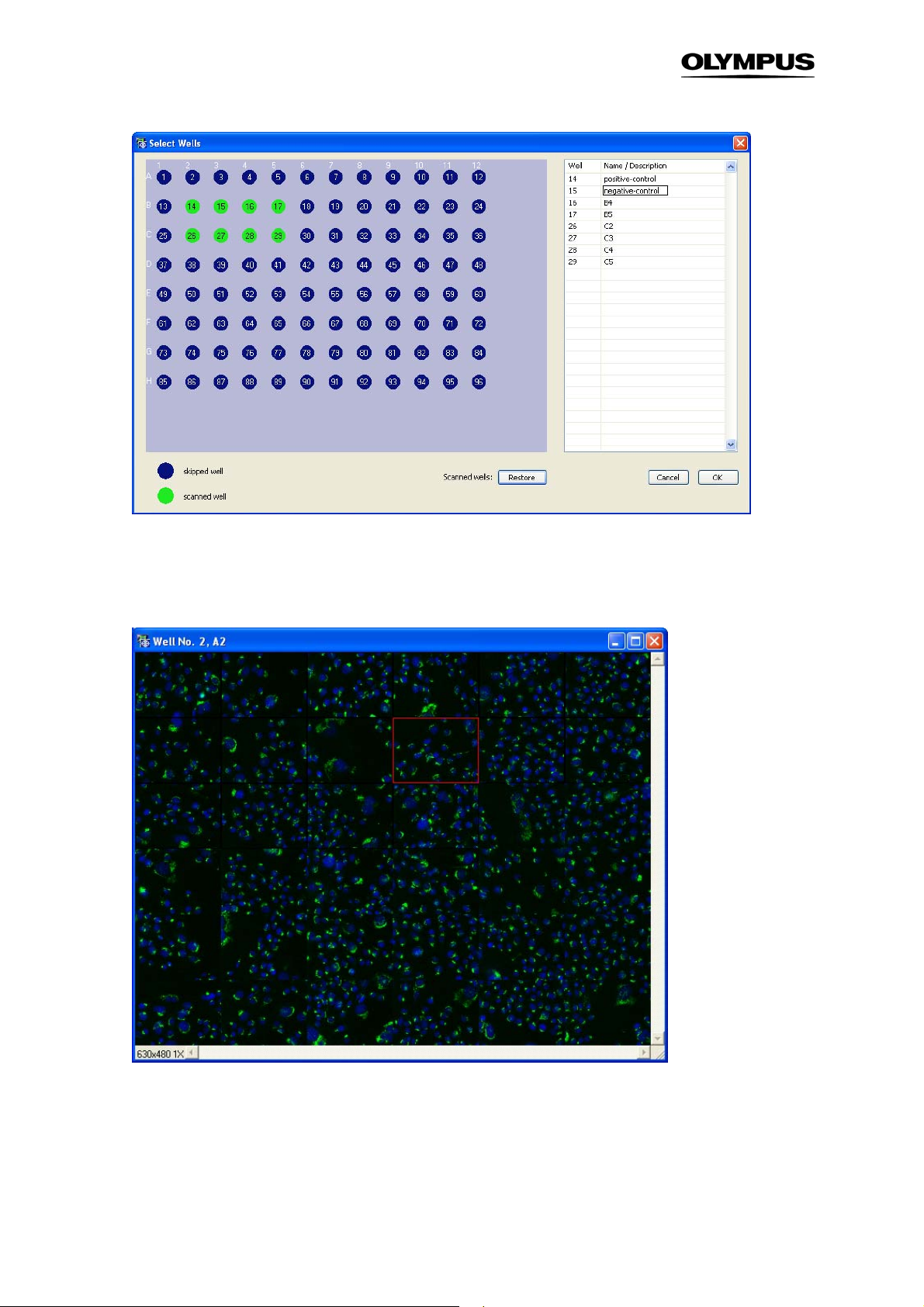
14 Chapter 2 – Main User Interface
Right-click on one of the scanned wells gives you the option Well Overview. For a time series you will
have to set the time point first. The Well Overview will display all the positions you recorded for one
well. Selecting one of the images will load the image in the main display on the front panel.
Well overview.
Page 21

Analysis Software Manual Chapter 3 – Assays 15
3 Assays
Assays are the recipes to extract the data of interest from the images of a scan. They define which
objects are to be recognized and how and which measurements are to be performed on the found
objects. This chapter explains in detail how assays are to be set up or modified.
3.1
General.......................................................................................................16
3.2 Object Finder: Detecting Main Objects.......................................................16
3.3 Sub-object Finder: Detecting Sub-objects ..................................................18
3.4 Object Finder Modules................................................................................20
3.4.1 Entire Image..............................................................................................20
3.4.2 Intensity Threshold....................................................................................20
3.4.3 Edge Detection.......................................................................................... 21
3.5 Measurement Parameters .......................................................................... 25
3.6 Derived Parameters.................................................................................... 26
3.7 Image Processing.......................................................................................27
3.7.1 Background Correction ............................................................................. 28
3.7.2 XY Shift.....................................................................................................29
3.7.3 Inversion....................................................................................................29
3.7.4 Cut Image..................................................................................................30
3.8 Virtual Channels.......................................................................................... 31
3.8.1 Simple Math..............................................................................................31
3.8.2 Spectral Unmixing..................................................................................... 33
3.9 Tracking – Analyzing Time-Lapse Data...................................................... 35
3.9.1 Tracking Configuration.............................................................................. 37
3.9.2 Track Analysis Parameters.......................................................................38
3.9.3 Trace Viewer............................................................................................. 43
Page 22

16 Chapter 3 – Assays
3.1 General
An assay defines all steps necessary to extract quantitative data from the acquired images. It usually
starts with some sort of image processing like background correction. Secondly, objects have to be
detected in the images. The analysis, for example different kinds of measurements, e.g. area, intensity,
shape…, is finally performed on these objects and results for the samples (e.g. wells) are generated.
The AssayEdit Assay command opens the Assay Settings window and allows applying or adapting
the assay to the loaded scan. The Assay Settings window contains six tabs: Main Object, Sub-objects,
Parameters, Derived Parameters, Image Processing and Virtual Channels. The tabs are arranged from
left to right, but you can always jump back and adjust the settings of former steps. However, background correction (in the Image Processing tab) should be performed prior to object detection as it
changes the intensity values.
scan^R distinguishes between two kinds of object types: an assay always defines one Main Object
type and up to four Sub-Object types connected to it. To give an example, main objects may be individual cells while their sub-objects are individual structures within them.
To represent this hierarchical structure, the Assay Settings window’s tabs Main Object and SubObjects are used to adapt different set the search algorithms for main and sub-objects in order to extract the structures of interest from the images. This is done by different Object Finder Modules that
implement different rules for object detection.
The Parameters and Derived Parameters tabs contain the information about the kind of information to
be extracted from the objects (e.g. area, shape,…) .
The Image Processing tab allows defining image processing steps that are to be executed before the
object detection and parameter extraction.
In the Virtual Channels tab new channels can be created as a result of post-acquisition image processing (e.g. spectral unmixing). To access the Virtual Channels tab you have to navigate through the tabs
to the right using the arrow buttons on the top right.
3.2 Object Finder: Detecting Main Objects
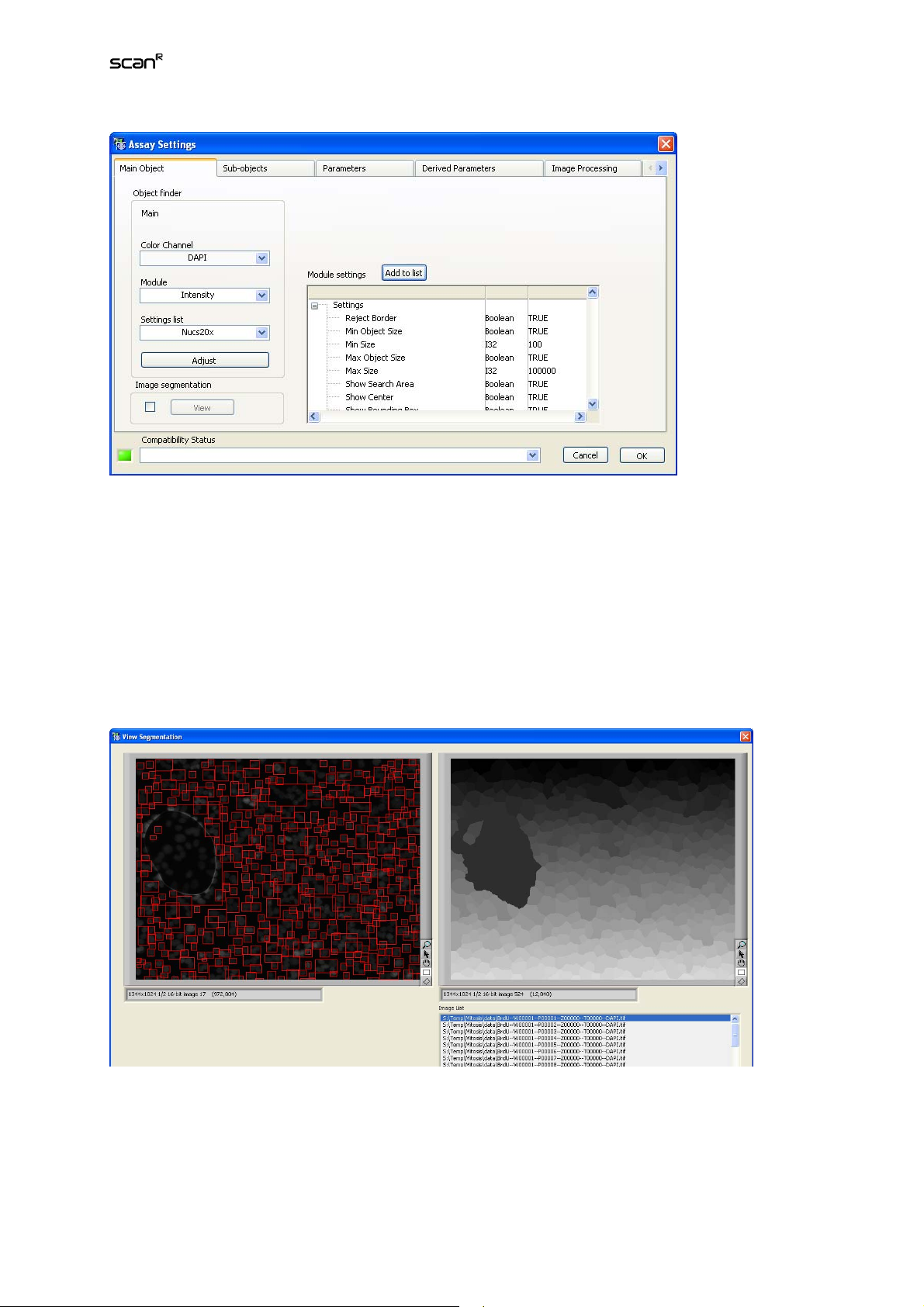
The Main Object tab of the Assay Settings window provides the commands to define the Main Object
detection.
Color Channel. Select the color channel on which the main object detection is to be performed.
Module. Select the method to detect individual Main Objects from the shortlist.
Page 23
Analysis Software Manual Chapter 3 – Assays 17
Settings list. Each Object Finder Module has a list of preset parameters. The lists can be modified and
stored at will. Individual modifications of these settings are marked as Modified. Select the settings of
choice from the shortlist.
Adjust. This command opens the configuration dialog of the selected Object Finder Module. (For
changing the list of Object Finder Modules see Chapter 3.4,
Object Finder Modules)
Module settings. This field lists the current settings of the selected Object Finder Module.
Add to list. This adds the modified settings to the Settings list. Click the button to open the Add Settings to OFL window (Object Finders Library, see Chapter 4.5,
Object Finders Library
) where you have
to give a New Settings Name for the modified settings list.
Image segmentation. This function divides – if activated via the check box – the entire image into segments: as many segments as there are Main Objects where each segment is assigned to the Main Object in its center. In other words, each image pixel is assigned to the Main Object it is closest to. All
pixels that are assigned to the same Main Object form one segment of irregular shape and size. The
Page 24
18 Chapter 3 – Assays
View button opens the View Segmentation window that contains on the left a display of the objectcircumscribing rectangles and on the right a display of the segments.
3.3 Sub-object Finder: Detecting Sub-objects
Sub-objects are structures that are directly linked to individual Main Objects. The search for Subobjects takes place on an image mask derived from the corresponding Main Object. This Main Object
mask can be adapted for each Sub-object type separately.
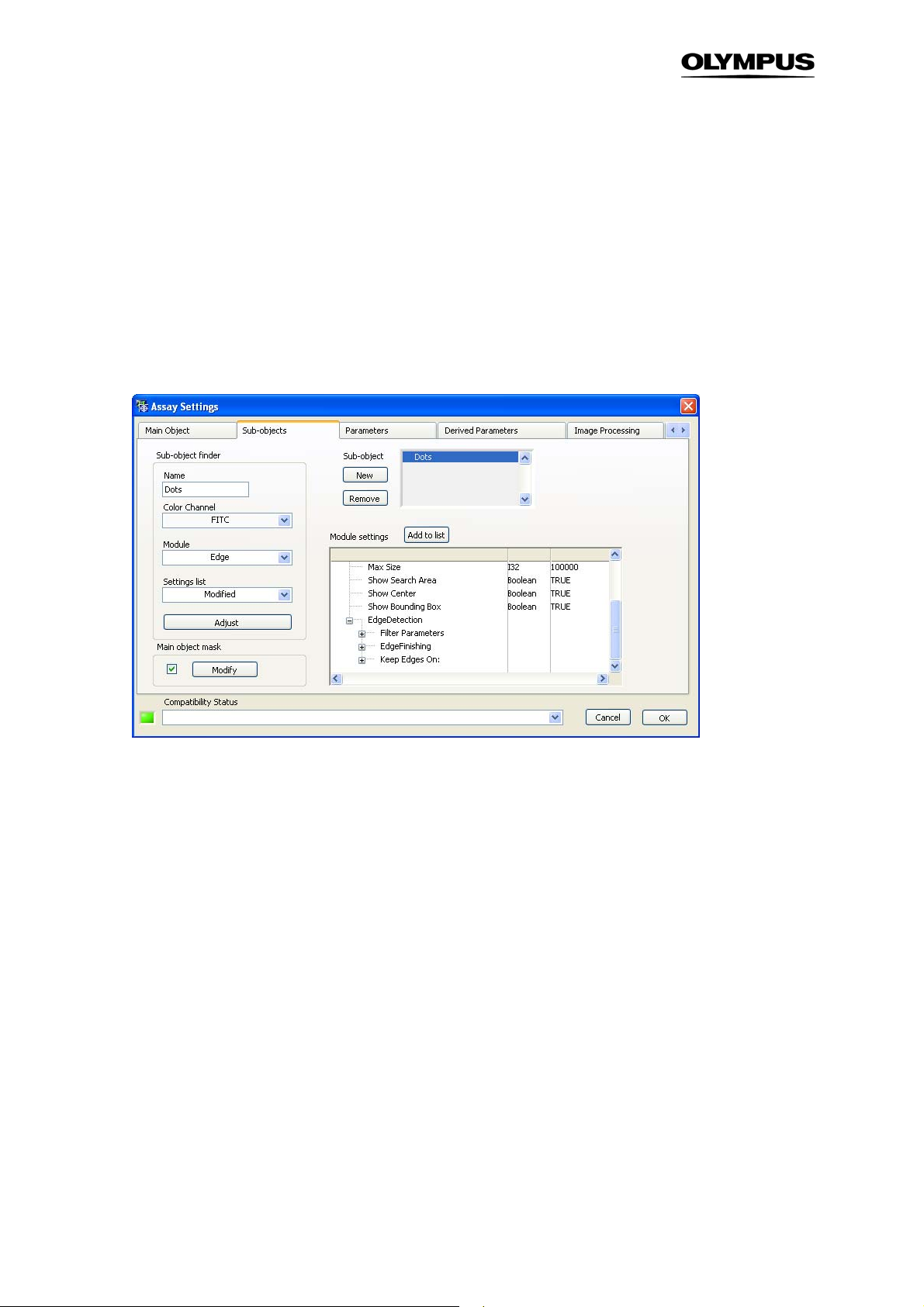
Sub-object finder: Color Channel, Module, Setting list, Adjust, Add to list. These functions are analogous to the ones described in the previous Chapter 3.2,
Name. Give a name to each new Sub object. The default name is Obj. 1.
Sub-object list. It gives an overview of the defined Sub-object types. The New and Remove buttons
allow the insertion and deletion of Sub-object types.
Main Object Mask. Each individual Main Object found in an image creates a mask. The individual Subobjects are associated with this mask rather then with the Main Object itself. Imagine a Main Object is
the cell nucleus and the Sub-objects are structures outside of it. In order to be detected, the original
Main Object Mask – which only covers the area on the nucleus – needs to be modified in order to enable the detection of the Sub-objects.
Click the checkbox to enable the image mask modification of the Main Object.
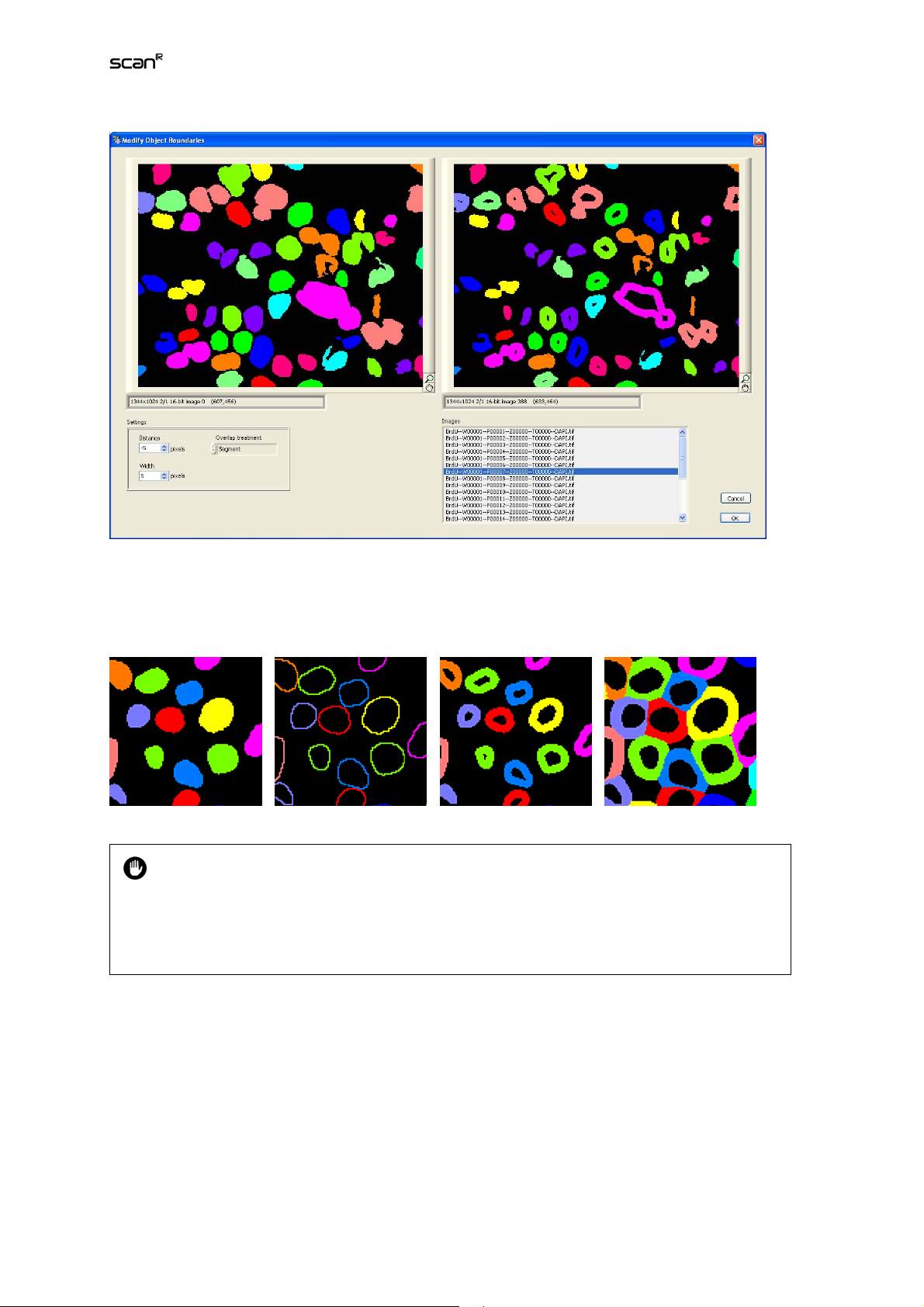
Modify button. Click here to open the Modify Object Boundaries dialog to adapt the main object mask
to the needs of the Sub-objects detection.
Distance. The distance is measured from the outer rim of the main object mask (positive and negative
values are valid)
Object Finder: Detecting Main Objects
.
Page 25
Analysis Software Manual Chapter 3 – Assays 19
Width. Extension of the sub-object mask
The examples below illustrate the effects of these parameters.
Overlap treatment. Options are Segment, Segment (slow) and remove.
Main object mask Distance 0, Width 1 Distance -5, Width 5 Distance 8, Width 8
When sub-objects are used for analysis, a further parameter becomes available in the Parameter tab (see Chapter 3.5,
for sub-objects is used, otherwise it would be
Measurement Parameters
subobjectsname
): Obj. 1 counts (if the default name
counts). This parameter
gives the number of sub-objects detected for each main object and is a parameter of the
main object.
Page 26
20 Chapter 3 – Assays
3.4 Object Finder Modules
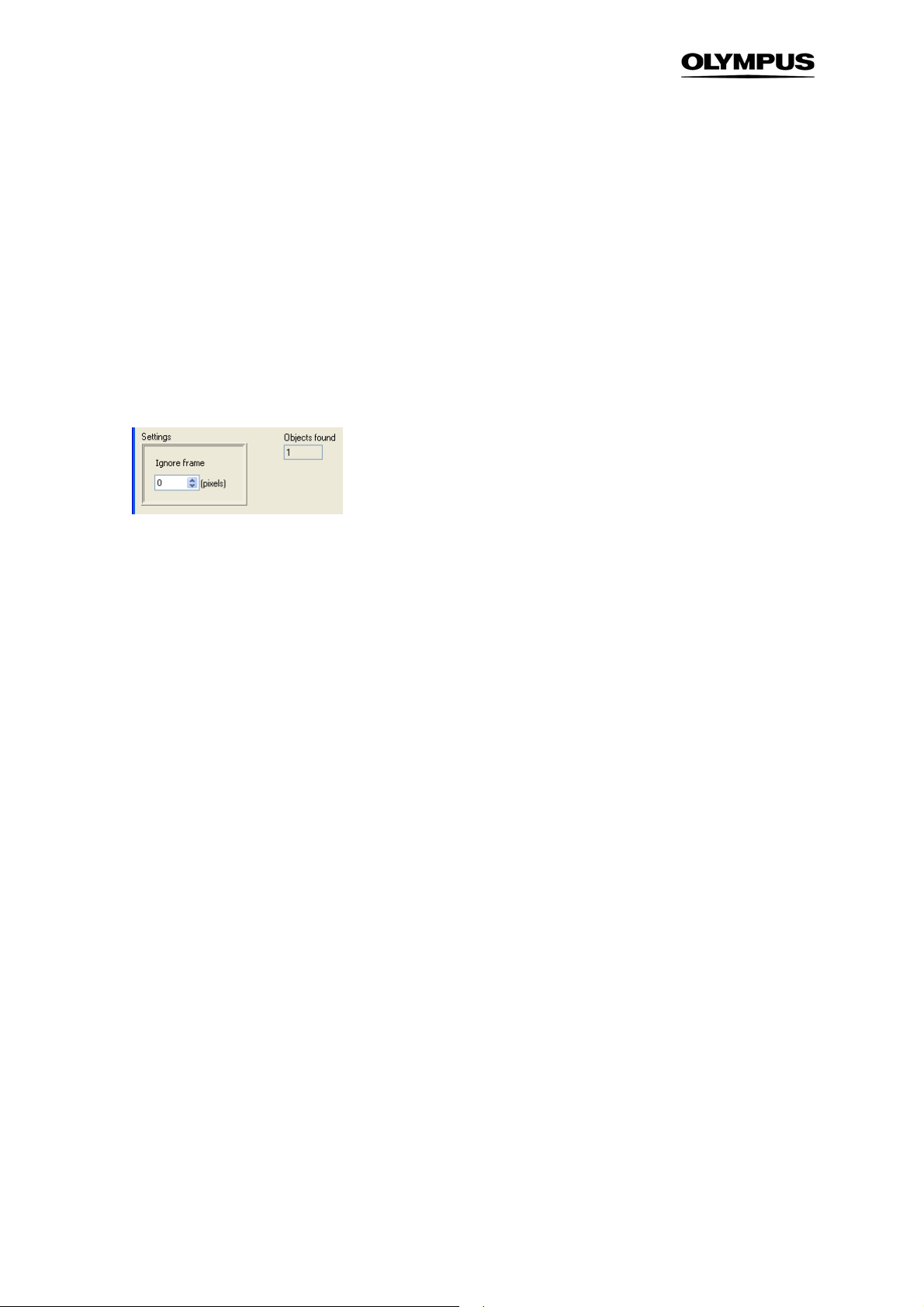
3.4.1 Entire Image
This is a very simple object definition: the entire image is used as object. You may use this for measurements of integral intensities of your sample, i.e., for each image and each parameter (see. Chapter
Measurement Parameters
3.5,
age.
) a single value is calculated, independent of the objects within the im-
Ignore frame. This is the only parameter to adjust: the size of a bordering frame to be ignored. The
default value is 0 (no bordering frame).
3.4.2 Intensity Threshold
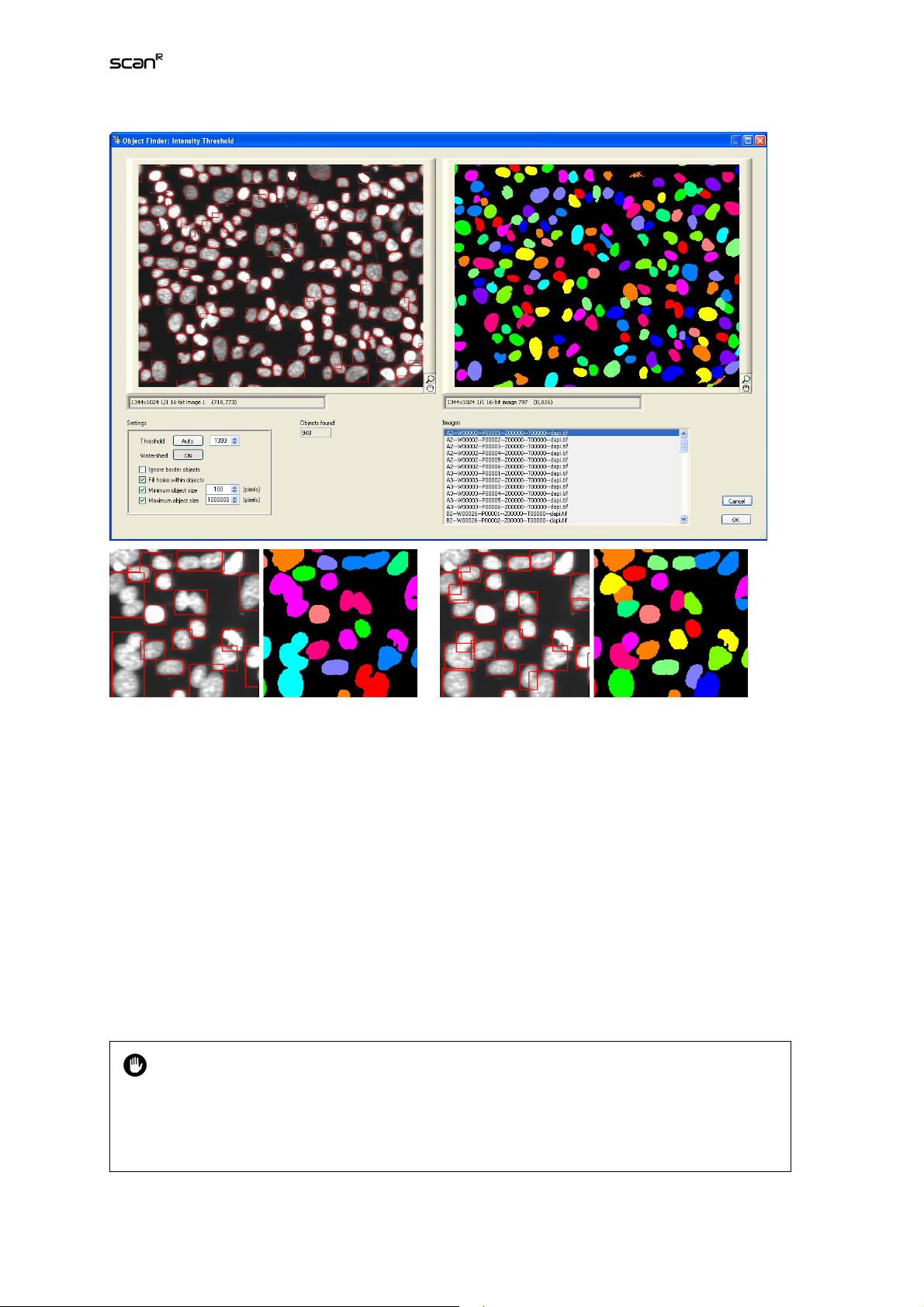
As the name says, the Intensity Threshold method is based on intensity values: pixels with intensities
above a predefined threshold will be united to one individual object.
The Object Finder: Intensity Threshold dialog has two image viewer displays. The left one shows the
gray value image including all detected objects marked with a red bounding box. The right one shows
the binary mask of each object.
Settings
The settings used when opening the window depend on the Settings list selected in the Main Object or
Sub-object tab (see Chapters 3.2,
Detecting Sub-objects
Finders Library (OFL)).
Threshold. This is the intensity cut-off for objects. Type in a value or use the arrows to adjust it.
) and are loaded from the Object Finders Library (see Chapter 6.3.2, Object
Object Finder: Detecting Main Objects
and 3.3,
Sub-object Finder:
Threshold: Auto. Click this button to automatically evaluate the image background and set a meaningful cut-off value.
Watershed. If neighboring objects are so close to together that thresholding does not lead to a clear
separation, they will be detected as one object. (See left image pair below.) The Watershed algorithm
separates these objects along the contractions of the detected masks. (See right image pair below.) Set
the toggle button to On to use this option.
Ignore border object. Check this box to ignore all objects that are cut-off by the image border.
Page 27
Analysis Software Manual Chapter 3 – Assays 21
Fill holes within objects. Check this box to fill the object mask in case it contains holes.
Minimum/Maximum object size. Check these boxes and adjust the values to apply minimum and
maximum size filters to the objects (in order to ignore objects that are outside these size limits).
3.4.3 Edge Detection
The EdgeSegmentation module is a general purpose edge based particle detector. The idea of the
algorithm is to find a closed contour around each particle. First the edges of the image are extracted.
For those edges which already form a closed contour the algorithm stops. Since the remaining open
edges may be part of a closed contour around a particle, the algorithm then tries to combine these
open edges so that they form a closed contour as well.
The edge detection algorithm yields better results when objects of strongly varying intensity
have to be detected. In these cases the threshold detection will either lead to clusters when
the threshold is set to a low value in order to detect also dim objects. If a higher value for
the threshold is set, then the dim particles will be missed. Furthermore, as edge detection
is intensity independent it is especially suitable for cell-cycle analysis.
Page 28

22 Chapter 3 – Assays
To reduce complexity, the process of finding the right settings is split up into three independent steps.
The three settings clusters in the Object Finder menu reflect these three steps. They are traversed from
left to right, but you can always jump back and adjust the settings of former steps. In each step, the
result of adjusting the current steps settings is shown in the image above.
1. Clipping. On the left side select an image of your choice from the list. Adjust the clipping ei-
ther by pressing Auto or manually by moving the green bars. If one or both bars are missing,
just press Auto once. Try to clip away any unwanted noise or artifacts while maintaining
good contrast between the particles you want to detect and the background.
2. Edge extraction. Click on the image or the settings cluster in the middle to get to the second
step. In this step the edges of the image are extracted. First grab the Maximum object size
slider and adjust it so that the largest particles you want to detect just match inside the yel-
Page 29
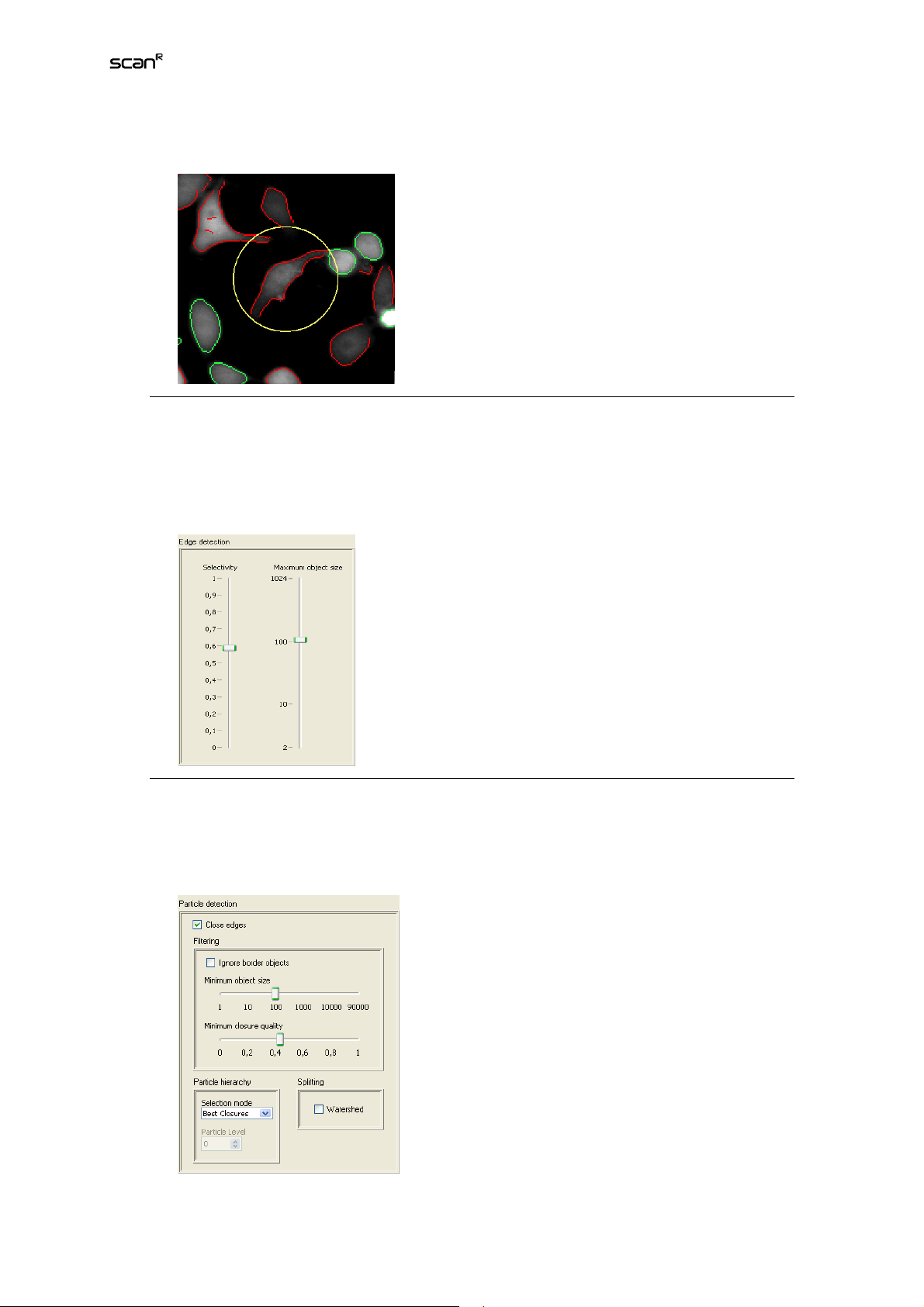
Analysis Software Manual Chapter 3 – Assays 23
low circle appearing in the images.
3. Play around with the Selectivity slider to get just the strong edges (up) or also the weak
edges (down). Try to increase the selectivity as much as possible, thereby removing edges
due to noise and artifacts, without letting gaps in the contour of wanted particles get too
large. As you can see in the image above, contours with closed edges are marked green while contours with open edges are red.
4. Edge Closing. Click on the image or the settings cluster on the right to get to the third step.
In this final step the open edges extracted in step two are now closed by combining them
with other open edges. You can then filter particles by size and closure quality, split them
with the watershed algorithm or select a hierarchy.
Page 30
24 Chapter 3 – Assays
5. First decide if there may have already been sufficient particles detected in step two (the
green ones). If you think so you can skip the closing process by unchecking Close edges,
thereby reducing processing time dramatically. All unclosed edges (red) are discarded then.
Generally the loss of particles is too high with Close edges being unchecked. If Close edges
is checked, you can see in the middle image that the open edges (red) are connected to a
closed contour by yellow lines.
6. In the Filtering cluster you can filter particles by location, size and closure quality. The clo-
sure quality is a rating attributed to each detected particle, which describes the quality/reliability of the respective closure. Particles whose contour has already been closed in
step 2 (the green ones) have closure quality 1. Especially when particle detection is difficult,
you can at least filter out most of the wrongly detected particles by moving the Minimum closure quality slider towards 1.
7. By checking the Watershed checkbox, you can split particles which have merged. The algo-
rithm inspects the shape of each particle, splitting it at constrictions. This can be extremely
useful when detecting nuclei.
8. Sometimes detected particles are nested into each other. E.g. spots inside nuclei or nuclei
inside the cytoplasm. Since overlapping particles are not allowed, the Particle hierarchy cluster provides options to select the nesting or hierarchy level you are interested in. See below
for the function of the Selection mode options.
9. Don’t forget to check other images of the scan to verify that your settings work with them as
well. When you are satisfied with your settings click Ok on the bottom right side.
Level 0 Level 1
The options at the Selection mode dropdown list are:
Min level. Selects all level 0 particles.
Max level. Selects all particles which do not have other particles nested inside.
Selected level. Selects all particles with the specified level.
Best closures. Selects for each nesting branch the particles which are best according to their closure
quality.
Page 31
Analysis Software Manual Chapter 3 – Assays 25
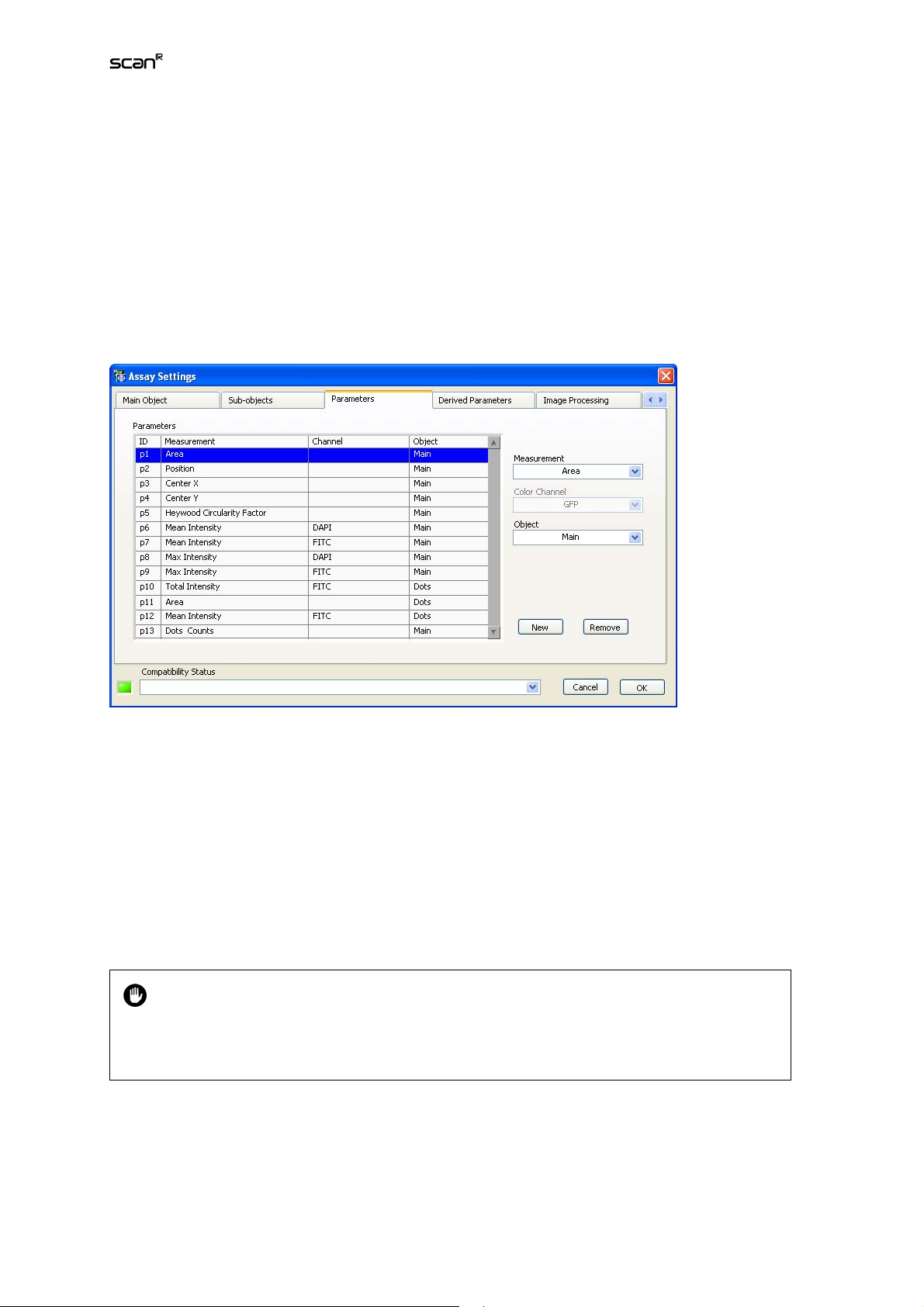
3.5 Measurement Parameters
The scan^R Object Analyzer modules in the standard configuration offer a large number of individual
parameters that may be measured for each recognized object. To limit memory utilization and save
CPU time, only the values for the parameters listed in the Assay SettingsParameters list will be extracted during the analysis.
Parameters list. Each Measurement is labeled with an ID (p1, p2,…) and is assigned to the Main Object
or a Sub-object and – depending on the type of measurement – may be assigned to a Color channel.
Measurement. Select a parameter from the shortlist. The available parameters can be adapted through
ModulesObject Analyzers (Chapter 6.3.1) .
Color channel. Assign a Color channel from the shortlist to the newly added parameter.
Object. Assign an Object type from the shortlist of available object types. The image mask of this object defines the parameter measurement area.
New. This allows inserting a new parameter into the list.
Remove. This deletes the selected parameter from the list.
A special parameter is available in the Measurement list when Sub-objects are used: Obj. 1
counts (if the default name for sub-objects is used, otherwise it would be
counts). This parameter gives the number of sub-objects detected for each main object
and is a parameter of the main object.
subobjectsname
Page 32

26 Chapter 3 – Assays
3.6 Derived Parameters
The Assay SettingsDerived Parameters tab allows performing calculations on the Parameters defined
in the Measurements list by the use of algebraic expressions (+, -, ×, ÷, sqrt, ^ etc. For a complete list
see Appendix 6.4). This is especially useful for parameters assigned to different color Channels.
Derived Parameters list. It lists the Derived Measurements to be carried out. Each Derived Measurement is labeled with an ID (D1, D2,…) and requires a formula that entangles parameter IDs from the
Measurements List.
Name. This is the input field for setting the new Derived Measurement's name.
Formula. This is the input field for the algebraic expression that connects the desired parameter IDs
from the Derived Measurements List.
Variable Selector. The parameters of the measurements list can be selected from this drop-down menu
and their corresponding ID is entered in the Formula-field. Vice-versa if you select a parameter in the
formula field you get the corresponding parameter name displayed in the Variable Selector field.
Special Operators
It is possible that a number of individual Sub-objects of the same type can be identified
within the mask of a single individual Main Object. A set of statistical operators allows
evaluating individual parameters of the entire Sub-object population of this type for the individual Main Objects:. These Operators are SUM(), STDV(), and MEAN().
Statistical Operators:
• SUM(ID). This operator sums up all values of the given parameter ID of the Sub-objects belonging to
each individual Main Object. that belong to the same Sub-object type.
Page 33

Analysis Software Manual Chapter 3 – Assays 27
• MEAN(ID). This operator averages all values of the given parameter ID of the Sub-objects belonging
to each individual Main Object that belong to the same Sub-object type.
• STDV(ID). This operator calculates the standard deviation of all values of the given parameter ID of
the Sub-objects of each individual Main Object that belong to the same Sub-object type.
The parameter resulting from a special operator expression is assigned to the Main Object.
New. This allows inserting a new Derived Measurement into the list.
Remove. This deletes the selected parameter from the list.
3.7 Image Processing
Images can be processed before being analyzed in an Object Finder Module. Thus, image quality can
be enhanced and the object detection improved and facilitated. A set of predefined image processing
modules is available.
A variable set of image processing steps can be assigned to each color channel. The configuration of
the individual steps takes place via the Image Processing window accessible via AssayEdit Assay.
Image processing changes the image data temporarily! The original image data will not be
lost but remain stored. However, in the further course of the assay execution the analysis
will always work on the processed data. In order to keep the original data accessible during
the assay, virtual channels have to be created instead, see Chapter 3.8,
Virtual Channels
.
Image processors list. It lists the Image Processor Modules, i.e., the processing steps, to be applied on
the different Color channels of the images. The order of appearance in this list also sets the order of
Page 34

28 Chapter 3 – Assays
execution. Moving the active entry up or down the list by using the arrow buttons changes the order of
execution.
Module. Select an Image Processor Module from this shortlist.
Color channel. Assign a Color channel from the shortlist to the active Image Processor Module.
Adjust. Click here to open the Image Processing window for the active Image Processor Module.
New. This allows inserting a new Image Processor Module into the Module list.
Remove. This deletes the active module from the Image processors list.
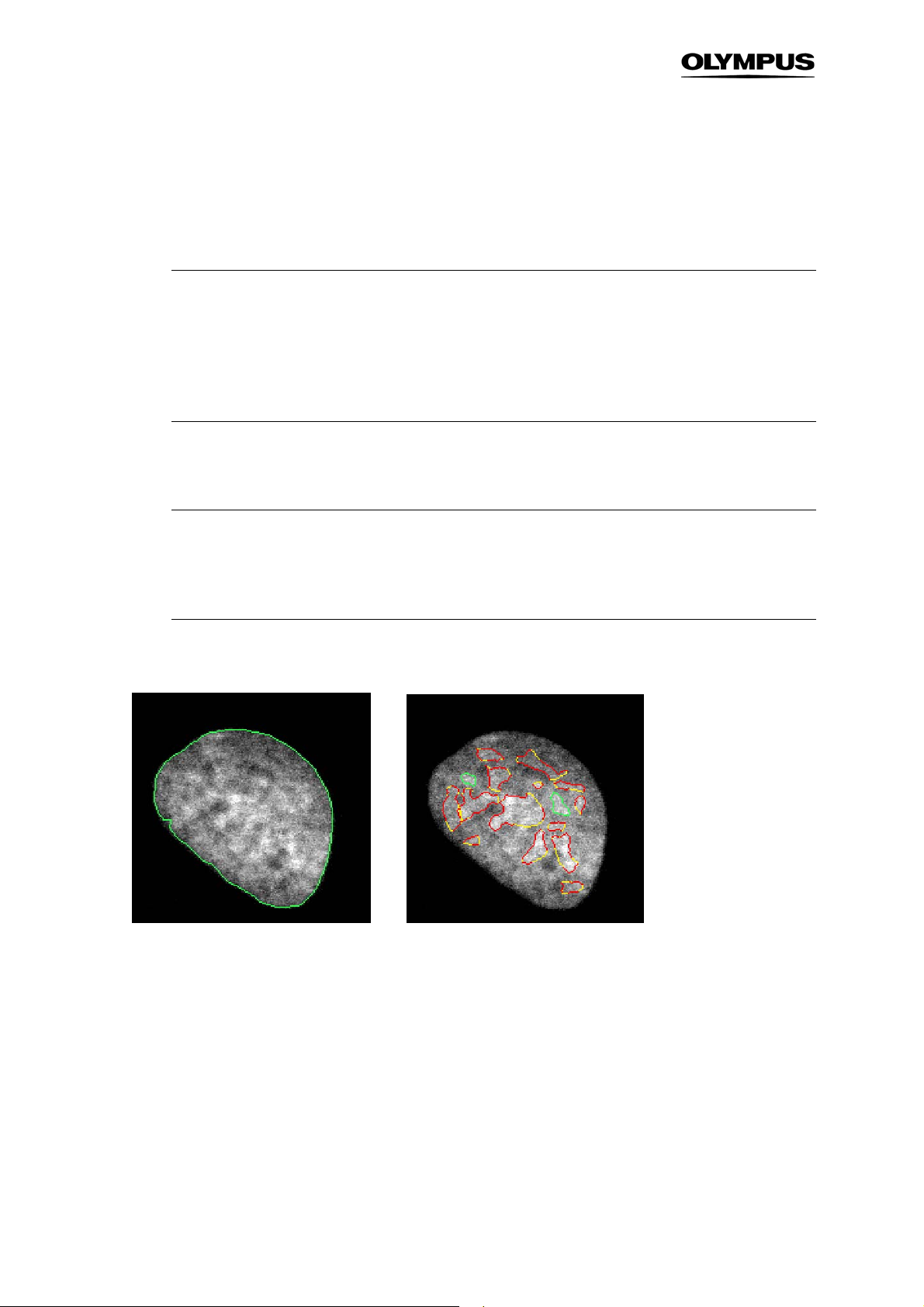
3.7.1 Background Correction
For applications involving quantification of intensities and in case of inhomogeneities it is always recommended to use background correction. The algorithm implemented is an intensity-conserving algorithm which ensures that the signals remain unchanged and can therefore be quantified (left image:
before background correction, right image: after background correction).
Size filter. This is the only parameter to be set.
For background correction a Size filter of 200 is the default value. For most cases when a
general background has to be removed it works fine. However, to better extract e.g. small
particles in the nucleus, a background correction filter size of 15 can be suitable (cf. example below).
Page 35

Analysis Software Manual Chapter 3 – Assays 29
Example for small Size filter for background correction.
3.7.2 XY Shift
In certain cases it may occur, that image channels are shifted relative to each other in the channel overlay. This is rather often the case if an observation emission filter wheel is used and images are acquired
with different emission filters. This function allows correcting the shift along the X- and Y-axes.
XY Shift. Set here the number of pixels the chosen channel is to be shifted along the X- and Y-axes
relative to the other channels.
You have to control the result in the main user interface by pressing the Processed button. It is useful
to zoom into the image so that individual pixels can be detected visually.
3.7.3 Inversion
This function serves to invert the intensities of an image channel, i.e., to convert a channel into its
negative image. This is necessary for example if objects are to be detected in transmission images. The
object detection tools of scan^R are designed to detect bright objects on dark background. The situation is reverse in transmission images. Thus they have to be inverted prior to the object detection.
Adjust Intensity. This defines the maximum intensity in the converted image. I.e., it is the intensity that
the originally darkest pixel will get in the converted image. 4096 is the default and corresponds to the
maximum intensity in a raw image taken by a 12-bit CCD-camera.
Page 36

30 Chapter 3 – Assays
3.7.4 Cut Image
This module allows defining regions of interest inside an image and setting all image parts outside the
region to the intensity 0. Different drawing tools are available.
Rectangle
Define a rectangle by mouse drag. Mouse dragging the center changes its position. The size can be
adjusted by dragging the corners.
Rotated Rectangle
This is similar to the rectangle tool. Upon mouse-over its central axes are displayed. Dragging the ends
of the axes turns the rectangle.
Polygon
Standard tool to draw polygons.
Freehand
Standard tool to draw freehand regions. The region is closed automatically once the mouse button is
released.
Circle
Standard tool to draw circles. Close the circle by double click.
Ring Segment
Standard tool to draw rings. Once a ring is drawn the inner and outer borders can be dragged to adjust
the thickness. The cutting line can be dragged to convert the ring into a ring segment.
Page 37

Analysis Software Manual Chapter 3 – Assays 31
3.8 Virtual Channels
Virtual channels are image channels that are not created via image acquisition during the execution of a
scan. Instead they are a result of post-acquisition image processing and added as new channels to the
original image data. These can then be used for further analysis steps, e.g. object detection.
To access the Virtual Channels tab in the Assay Settings window you have to navigate
through the tabs to the right using the arrow buttons on the top right.
New. Click here to create a new entry in the VC Process List.
Virtual Channels list. Default names for the virtual channels resulting from the processing are automatically created. It can be changed manually.
Remove. Click here to delete the active entry from the VC Process List.
Module. Background Correction, XY Shift, Inversion and Cut Image are the same functions as described in Chapter 3.7, Image Processing.
Adjust. Click here to open the dialog window of the selected module.
3.8.1 Simple Math
The Simple Math module serves to perform calculations on the image through basic arithmetic operations: addition, subtraction, multiplication and division.
Page 38

32 Chapter 3 – Assays
Module. Select Simple Math from the Module pull-down list to set this module as active entry in the VC
Process List.
Input Channels list. All color channels of the images are possible Input Channels for the Simple Math
module. The channel selected as first Input Channel is always the first source of the arithmetic operation. The other channels are possible second sources.
Adjust. Click here to open the Simple Math dialog window. It contains displays of the overlay of the
Input Channels and of the Virtual Channel that is created by the selected arithmetic operation.
Settings
Channel 1, 2, 3. Each of the Input Channels can be weighed prior to the arithmetic operations by using
the sliders or entering a multiplication value between 0 and 10 into the respective boxes.
Arithmetic operations selectors. Select the operation (+, –, ×, ÷) that is to link Channel 1 and Channel 2
from the first pull-down selector (e.g. DAPI+FITC). Select the operation that is to link the result of
Channel 1 and 3 with Channel 3 (e.g. (DAPI+FITC)×TxRed) from the second pull-down selector. Select
Skip to deactivate Channel 2 or Channel 3, respectively.
3.8.1.1 Example: Cytoplasm not detectable on a single color channel
Page 39

Analysis Software Manual Chapter 3 – Assays 33
In this example it is not possible to detect the cytoplasm on a single color channel because the area of
the nucleus has very little cytoplasmatic staining:
Problem. The detection is incomplete because staining is missing on the nucleus area.
Solution. The nucleus staining and the cytoplasmatic staining are added as VC.
Result. Full Cell Segmentation of the complete cell can be performed on the calculated new channel.
3.8.2 Spectral Unmixing
A major problem in live cell imaging arises from the use of different fluorochromes with overlapping
spectra in one multi-labeled sample, impairing a number of applications. Even with the use of high
quality optical filters it is not satisfactorily possible to separate the spectral information. The consequence is the excitation and imaging of structures that are labeled with one of the present fluorophores
when using a filter set that is actually chosen to excite and image another fluorophore.
With the Spectral Unmixing module it is possible to separate and resort the contribution of different
fluorochromes to the total signal in each color channel and redistribute the different color intensities. It
thus improves the spectral resolution of the channels considerably and facilitates for example colocalization studies.
Get from ROI. Takes the mean value within the marked region as Stain/Background.
Ignore Stain. Click here to ignore the third channel in order to properly unmix two-channel images.
Background. Spectral Unmixing yields quantitatively meaningful data only if a background subtraction
is performed prior to it.
Page 40

34 Chapter 3 – Assays
Show details. Displays the matrix created by the selection of the stains. This matrix is used for the
processing of the images. The entries of the matrix can be changed manually. A graphical representation is displayed.
Output channels. Select the stains to see the result of the spectral unmixing.
1. In the Module shortlist select Spectral Unmixing and press New.
2. Press Adjust to start the 3x3 Spectral Unmixing menu.
3. Click on the Background button.
4. Mark a background area using the drawing tool (button on the bottom right corner of the im-
age displays).
5. Click the Get from ROI button.
6. Identify a structure that contains just the first fluorophore.
7. Mark the structure using the drawing tool (button on the bottom right corner of the image
displays).
8. Click on the Stain 1 button.
Page 41

Analysis Software Manual Chapter 3 – Assays 35
9. Click the Get from ROI button.
10. Repeat steps 4 – 7 for the other fluorophores (stains).
In order to perform the spectral unmixing the software has to determine the contribution of
the fluorescence of different fluorophores to the different color channels. To do so, ideally
series of mono-labeled reference samples would be used. In case such samples are not
available for each of the fluorophores, molecular structures have to be identified by the
user that are certain to contain just only one of the fluorophores and that do not spatially
overlap with structures containing other fluorophores.
The result of the spectral unmixing is shown in the right display of the 3x3 Spectral Unmixing dialog
window.
Unmixing dual-labeled samples. In the case of two-channel images the third input channel
will be occupied by one of the two available channels. Click Ignore stain to ignore the third
channel in order to properly unmix two-channel images.
3.9 Tracking – Analyzing Time-Lapse Data
The tracking function of scan^R Analysis allows the analysis of objects over the course of time, i.e., in
experiments that consist of time-lapse acquisitions. It relates any object detected in an image to the
same object in the previous and subsequent images in the time-lapse series acquired at the same
stage position. Thus the change of the parameters that are measured according to the assay settings
can be followed over the course of time.
To enter the Trace View select Trace in the lower left part of the front panel or in the menu bar select
TrackingTrace view. The front panel display changes such, that the displayed objects change from
Main to Trace and also the available X/Y-parameters in the histograms are changed according to the
parameters that are defined in TrackingDefine Parameters (See Chapter 3.9.2.1,
Original Parameters
).
Page 42
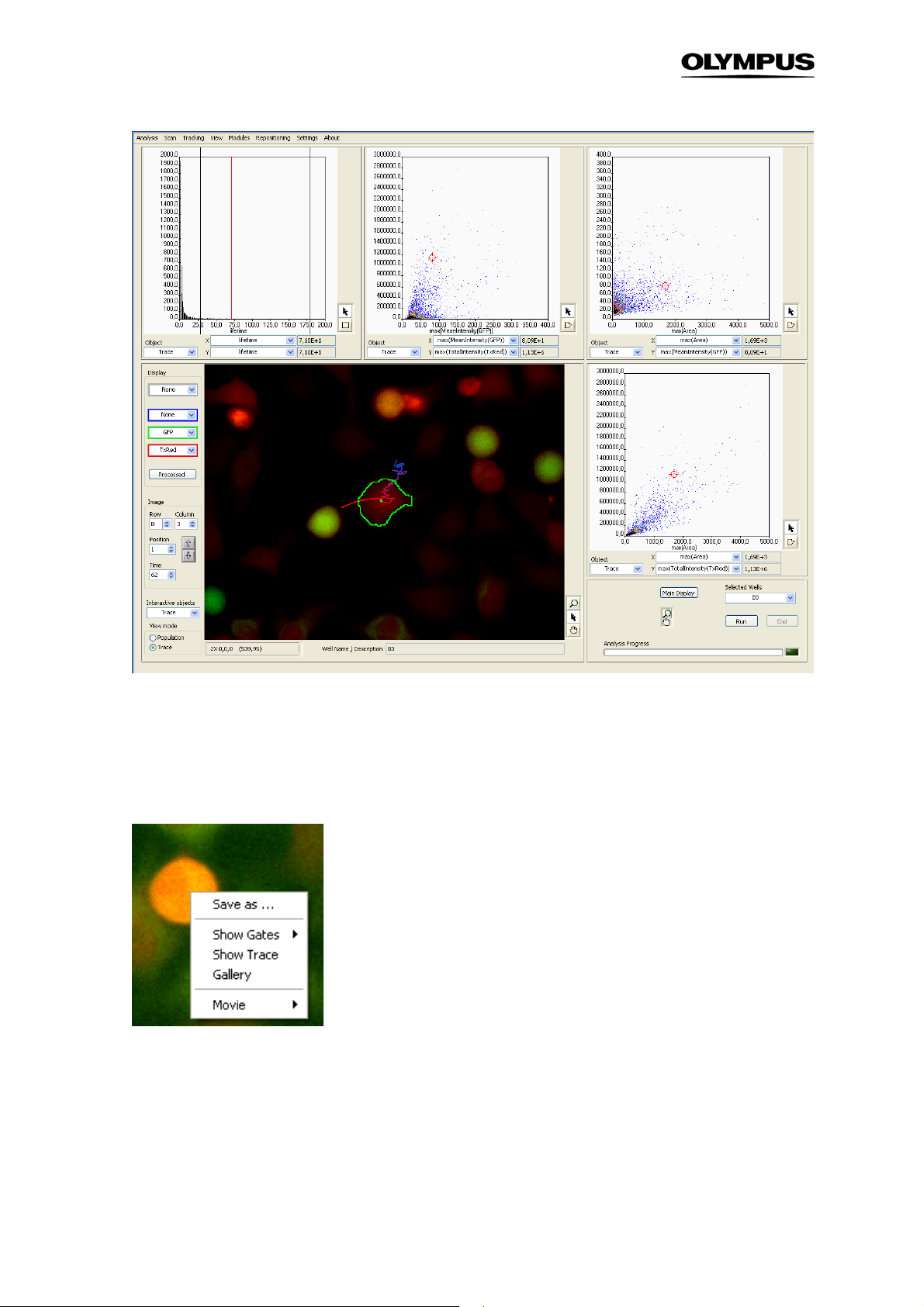
36 Chapter 3 – Assays
When clicking on one tracked object in the image now not only the object is highlighted with a green
border but also the trace it covered during acquisition will be displayed. With time the object moves
from the blue end of the line to the red end. Like in the Population View the detected object and the
corresponding data points in the histograms are directly linked.
Right-clicking on an object in the image yields the following context-menu:
Save as … This allows you to store the displayed image separately.
Show Gates. It has the same function as in the population view. When you select a gate from the list,
the objects that fall into the gates are marked with a box in the front panel image.
Show Trace. This opens the Trace Viewer which displays the time-curve for the selected object. (See
Chapter 3.9.3,
Trace Viewer
)
Page 43
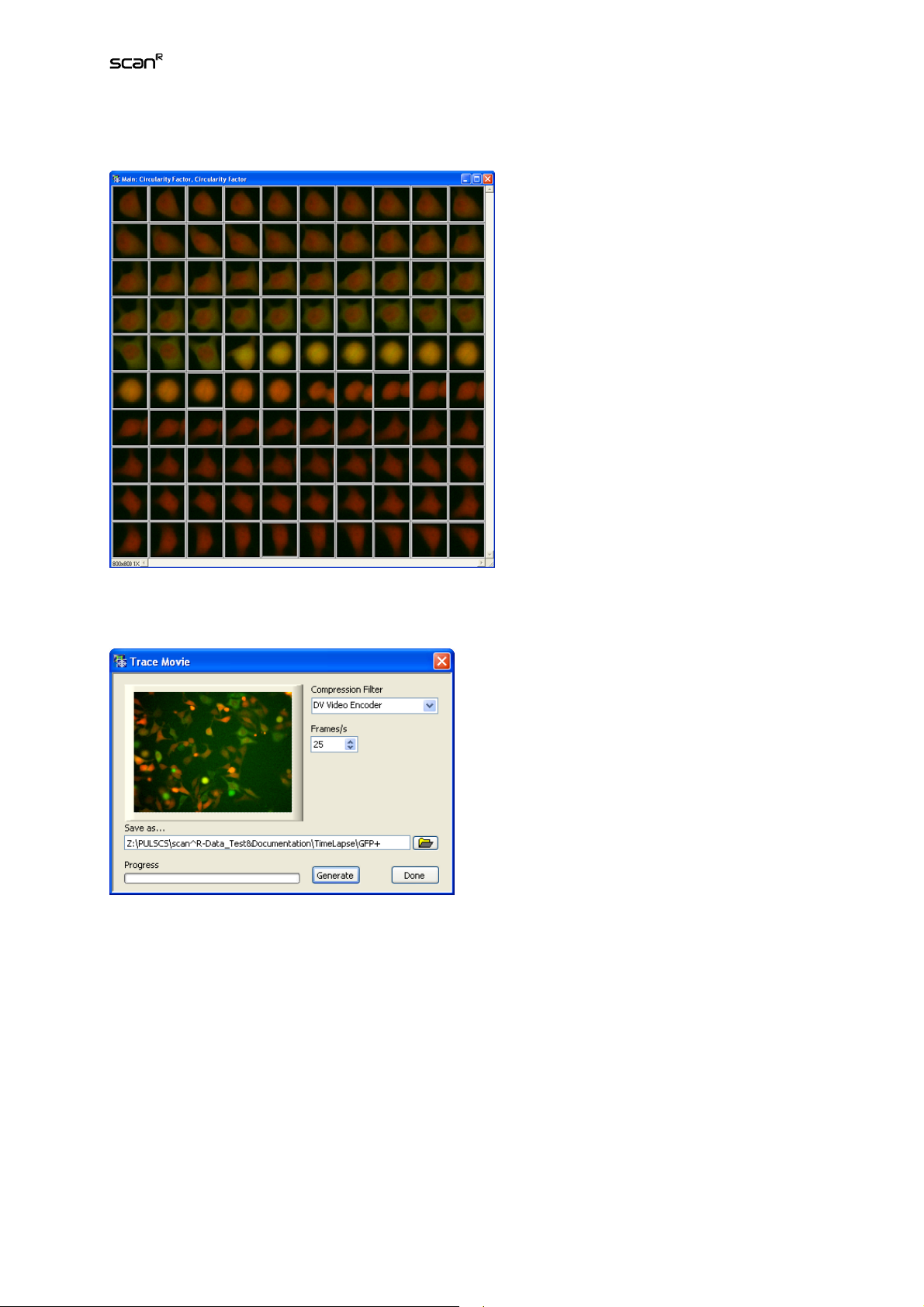
Analysis Software Manual Chapter 3 – Assays 37
Gallery. it displays a time-gallery of the selected object.
Gallery display in time-lapse mode
Movie. This opens the menu Trace Movie which allows you to export a movie of a single trace or a
complete position.
3.9.1 Tracking Configuration
The TrackingConfigure Tracer command opens the Tracker Settings window. It also opens automatically when the View mode: Trace is activated without any tracking parameters being set already.
Tracked Object Type. Select here the kind of objects to be tracked, i.e. main objects or any of the subobjects – if such are defined in Assay SettingsSub-objects. (See Chapter 3.3,
Detecting Sub-objects.
)
Sub-object Finder:
Page 44

38 Chapter 3 – Assays
Range (Pixel). Set here the maximum difference that is allowed to change between two frames in order
to maintain a track. If the difference exceeds the Range an object will NOT be related to similar objects
in the previous and subsequent images of the time-series.
OK. Click here to start the tracking. The advance can be followed in the status bar at the bottom right
corner of the scan^R Analysis main interface. Once it is completed the Trace Parameters window
opens.
3.9.2 Track Analysis Parameters
During the tracking is run not only the X/Y-positions of an object over time are calculated, but also the
time course of the parameters previously determined in AnalysisEdit AssayParameters (see Chapter 3.5,
displayed in Trace ParametersOriginal Parameters. This menu allows defining which kinetic parameters of these curves are extracted, e.g. the maximum intensity, the time of maximum intensity or the
duration of an increase in signal…
3.9.2.1 Original Parameters
Open the Trace ParametersOriginal Parameters window directly via TrackingDefine Parameters. At
startup it shows the kinetics graph of one of the measured parameters of the first track in first stage
position of the first well acquired and selected for analysis. (See Chapter 2.7,
sis.
To navigate through the curves use the Well, Position, and Trace selectors on the right. Each Trace
represents the object in subsequent time frames that the tracer, according to the settings in TrackingConfigure Tracer detected as belonging together. In the Original Parameter menu you can set the
Operators that are applied to the time curves to quantify the time curves). By applying an Operator
(min, max, std, first, etc.) the time-curves again are reduced to a single values per curve that in turn can
be displayed in a 1-D or 2-D histogram. For example when TotalIntensityGFP was selected in the Parameter tab of Edit Assay, the time curve of TotalIntensityGFP can now be plotted. By applying the
Operator max on this time curve, for all traces the maximum of TotalIntensityGFP will be calculated.
Measurement Parameters
)
) is extracted of the data. The curves for each individual object are
Selecting Wells for Analy-
Page 45

Analysis Software Manual Chapter 3 – Assays 39
Trace parameters.
Well, Position, Trace. You can navigate through the set of curves using these functions and their arrow
buttons.
Add Range. Click here if you want to analyze just a part of the trace. Two vertical blue bars appear in
the graph to define the upper and lower limit of the range. They can be moved via mouse-drag. The
analyzed part of the curve is displayed solid. The remaining part is displayed as a dashed line. Click
Add Range several times to create multiple ranges.
Two ranges applied to a trace
Add Threshold. Click here to open a menu which allows you set an upper or lower bound for the curve.
A blue bar appears that marks the upper or lower part of the curve, respectively. The bar can be moved
via mouse-drag.
Remove. Click here to remove the Range limits.
Page 46

40 Chapter 3 – Assays
Parameter. Select the measurement parameter of which the time-curve is to be shown in the graph
display.
Derivative. Check this option to calculate and display the first derivative of the kinetics time-curve of
the selected Parameter.
Smoothing (Sigma). This function applies a smoothing filter on the track data. Set the strength of the
smoothing with the slider. The graph display is being updated immediately.
Definitions. This lists the names of the values to be determined from the curves as well as the operators
used. In the Result column the values for the selected trace for all parameters are listed.
Operator. Select an operator from the shortlist. This will be applied on the currently selected Parameter.
For example, if
mean
is applied on the
Mean Intensity
of the tracked objects the time average of the
mean intensity of each object will be calculated.
The available Operators are:
– Lifetime. Gives the number of contiguous time points the particle is detected
– Sum. Calculates the sum of the parameter value over the time
– Mean. Calculates the mean value of the parameter
– Min. Takes the minimal value the parameter reaches
– Max. Takes the maximal value the parameter reaches
– Std. Calculates the standard deviation of the values
– First. Takes the value of the parameter at the first point of the curve
– Last. Takes the value of the parameter at the last point of the curve
– T_max. Takes the time point when the parameter is maximal
– T_min. Takes the time point when the parameter is minimal
– T_first. Gives the time point when the trace starts
– T_last. Gives the time point when the trace ends.
– Num_zero_crossings. Gives the number of zero crossings of the curve
– Num_local_max. Gives the number of local maxima of the curve
– Num_local_min. Gives the number of local minima of the curve
Name. The name of the analysis function is set automatically as <Operator>(Parameter). It can be
changed manually.
New. Click here to create a new list entry.
Remove. Click here to delete the selected list entry.
Advanced. Gives advanced options. The Derivative checkbox is replaced by a menu that allows you to
apply Anti(-)/Derivatives(+) of higher order. In the Operator shortlist a new option becomes available:
the curve-fit.
Curve-fit. This Operator allows you to fit certain models to the parameter curves. Upon selecting curvefit from the Operator drop-down menu the Define-button becomes accessible.
Page 47
Analysis Software Manual Chapter 3 – Assays 41
Define. This opens a menu to set the options for curve fitting.
As objects might move in or out of the focal plane, or the value entered for Range might be
set too small, there might be a large number of short traces. Therefore Lifetime is a useful
parameter as it allows setting a gate on long traces.
3.9.2.2 Curve Fitting
Model type. Select the type of model that is to be fitted to the curves. You can select Polynomial, Exponential, General ls linear and non linear.
Polynomial. The only parameter to be set is the Polynomial order. E.g., the model function is
f(t)=a0+a
2
·
t+a
·
t
for Polynomial order of 2. In this example the parameters
1
2
a0, a
and
1
a2
are fitted to the
curves.
Exponential. No parameters have to be set. The model function is
c
are fitted to the curves.
f(t)=a·exp(ct)
. The parameters a and
General ls linear. General linear least-squares fit. Here you can enter arbitrary Basis functions (linear,
exponential, trigonometric,…) which will be combined linearly. E.g. set f
will be combined linearly to
linear combination of a line and a Gaussian. The parameters
f(t)=a
·
f1(t)+ a
0
·
f2(t)= a
1
·
(17·t+4)+ a
0
·
exp(-(t+2)^2)
1
a
and
0
(t)=17·t+4 and f
1
a
are fitted.
1
(t)=exp(-(t+2))^2
2
. This function represents a
Non linear. Here you can enter an arbitrary model function to be fitted with up to 6 parameters. You
have also to enter the start values as First guess. E.g. if you want to fit a Gaussian you set
f(t)=a
·exp(-(t-b)2/(2c2)) and enter start values for a, b and c. You can also set the number of iterations
to be run by entering a value for Max iterations.
a0,, a1,
The parameters that are fitted (e.g.
…) and the mean squared error of the fit (mse) are listed in the
derived Parameters tab in the following representation: curve_fit<
curve_fit<
modelfunction
>(Parameter).a1, …, curve_fit<
modelfunction
>(Parameter).mse.
modelfunction
>(Parameter).a0,
Page 48

42 Chapter 3 – Assays
3.9.2.3 Derived Parameters
The Trace ParametersDerived Parameters tab allows to perform calculations with the parameters
listed in the Definitions list described above by the use of basic algebraic expressions (+, -, ×, ÷, sqrt,
^ etc. For a complete list, see Appendix 6.4).
Original Parameters. This is the list as created on the Original Parameters tab and includes also the
fitted parameters when the operator curve_fitting is applied.
New. Click here to create a new list entry.
Name. Set the name of the new Derived parameter here.
Formula. Set the formula here using the ID of the original parameter(s) and the algebraic expression.
Example.
Imagine you want to measure the duration of a peak. It starts with a pronounced increase in intensity
and ends with a pronounced decrease. Mathematically this would be the time between a maximum and
a minimum in the first derivative of the Mean Intensity curve. The Original Parameters needed are thus
time of the maximum (t-max) and time of the minimum (t-min) of the Parameter Mean Intensity with the
option Derivative. The formula for the Derived Parameter would thus be p4-p3 when using the entries
as shown in the screenshot.
Page 49

Analysis Software Manual Chapter 3 – Assays 43
3.9.3 Trace Viewer
Open the Trace Viewer by selecting TrackingShow Traces in the main menu. Alternatively you can
right-click on one detected object in the image in the Trace View and select Show Trace to see the
trace of this individual object. Set the number of displayed traces with the slider on the right. To display
only objects of a certain gate that was previously defined in the Trace View this gate can be selected in
the Gate drop-down menu.
The results of the tracking are displayed in the Trace Viewer window once the tracking has been carried
out.
Max displ. traces. Use the slider to adjust the number of displayed. The traces will be taken from a
random selection of the specified gate.
Single mode. Use this option to have just one trace displayed. If a trace is selected via mouse click it
will be this one that is being displayed.
Gate. Select a gate to have only the corresponding traces displayed.
Time Scale.
of the time-lapse series. When
Real Time
is the default and causes that each point in time is set relative to the beginning
Trace Time
is selected each point in time is set relative to the beginning
of each track when t_first is selected, which may be important if an object was not tracked right from
the start of the time-lapse series and one is interested especially in the data at the beginning of each
track. Alternatively, when applying the operator t_max e.g. on the parameter MeanIntensity(GFP) the
time point when the mean intensity of GFP is maximal is set to be 0. This way the time-curves can be
displayed “synchronized”.
Page 50

44 Chapter 3 – Assays
Parameter. Select parameter of which the time-curve is to be shown in the graph display.
Derivative. Check this option to display the first derivative of the curve of the selected Parameter.
Smoothing (Sigma). This function applies a smoothing filter on the .time-curve. Set the strength of the
smoothing with the slider. The graph display is being updated immediately.
Selecting objects and navigating along tracks: Tracks in the Trace Viewer and the objects
in the images they derive from are directly linked. Upon clicking on a time-curve the corresponding image with the marked object will be shown in the image display of the main
scan^R Analysis window. Also the data points marked with a red circle in the histograms
are linked to the time-curves. Likewise, upon clicking on an object in the image display or
in a scatter plot the corresponding time-curve will be shown in the Trace Viewer.
A dashed vertical line marks the current point in time in the Trace Viewer. Upon moving the
cursor left and right while keeping the mouse clicked one can thus scroll back and forth
through the "movie" of the time-lapse series.
Page 51

Analysis Software Manual Chapter 4 – Analysis Results 45
4 Analysis Results
This chapter describes how an analysis is executed, how the results can be grouped into object populations and how these can then be analyzed and statistically evaluated.
4.1
Running an Analysis...................................................................................46
4.2 Managing Gates.......................................................................................... 47
4.2.1 Gates and Regions. .................................................................................. 47
4.2.2 The Gate Manager.................................................................................... 47
4.2.3 Color Gating.............................................................................................. 49
4.3 Well Results................................................................................................ 52
4.3.1 Measurement Results...............................................................................52
4.3.2 Well Results: Populations .........................................................................53
4.3.3 Export Definitions...................................................................................... 54
4.3.4 Export results of individual objects............................................................56
Page 52

46 Chapter 4 – Analysis Results
4.1 Running an Analysis
1. Execute ScanOpen… and select the Experiment Descriptor.xml file in the storage folder of
the scan. By doing so the experimental settings are loaded and the first image of the scan
(taken at the first position of the first well) will be displayed. If you want to modify or revisit an
analysis execute AnalysisOpen… instead.
2. Execute AnalysisLoad Assay… to read in a *.say assay file stored in the scan^R Analy-
sis/Assays folder. If you want to modify the assay file of the current analysis execute AnalysisEdit Assay… instead.
3. Set or modify the parameters in the assay as described in Chapter 3,
4. If you want to perform the analysis on a subset of the wells, open the Select Wells window
via ScanSelect Wells… and select the wells of interest.
5. In order to observe the analysis results online prepare the four histogram windows by select-
ing the objects and measurement parameters to be displayed.
6. Click the Run button (or execute AnalysisRun) to start the analysis. You can follow the
progress of the analysis in the window at the bottom right of the main scan^R Analysis interface.
The window shows the Analysis Progress and displays the image that is currently being analyzed in the
thumbnail display. Click the Main Display button to have this image displayed in the main mage window. You may Pause or End the analysis by clicking the respective buttons.
Assays.
Once the analysis is completed store the results via AnalysisSave as…
Page 53

Analysis Software Manual Chapter 4 – Analysis Results 47
4.2 Managing Gates
scan^R Analysis detects objects in all images acquired during a scan and performs measurements and
analyses on each object found. The data can be displayed in form of histograms where each colorcoded data point represents the results of one object – or of several objects that happen to have identical results. The results – and thus the objects – can be grouped by Gates and Regions. The Gates
allow classifying the objects according to their properties, i.e. the parameters that were extracted for
these objects. Once these gates are defined, they can be applied to all further measurements for automatic quantification. The gates are administered in the Gate Manager, here you find also access to the
Well Results menu which contains the detailed results for all wells.
4.2.1 Gates and Regions.
scan^R distinguishes between Gates and Regions. Regions define classification rules in form of polygons or ranges in histograms. Gates are composed of one such region or of several regions that are
linked with Boolean operators (AND, OR,, AND NOT). Once a Gate is set, only data points within its
boundary are being considered in further steps.
Gates and Regions can be created using the drawing tool beside each scatter plot. A Region can be
defined as Gate by selecting Set Gate in the histogram context menu or by defining a gate in the Gate
Manager. (See Chapter 2.4,
By clicking on the borderline the Region can be displaced. By clicking one corner of the region this
corner can be moved in order to include or exclude data points.
Right-clicking on the borderline of a Region opens the Region context menu. (See Chapter 2.4.2,
Region Context Menu
Managing Histograms and Scatter Plots.
).
)
The
4.2.2 The Gate Manager
The Gate Manager allows converting Regions into Gates and to combine Gates by Boolean operations.
The Gate Manager also gives an overview of the results in the histograms (H1 to H4) and the applied
Gates.
The command AnalysisAssay Gating opens the Gate Manager window that allows administrating the
Gates and Regions.
Regions. This lists all regions drawn into the histograms.
Show. Click here to display the selected Region in the histogram selected in the Gate application list.
Page 54

48 Chapter 4 – Analysis Results
Remove (Region Box). This command removes the selected entry from the Regions list and deletes the
defined Region.
Gates. It lists all gates with their Names and Definitions.
Name. Set the name of a Gate.
Color. Click into the box to open the dialog window to select the color of the object boundaries when
displayed in the image viewer. This color is also used for color gating. (Compare Chapter 2.4,
Histograms and Scatter Plots
.)
Managing
Definition. A Region or a Boolean combination (via “AND”, “OR” or “AND NOT”) of Regions can be set
to define the Gate.
New. Click here to add a new Gate. The default Name and default Definition will be that of a region in
the Regions list. The Name can be modified at will; the definition must be a region or a logical combination of regions.
Remove. This command removes the selected entry from the Gates list.
Gate Application. This table lists all histograms with the applied Gates and statistical information about
the gated data. (Number of Objects (Count), Percentage of total number of detected objects (% Tot.),
Mean Value, STDV and CV% for the parameters plotted along the X and Y-axis)
Histogram. Number of the histogram window in the scan^R Analysis front panel, H1 to H4, respectively.
Gate Name. Gate applied to the histogram.
Count. Number of objects in the histogram.
%Tot. Amount of objects in percent of the total objects.
xMean (yMean). Mean value of the abscissa (ordinate) parameter of all objects in the histogram.
xCV (yCV). Standard deviation of the abscissa (ordinate) parameter of all objects in the histogram
Page 55

Analysis Software Manual Chapter 4 – Analysis Results 49
xCV% (yCV%). Coefficient of variation of the abscissa (ordinate) parameter of all objects in the histogram
Applied Gate. This command allows changing the applied gate in the selected histogram. The pulldown menu lists all gates from the Gates table.
Gallery. This command generates an image gallery of all objects in the selected histogram. The number
of images is by default limited to 100. If the histogram contains more objects the 100 objects closest to
the center of gravity of the region will be shown by default. (See Chapter 2.2, General Settings.)
Export Data: This command exports the data of the selected histogram as tabulator delimited table in
txt format.
Well Results. This opens the Well Results window, see Chapter 4.3,
Well Results
.
4.2.3 Color Gating
Color Gating shows different populations as defined by the Gates in different colors in the one- and
two-dimensional histograms. Color Gating is a property of each individual histogram displayed; thus,
histograms with and without Color Gating can be displayed simultaneously in the different displays.
In order to activate Color Gating right-click in the histogram and from the context menu (right click)
either activate the option Color Gating directly or open Settings... and there activate the checkbox
Color Gating.
In the Histogram Properties window that opens via Settings… select Show Legend. This way you can
additionally show the legend of the colors in the histogram. The population percentages of the displayed gates are also displayed.
The following rules apply to decide which color is shown in Color Gating:
– If there is only one object located on a pixel (Bin) the gating color of that object is shown.
– If there are multiple objects on a histogram pixel the gating color of the relative majority of the
objects will be shown. Exception: If there is a Gate applied to that Histogram that Gate will be
automatically in the background as 100% of the objects will belong to that Gate.
Page 56

50 Chapter 4 – Analysis Results
– Objects not belonging to a gate are shown in black.
– If there is no majority Gate the Color order of the Gating list will be used (AnalysisAssay Gat-
ing…; see Chapter 4.2.2,
buttons in that window.
– If a histogram is gated (context menu: Set Gate) the objects will be shown in the color of the
sub-gate they belong to.
The Gate Manager
). The "ranking" can be changed with the arrow
The order mechanism of Color Display (1-D):
– In 1-D histograms the applied Gate is always in the background.
– The order of display is organized by areas. If one of the Gates has less area above the other
one it will be shown in the foreground.
The selector line is now position sensitive in 1-D Histograms as well. Thus if you click on a
green area you will get an object from the green Gate, if you click on gray you will get one
from the gray Gate.
Examples:
1. The example below shows a cell cycle with G1 and G2 defined as gates with different colors.
The histogram is not gated, thus all detected objects are shown. Some of the pixels represent more than one object. In this case all pixels that have at least one object that belongs to
the gate
jority of G1 or G2 because G1 and G2 are subgroups of
Manager is
Selected
Selected
are shown in red. Only the red color is shown because there is never a ma-
Selected
and the order in the Gate
above G1 and G2. The other objects are shown in Black because they
do not belong to any Gate.
Page 57

Analysis Software Manual Chapter 4 – Analysis Results 51
2. In this example the situation is identical to the first example, but the order has been changed.
All Pixels having a majority of G1 will now be shown in a green. All G1 objects also belong to
Selected
the data point is determined by the list order and now G1 has a higher priority than
but when the same number of objects belongs to G1 and to
Selected
the color of
Selected
.
3. In the last example the order is changed again, but now also the gate
the histogram. Now the exception Rule 2) applies such that the
Selected
Selected
is applied to
is not dominant.
Page 58

52 Chapter 4 – Analysis Results
4.3 Well Results
The Well Results window (via AnalysisAssay Gating) shows the results of a complete scan analysis.
Statistical results of different parameters are listed for gated populations of the individual wells or of
groups of wells and are displayed graphically in a histogram. Additionally the results can be exported
as txt-file for further analysis with other software.
4.3.1 Measurement Results
The Measurement Results tab lists all the wells recorded in one measurement and displays the results
(Number of Objects, % of tot., Mean, Error, Error % and CV) of the measurement parameter which is
selected in the drop-down menu Measurement. The result for a combination of several wells is given in
the bottom line when several wells are selected (press shift and click the lines you want to combine).The results can be exported by Export Table. The graph on the right gives a graphical representation of the results.
Well/Group results. This table gives a statistical analysis of the measurement results for each well or
group of wells. It lists the mean value of a parameter for the gated population in each well or group of
wells as well as its error and coefficient of variation.
Objects. Number of objects belonging to the population of the selected Gate
% of tot. Relative size of the Gated population
Mean. Mean value of the selected Parameter. It is shown in red in the histogram.
Error. Absolute error of Mean. It is shown in green in the bar plot.
Error %. Relative error
Page 59
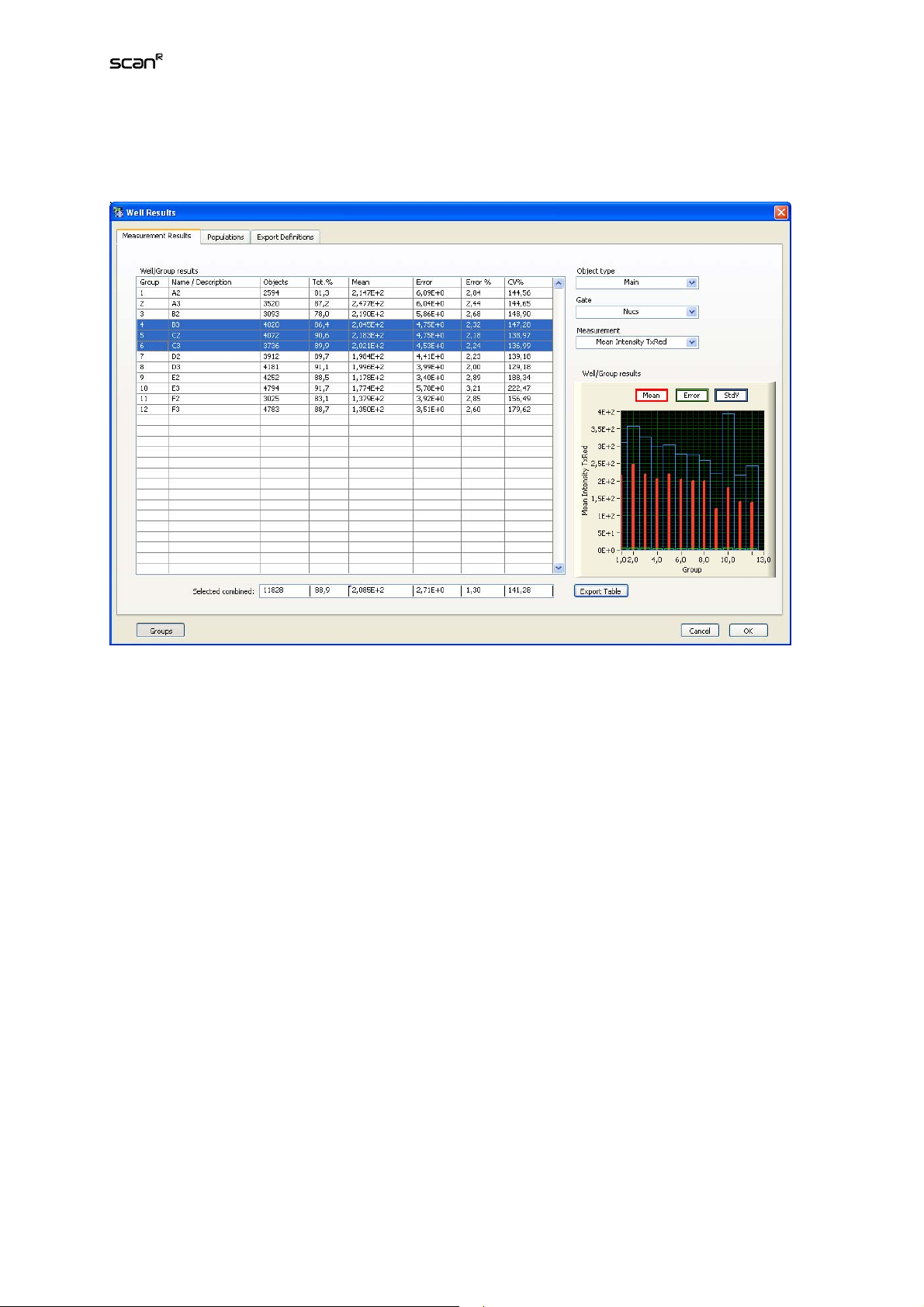
Analysis Software Manual Chapter 4 – Analysis Results 53
CV%. Coefficient of variation of Mean.
StdV. Standard deviation of Mean. It is shown in blue in the bar plot.
Object Type. Select the object type from the shortlist (Main/Sub-objects).
Gate. Select a gate from the shortlist in order to get the values for the corresponding gated population.
Measurement. Select the parameter of interest from the shortlist.
Selected combined. Select a number of Well/Group entries (press shift and click in the lines you want
to combine), their combined results will be given here.
Export Table. This function exports the data as tabulator delimited table in txt format.
Wells / Groups. Use this button to select if the values are to be given for individual wells or entire
groups of wells. All wells that have the same Name / Description entry form a group. Wells that do not
pertain to a group will be listed independently. The Name / Description is set in ScanSelect Wells
(see Chapter 2.7,
Selecting Wells for Analysis
).
4.3.2 Well Results: Populations
In the Populations tab the number of objects (absolute and relative) in all defined gates are listed. A
reference gate can be set to the number of objects in all other gates to the reference gate. The histogram on the right gives a graphical representation of the results.
Size comparison. The table gives the amount of objects in absolute numbers or as percentages that
pertain to each of the Gates defined in the Gates Manager. (See Chapter 4.2.2,
The Gate manager.)
Page 60

54 Chapter 4 – Analysis Results
Display Counts / %. This toggle button switches between the listing of total numbers and the percentage of objects.
Object type. Select the object type from the pull-down list (Main/Sub-objects).
Reference gate. The population of the selected Reference gate is set relative to the populations of the
other gates in the Size comparison table.
Export Table. This function exports the data as tabulator delimited table in txt format.
Wells / Groups. Use this button to select if the values are to be given for individual wells or entire
groups of wells. All wells that have the same Name / Description entry form a group. Wells that do not
pertain to a group will be listed independently. The Name / Description is set in ScanSelect Wells.
(See Chapter 2.7,
Selecting Wells for Analysis.
)
4.3.3 Export Definitions
In the Export Definitions tab the parameters that are to be exported can be defined. This is especially
useful for batch analysis and if not the complete results tables given in the Populations or the Measurements tab is to be exported.
Select New to create a new export file. The parameters to be exported have to be selected in the other
tabs, i.e. in the Populations tab or in the Measurement Results tab. When you right-click in these two
tabs on the column names a context menu will open which allows you to export the selected column to
a tab-delimited .txt-file.
Page 61

Analysis Software Manual Chapter 4 – Analysis Results 55
New. Creates a new tab-delimited .txt-file for export.
Export file name. Enter the name for the txt file here. The default is Result0*.txt
Remove. Removes the selected File and its selection
Export. Allows setting set the folder where the .txt-files are stored manually. Otherwise the folder given
in Settings (see Chapter 2.3,
General Manager
) is used.
Wells / Groups. Use this button to select if the values are to be given for individual wells or entire
groups of wells. All wells that have the same Name / Description entry form a group. Wells that do not
pertain to a group will be listed independently. The Name / Description is set in ScanSelect Wells.
(See Chapter 2.7,
Selecting Wells.
)
Page 62

56 Chapter 4 – Analysis Results
4.3.4 Export results of individual objects
AnalysisExport Table: exports the values determined for each detected object to a spread sheet. The
values exported depend on the active view. When also sub-objects are detected not only one file is
exported but for every sub-object a separate list is exported. For the “population view” the list contains
for example:
ParameterData_Main.txt
ObjectID MeanIntensityDAPI Obj.1Counts ParentObjectID ParentTraceID R01
0 174,52045 NaN -1 0 0
1 295,50986 2 -1 1 1
2 220,76321 1 -1 2 1
3 161,89612 NaN -1 3 0
ParameterData_Obj. 1.txt
ObjectID Total Intensity GM130 Area Parent Object ID ParentTraceID R01
0 201294 608 1 1 0
1 2504 12 1 1 0
2 2232 12 2 2 0
3 41191 108 4 4 0
4 4860 18 4 4 0
• Object ID. ID of the detected object
• Parameters. (e.g., MeanIntensityDAPI, Area,….)
• Derived Parameters.
• Parent Object ID. -1 if it is the main object, if not the ID of the main object is given
• ParentTraceID. if tracking was performed the ID of the Trace the detected object belongs to is
given. If no tracking was performed the Trace ID is set to -1.
• Gates. If the object belongs to the listed gate the value is set to 1, if not it is set to 0.
For the “Trace-view” the list contains:
TraceID lifetime max(Area) TrackedObjType TraceLength FirstParticleID LastParticleID R01
0 1 31 0 1 0 0 0
1 15 78 0 15 1 1940 0
2 8 626 0 8 2 1041 0
3 19 849 0 19 3 2640 0
• Trace ID. The ID of the Trace
• Parameters of the Trace (e.g. lifetime, max(Area),…)
• Derived Parameters.
• Traced Obj. Type. (0=main object; 1, 2, 3,…. Sub-object type)
• Trace Length. Number of objects in subsequent time frames belonging to the Trace
• First Particle ID. ID of the first object belonging to the Trace
• Last Particle ID. ID of the last object belonging to the Trace
• Gates. If the object belongs to the listed gate ate the value is set to 1, if not it is set to 0.
Page 63

Analysis Software Manual Chapter 5 – Example Assay – Step by Step 57
5 Example Assay – Step by Step
This chapter guides you step by step through the setup and execution of an assay and the analysis of
the results.
Setting-up and Executing an Assay............................................................ 58
5.1
5.2 Analyzing the Data......................................................................................61
5.3 Time-Lapse Analysis................................................................................... 64
Page 64

58 Chapter 5 – Example Assay – Step by Step
5.1 Setting-up and Executing an Assay
The example scan which is used here to guide you through the setup and execution of an assay consists of images taken from cells labeled with DAPI to mark the nuclei and with FITC to mark the microtubules. A repair protein which is active in late G2 phase stained with TexasRed.
1. Execute ScanOpen…, navigate to the folder where the demo data are stored, select the
experiment_descriptor.xml file ( /…/scan^R_Demo_Data_DVD_20070216/Slabicki_Buchholz
_MPI-CBG/H2AX_PFA_002/experiment_descriptor.xml; sample data kindly provided by F.
Buchholz, MPI-CBC, Dresden) and open it with OK.
scan^R Analysis loads the first image of the scan, i.e., the image acquired at the first position
of the first well in the main window of the font panel. Only the first channel of the image will
be shown at first and in gray scale. Move the cursor over the image as a test and observe the
status bar at the lower left. The channel intensity of each channel at the pixel positioned at
the tip of the cursor arrow. Is shown in the status bar. The intensity values of the objects are
usually much higher than the camera offset; the dark intensity of the camera.
Memorize the intensity values of the brighter structures in the image for the subsequent display adjustment.
2. Set the grayscale selector to None, and load the dapi channel in the blue selector and the
TxRed channel in the red selector.
3. Execute ViewLayout and adjust the display as explained in Chapter 2.5,
Viewer
.
4. The first step in setting-up the assay is to find the best settings for the object detection. If
you would have opened an old analysis file (*.sca), rather than the scan data only, the histograms would already show the data points of the analysis that was run before. In order to get
these results from-scratch you first have to set up a new assay to perform a background correction, determine main (and sub-) objects, and the parameters you want to extract from the
data.
Using the Image
5. Execute AssayEdit Assay to open the Assay Settings window.
Page 65

Analysis Software Manual Chapter 5 – Example Assay – Step by Step 59
6. The first step in Analysis is to select the
Background Correction
Image processor Module on
the Image Processing tab for each of the image channels. Press Adjust and observe in the
new window the background in the image before (left) and after (right) background correction. When moving the mouse over the pixels the grey value should be reduced in the corrected images.
7. The next step is to determine the main objects. Therefore open the Main Object tab in the
Assay Settings. Here you have to select the channel in which you want to use for image
segmentation. In many typical cases this will be the channel with the DAPI-labeled nuclei, the
shape of which makes them rather ideal targets for object detection.
8. For image segmentation use the Intensity module. Both the Edge and the Intensity modules
usually give good results when detecting nuclei. In other cases this is mostly a question of
experience and testing. Click Adjust to open the Object Finder window. Here you can check
and adjust the settings of the intensity detection algorithm. You will already get a good result
by simply pressing the Auto button for determining the right threshold value and turn the Watershed algorithm ON. You can observe the effects of the parameters in the right panel where
the result of the segmentation algorithm is shown.
9. If Sub-objects are associated with the Main Objects, check and adjust their detection in the
same way on the corresponding tab in the Assay Settings window. In this example skip the
Sub-objects tab.
Page 66

60 Chapter 5 – Example Assay – Step by Step
DAPI-labeled nuclei Detected objects
10. In the Parameter tab you can choose which parameters will be calculated during the analy-
sis, i.e. you need to select the measurements that are to be carried out. Typical measurements are area, mean and total intensity and the circularity factor. Area (size) and circularity
factor are extremely helpful as they allow to discriminate between more or less round, single
nuclei and impurities or objects that consist of clustered, non separable nuclei. The latter are
usually much larger and less circular. If sub-objects are to be detected it is useful to add the
parameter
Obj. 1 Counts
each main object.
to the list to determine how many sub-objects are connected to
11. The Derived Parameters tab allows you to perform calculations on the previously defined pa-
rameters, e.g. here you can calculate rations between different color channels. (For example
you could evaluate mean Intensity Dapi / mean Intensity TxRed).
Page 67

Analysis Software Manual Chapter 5 – Example Assay – Step by Step 61
12. Now you have set up the analysis. Press OK to exit the Assay Settings menu. You can save
your assay with Analysis Save assay…
13. Start the analysis by clicking on the Run button.
14. Prepare the histograms in the main window if you want to follow the analysis online. It is
suitable to select the parameters Area and Circularity Factor in the first histogram and to display Total intensity dapi vs. Mean intensity TxRed in the second histogram. The small image
window left to the main window shows the main object recognition in the image that is currently being analyzed. By clicking on the Main Display button you can have it displayed in the
largeimage window.
5.2 Analyzing the Data
1. The first thing to do is to get an overview of the detected main objects, in our example the
cell nuclei. The nuclei should all have a similar size and a more or less round shape, i.e. a
Circularity Factor (CF) close to 1.0.
Set Area as X-axis parameter and Circularity Factor as Y-axis parameter and examine the
size/shape distribution of object population in the scatter plots. The bulk of the population of
the example has a CF of <1.1 and an area of around 300 pixels in images taken with 4x
magnification.
Clustered cells
CF = 1.17
Area = 1590 pixels
Objects that are larger and have a larger Circularity Factor may for example consist of clustered cells that could not be separated by the watershed algorithm.
Page 68

62 Chapter 5 – Example Assay – Step by Step
2. For further analysis only consider those objects that do not differ significantly in size and
shape from the bulk. To do so, use the Region tool and draw a polygon around the
most densely populated part of the scatter plot. (Close the region by double clicking).
Right-click in the scatter plot to open the context menu and select Gate. By doing so, the region R01 is converted in a gate of the same name (R01) and you can apply this gate to all
other histograms to exclude all objects outside this gate. Right-click again and select Set
Gatenone to get back all objects.
Right-click on the border of the polygon and select Gallery to display a selection of objects
inside the gate; this allows you to crudely evaluate if your gating was set in a useful way.
3. In the next histogram display the Total Intensity Dapi vs. the Mean Intensity TxRed. To in-
clude only the objects in R01 right-click in the histogram and select Set GateR01. You will
observe a cluster around the total Intensity Dapi of ~9x10
6
. The left cluster consists of the nuclei in G1, whereas the cluster with twice the dapi
1.8x10
intensity contains the nuclei in G2. (With G2 cells having twice the amount of DNA they will
have twice the amount of DAPI-labeling and thus roughly twice the fluorescence intensity.)
Draw a polygon around the cluster on the left with the Region tool and keep in mind the
name (R02). Execute AnalysisAssay Gating to open the Gate Manager window.
5
and another cluster around
Click new to create a new gate. You can give the new gate a meaningful name (e.g. G1) and
also select a color. In the Definition line enter R01 AND R02 to select only the objects which
are contained in both regions. Rename the Gate R01 to Nuclei; these are the objects defined
in step 2. Then Close the Gate Manager.
4. Next draw a region around the right cloud, i.e. the nuclei in G2 and repeat the steps de-
scribed in step 3 and name this gate G2.
X
5. In the two remaining histograms choose Mean Intensity TxRed for the
and Y axis. This will
give the distribution of objects with a certain amount of TexasRed staining in all nuclei. To
find out if the distribution of TexasRed is different in the G1 and G2 phase right-click and select Set GateG1 in the context menu in one histogram and Set GateG2 in the other histogram.
Page 69

Analysis Software Manual Chapter 5 – Example Assay – Step by Step 63
6. You will observe that the distributions in G1 and G2 look different. In G1 there are only nuclei
with a low mean intensity in TexasRed, whereas in G2 there are two populations: one with a
lower mean intensity in TexasRed and one with a higher intensity in TexasRed. Again select
the polygon tool to define a region around the two populations in G2 and create two gates
according to step 3. We name the nuclei with a high concentration of Texas Red G2_active,
and the nuclei with a low concentration G2_passive. The result in the Gate Manager may
look as follows:
7. Leave again the Gate Manager window. By right-clicking on the main window you can high-
light the nuclei in the different gates with boxes when you activate them (e.g. Show
GatesG2_active). Another option to compare the results is to right-click on the border of
the gate and to choose Region Gallery. Plot a region gallery of G2-Active and G2-passive to
compare the results of the gating.
8. In the lower part of the Gate Manager you will also find statistics of the displayed histograms.
For more detailed results click Well Results in the gate manager and go to the tab Populations. Here you will find the results for all wells and all gates. You can choose if you want to
display the total counts or the results as percentages. You can export these results for further analysis. A graphical display allows you to directly compare the results for different
wells. Select G2 as reference gate and G2, G2_active and G2_passive as gated populations.
Page 70

64 Chapter 5 – Example Assay – Step by Step
You will notice that in the first 8 wells the numbers of active and passive nuclei are almost
identical. However in the last 4 wells the number of active cells in G2 is strongly decreased.
9. To export the results either select Export Table in the Populations or Measurement Results or
select the results to be exported via Export Definitions as described in Chapter 4.3.3,
Definitions
.
Export
5.3 Time-Lapse Analysis
In this example a time-lapse series is analyzed. The images of the example show cells during mitosis.
The cells are marked with two stains (GFP and TxRed). The images left and right show images from the
same well and position, taken at two different time points (t=10, t=50). In order to analyze time-lapse
data it is necessary to segment the images properly and to track the moving cells from frame to frame.
t=10 t=50
Page 71

Analysis Software Manual Chapter 5 – Example Assay – Step by Step 65
1. Execute ScanOpen…, navigate to the folder where the demo data are stored, select the
experiment_descriptor.xml file ( /…/scan^R_Demo_Data_DVD_20070216/Slabicki_Buchholz_MPI-CBG/CMV-mRed+cyclineB1GFP/GFP+mRed_001/experiment_descriptor.xml;
sample data kindly provided by F. Buchholz, MPI-CBC, Dresden) and open it with OK.
2. Execute AssayEdit Assay to open the Assay Settings window.
3. Go to the Image processor module in the Image Processing tab to set Background Correc-
tion for all channels (TxRed and GFP). Press Adjust and observe in the new window the
background in the image before (left) and after (right) background correction. When moving
the mouse over the pixels the grey value should be reduced in the corrected images..
4. Open the Main Object tab in the Assay Settings. Here you have to select the channel which
you want to use for image segmentation. Select the TxRed channel (which has the stronger
signal) for image segmentation and use the intensity module. Click on Adjust: The Object
Finder: Intensity Threshold menu opens. Press Auto to set the threshold and use the watershed algorithm.
5. Skip the sub-object tab.
6. Go to the Parameters tab. Select here the parameters you want to detect for the main object.
Select Area, Center X, Center Y, Position, Well and Circularity Factor, which are determined
on the channel of the main object (i.e. TxRed). For the GFP Channel select Mean Intensity
and Total Intensity. Note that the intensities of the GFP channel are also detected on the
main object (which was originally segmented using the TxRed channel).
Page 72

66 Chapter 5 – Example Assay – Step by Step
7. Confirm the Assay Settings with OK and Run the analysis.
8. After the analysis you can plot the Histograms and Scatterplots as described in Chapter 5.2,
Analyzing the Data
.
9. The next step after the image segmentation and the extraction of the parameters is the track-
ing of the objects. Go to Tracking Configure Tracer. Check the Enable Tracking box and
select Main as tracked object. Set a range of 20 pixels. Confirm with OK. When leaving this
dialog the tracking will start. The progress of the tracking is shown in the status bar.
10. After the tracking is finished the Trace Parameters menu will open. Here you can browse
through the traces and determine the kinetic parameters that will be extracted from the
curves. Select the operators which will be applied to the kinetic curves of the selected Parameters. Let’s say we want to find all the cells that undergo mitosis during the measurement. When undergoing Mitosis the cells show an increase in the mean intensity of GFP.
First set the smoothing to ~1 to obtain smoothed curves. In order to find out the mitotic cells
in a simple approach set the following definitions: We need the max(MeanIntensity(GFP)) to
find out the maximum intensity of the curve. Then we need the lifetime to gate on the cells
that are tracked over all images. We are also interested in the time point when the maximum
mean intensity is reached. Therefore select also max(MeanIntensity(GFP))
Page 73

Analysis Software Manual Chapter 5 – Example Assay – Step by Step 67
11. Close the menu with OK. The analysis of the curves is performed. In the lower right of the
front panel the status bar displays the progress of the analysis.
12. When the analysis is finished you can plot a lifetime histogram in the first histogram panel. In
order to select the cells that were detected in all timeframes draw a rectangle around the
data points at 140. Right-click on the rectangle and select Gate to convert the region into a
gate.
13. Go to Analysis Assay Gating… and rename the Gate R01 to longtraces. Leave the menu
with OK.
Page 74
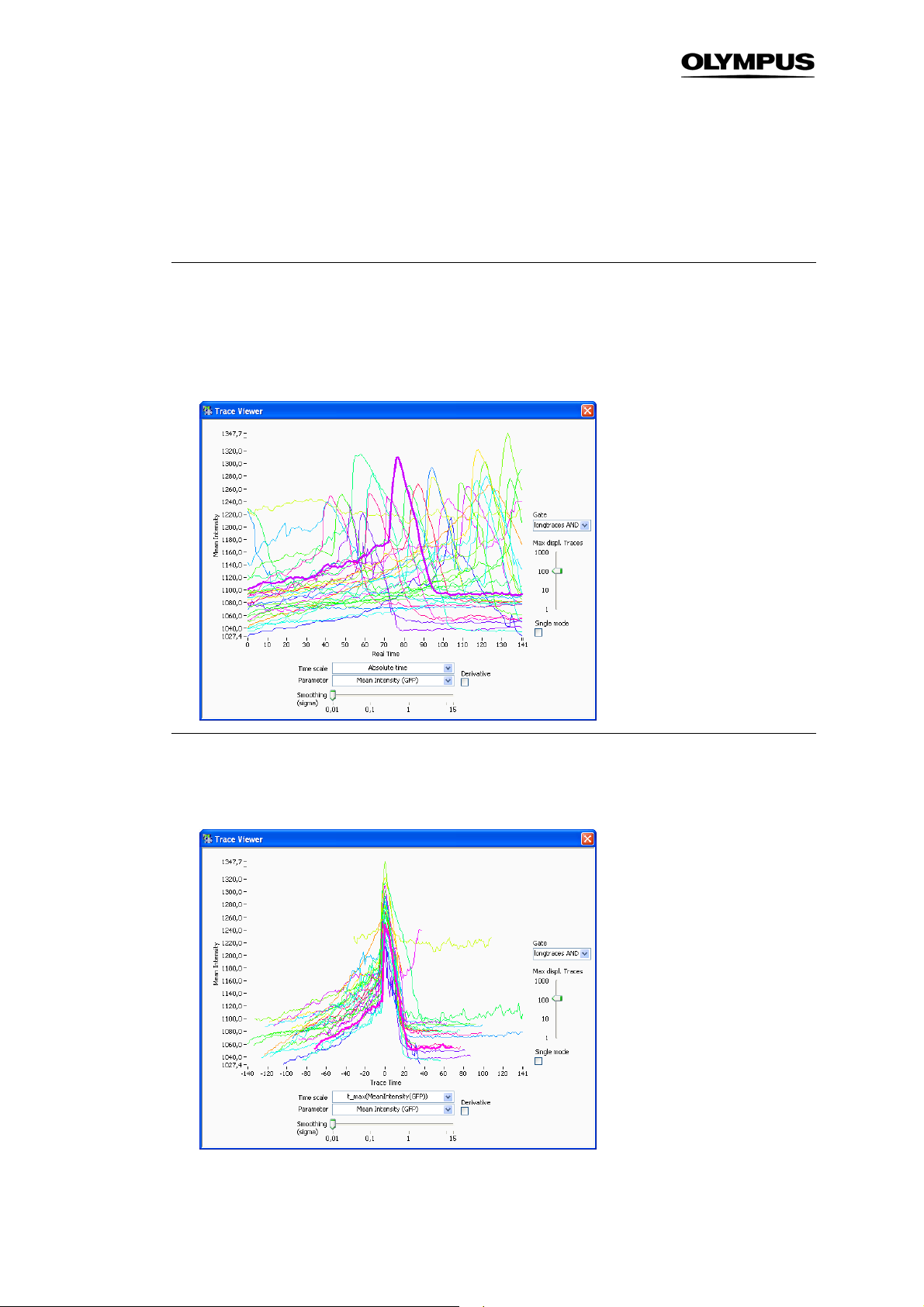
68 Chapter 5 – Example Assay – Step by Step
14. In the second histogram display, plot max(MeanIntensity(GFP)). Right-click in the histogram
and select Set Gate Longtraces in order to include only the cells that were detected in all
images into the analysis. Draw a rectangle around the datapoints on the right. These are the
cells which show a high mean intensity of GFP. Right-click on the border of the rectangle
and select Gate. The new gate longtraces AND R02 is created.
15. Mark one of the datapoints and the corresponding cell is shown in the image display with a
green outline. Right-click on the image of the cell and select show trace from the context
menu. The Trace Viewer opens and displays the curve of the selected cell. In order to show
all the traces of the gated cells, deactivate single mode and for Gate select longtraces AND
R02. The curves show all the same characteristics.
16. As default the Time scale is set to Absolute time but it is also possible to display and analyze
the curves regarding to a certain timepoint, e.g. the time of mitosis. Therefore select
t_max(MeanIntensity(GFP)) as Time scale.
Page 75
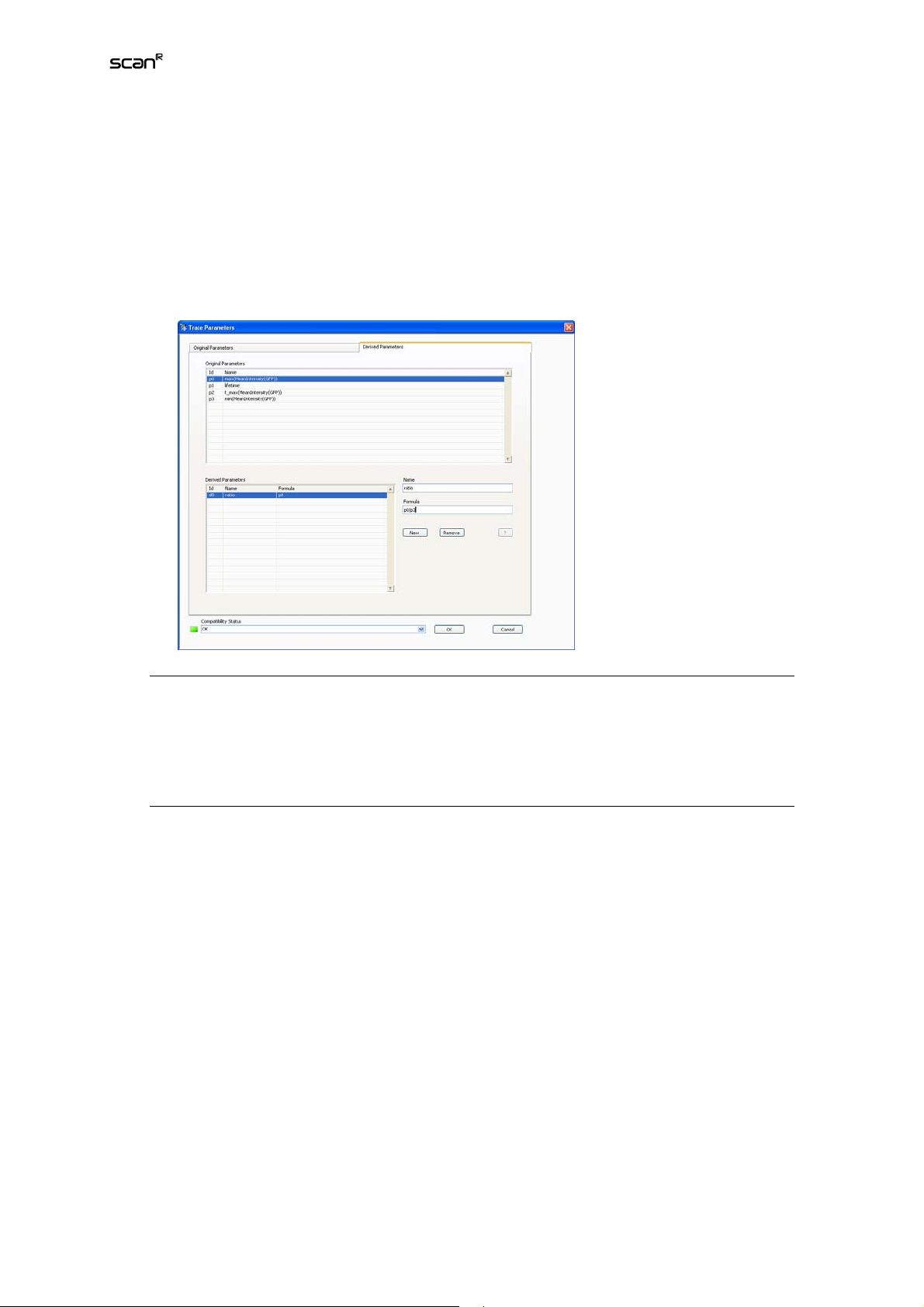
Analysis Software Manual Chapter 5 – Example Assay – Step by Step 69
17. As you can see, the green curve shows no mitosis but has a high mean intensity in GFP all
the time. (When you select the green curve and observe the corresponding object in the images you see that this is probably an apoptotic cell). In order to avoid this you can set up a
more advanced analysis, e.g. you can set the parameters max(MeanIntensity(GFP)) and
min(MeanIntensity(GFP)) in the Trace Parameters menu (therefore go back again to Tracking
Define Parameters). Go to the Derived Parameters tab In the Trace Parameters menu and
set as new derived parameter the ratio of max(MeanIntensity(GFP)) and min(MeanIntensity
(GFP)). Name this new parameter ratio.
18. After running the trace analysis plot a histogram of ratio. Set again longtraces as gate. The
data points around 1 show about the same minimum and maximum mean intensity of GFP.
Whereas data points >1 are the ones that undergo mitosis and have a higher maximum mean
intensity. Draw a rectangle around the data points with ratio >1. Right-click on the border of
the rectangle and select Gate. The new gate longtraces AND R03 is created.
19. Mark one of the data points and the corresponding cell is shown in the image display with a
green outline. Right-click on the image of the cell and select show trace. In order to show all
the traces of the gated cells, deactivate single mode and as gate select longtraces AND R03.
You will see that this parameters lead to a better discrimination of mitotic cells than the more
simple approach used before.
Page 76

70 Chapter 5 – Example Assay – Step by Step
Page 77

Analysis Software Manual Chapter 6 – Appendix 71
6 Appendix
File Conversion........................................................................................... 72
6.1
6.1.1 Images List................................................................................................72
6.1.2 Image Channel Definition..........................................................................73
6.1.3 Scan Settings............................................................................................ 74
6.1.4 View Conversion Results..........................................................................74
6.2 FCS Export Functionality............................................................................75
6.3 Libraries......................................................................................................77
6.3.1 Object Analyzers Library (OAL) ................................................................ 77
6.3.2 Object Finders Library (OFL) .................................................................... 78
6.3.3 Image Processing Library ......................................................................... 78
6.3.4 Virtual Channel Library.............................................................................. 79
6.4 Valid functions for derived parameters.........................................................80
Page 78
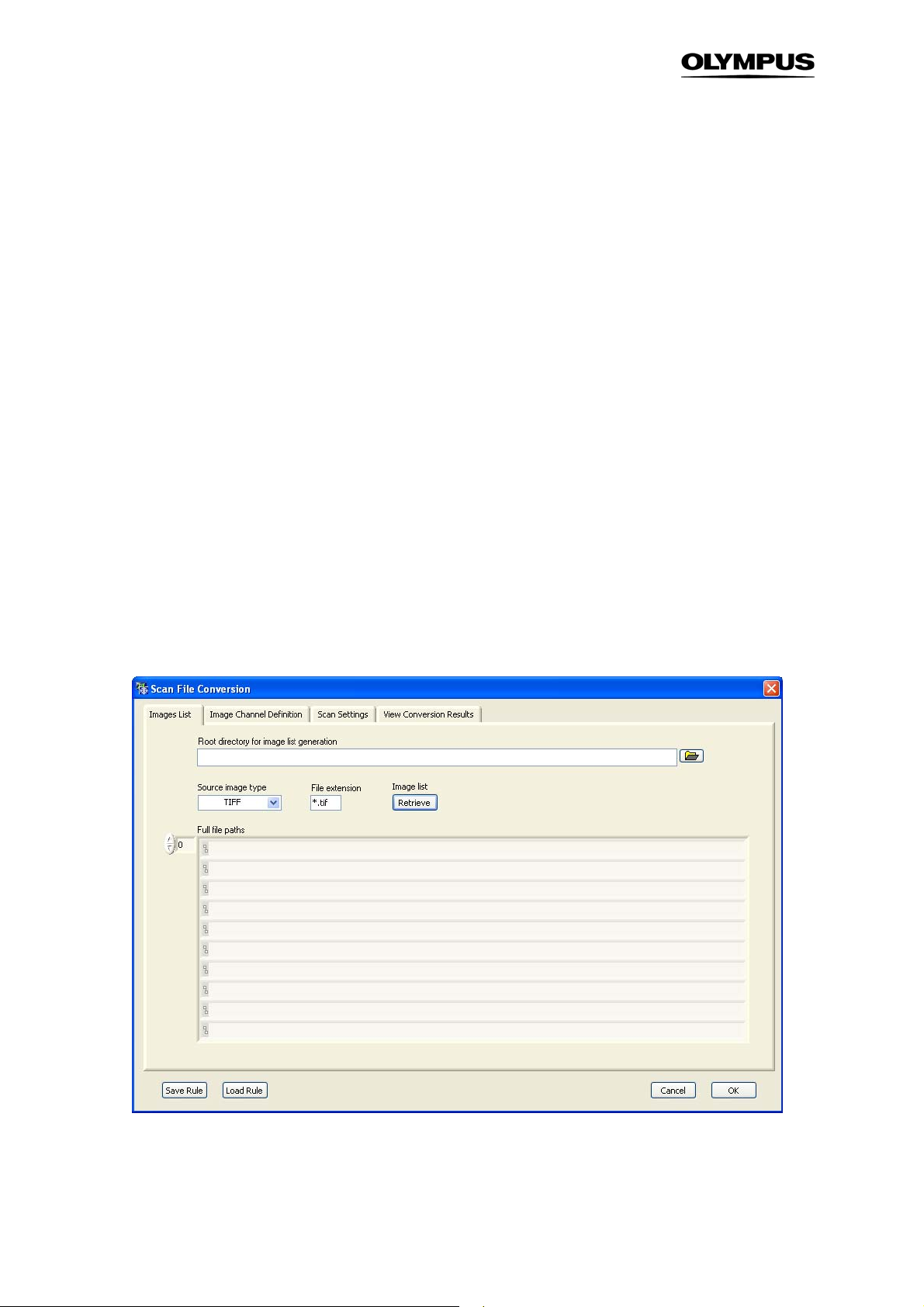
72 Chapter 6 – Appendix
6.1 File Conversion
scan^R Analysis is able to analyze image data acquired with a third party imaging system as long as
the data are stored as (monochromatic) 16-bit single TIFF images. The file names must allow the assignment of the images to individual positions, time stamps and color channels.
The command ScanCustom conversion opens the Scan File Conversion dialog that allows setting up
conversion rules. The creation of conversion rules is a four-step process, which will be described, in the
following chapters.
Save rule. This function saves the current conversion settings as a Custom Pattern file (*.cr).
Load rule. This function allows loading a stored set of conversion settings.
6.1.1 Images List
Root directory for image list generation. Click on the folder button on the right of the box to open the
navigation window. Navigate into the storage directory and click Select Curr. Dir.
Source image type. Select the desired image type (TIFF, JPEG, PNG or BMP) from the shortlist.
File extension. This is set automatically when the Source image type is chosen. The file extension
serves to filter the content of the selected directory
Page 79
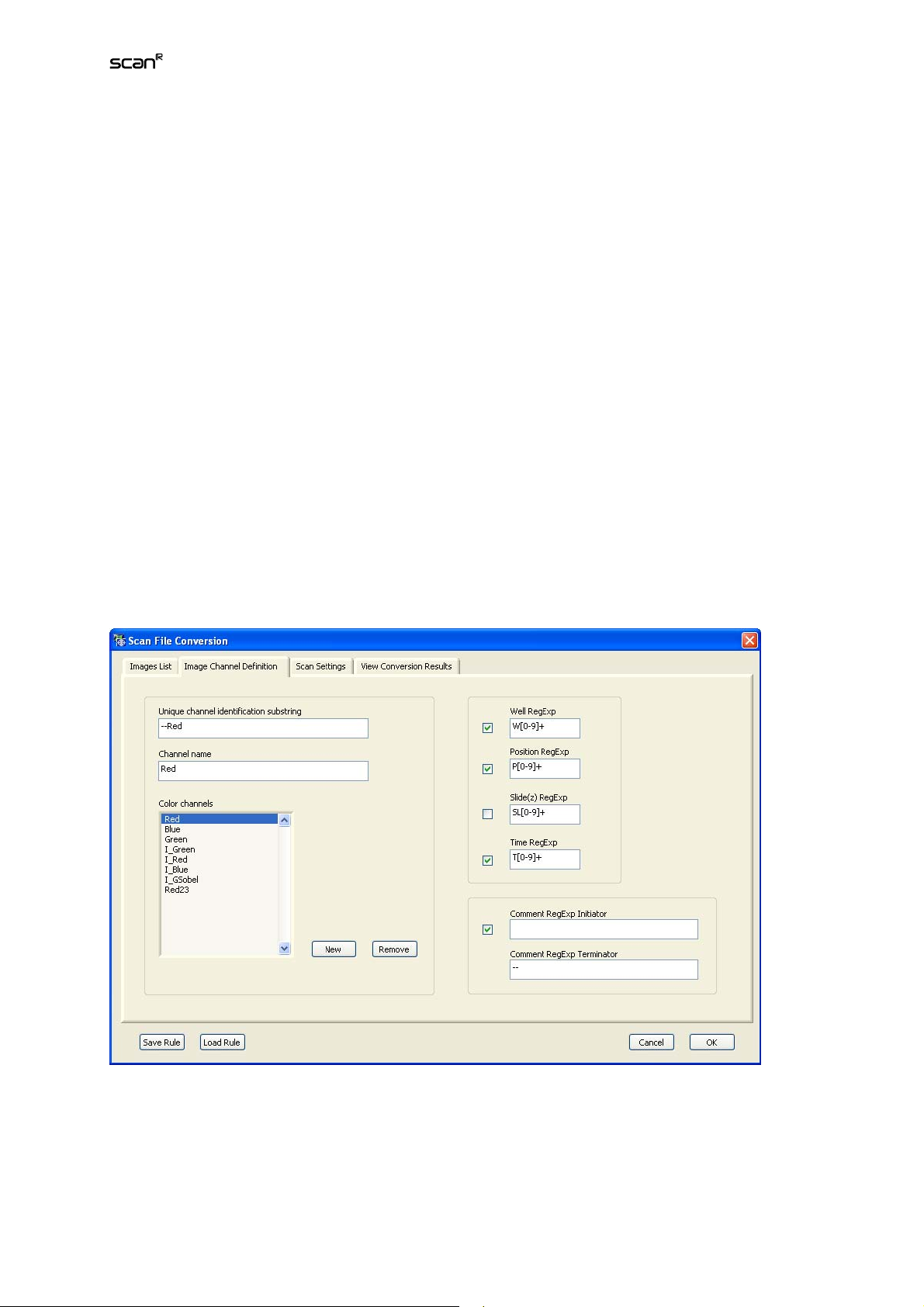
Analysis Software Manual Chapter 6 – Appendix 73
Image list: Retrieve. This function reads the matching files into the Full file paths list. The arrow buttons
allow easy browsing and verification of retrieved image data.
6.1.2 Image Channel Definition
If the file names of the third party images contain codes for Well, Position, Z-layer and Time, scan^R
will sort them automatically upon conversion into scan^R format. The Image Channel Definition tab is
used to define the conversion rules for this.
Unique Channel Identification String. If the original file names contain a certain expression – or Identification String – that classifies them as belonging to a certain color channel, enter it here.
Channel Name. Enter the Channel Name to be used in scan^R here.
New. Click here to add the newly defined channel to the Image Channel list.
Well, Position, Slide (Z), Time RegExp. If the file names contain information about Well, Position, Z-layer
and Time in form of regular expressions (RegExp), scan^R can group them when the corresponding
code is entered here.
An overview of the syntax of regular expressions can be found under
http://www.regular-expressions.info/tutorial.html.
Comment RegExp Initiator / Terminator. If the file names contain information about wells, scan^R can
use this as Well Description, see for example Chapter 2.7,
Selecting Wells for Analysis
. Enter the first
(Initiator) and last (Terminator) alphanumerical digit(s) in the respective boxes.
Page 80

74 Chapter 6 – Appendix
6.1.3 Scan Settings
Scan settings are used to reconstruct a minimal configuration file for the data conversion. scan^R
needs information about the used multi-well plate and the arrangement of the images within the single
wells in order to represent the data properly in the graphical user interface
Result Dir. Click on the folder button on the right of the box to open the navigation window. Navigate to
the storage directory and click Select Curr. Dir.
Positions: Columns/Well and Rows/Well. Set the number of image columns and rows, respectively, per
well.
Wells: Columns and Rows. Set the number of columns and rows, respectively, of the multi well plate.
Time-lapse cycles. Set the number of time points for this scan.
6.1.4 View Conversion Results
Test. Upon clicking this button, scan^R converts the file names and lists them in the New File Name
list. This allows checking whether the conversion rules yield the desired result.
Start. Click here to start the file conversion process. The Start button becomes active once a conversion rule is defined or loaded. It initiates the file conversion. This may be time consuming because an
entire new set of images is generated and stored.
Page 81

Analysis Software Manual Chapter 6 – Appendix 75
6.2 FCS Export Functionality
scan^R is able to export single cell data to ISAC FCS 3.0 Standard files. Please note, that this standard
is not fully implemented by most of the programs that specify FCS 3.0 import support. Therefore some
settings modifications for export might be necessary depending on the specific limitations of these
programs. In practice especially the FCS 3.0 requirements for double and integer 32 bit data support as
well as multi dataset FCS files might not be implemented because most flow cytometers produce integer 16 bit data in single data sets. It is therefore recommended to test the FCS 3.0 compatibility before
deciding which FCS program is to be used for the analysis. Most manufactures provide demo programs for testing.
In order to export FCS data open the Settings menu from the menu bar.
Page 82

76 Chapter 6 – Appendix
Export Data Type. Here you can choose the data types exported (Integer 16 / Integer 32 / double (64bit floating point) according to FCS 3.0 standard. It is recommended to use the highest precision that is
supported by the program intended to import the fcs files.
Scale Export Scaled (On/Off). This button activates the rescaling procedures during FCS export. Switch
off the scaling if you want to compare datasets from two separate export procedures quantitatively. If
activated, the scaling settings (Min/Max/Clipping) will be used.
Export Min / Export Max / Clipping %. Settings used for rescaling the original data according to:
x_fcs = x*PnG + PnOffset, with:
x_fcs exported data points
x the original data points and
n parameter number
PnG automatically determined linear scaling factor (Gain)
PnOffset
The resulting scaling offset and the scaling factor are stored in the FCS file header in the keywords
automatically determined scaling offset
PnG and PnOffset. If you switch on the scaling then the range of the original data will be scaled to fit
into the given export range.
settings according to:
PnG and PnOffest are calculated from the original dataset and the scale
Export Range = Export Max - Export Min
Original Data Range = Original Data Set Max Value - Original Data Set Min Value
Clipped Range = Original Data Set Max Value
Set Min Value
(ignoring the smallest clip %)
(ignoring the largest clip %) - Original Data
PnG = Export Range/Clipped Range
PnOffset = Export Min - PnG*(Original Data Set Min Value
%)
)
Analysis Export Wells (FCS Settings):
Merge Well Data. All Wells are taken together to a single dataset
Single File. Exports Wells into separate datasets in a single FCS file according to FCS 3.0 standard (this
Multi-dataset format is not supported by all FCS programs)
Separate Files. The export table function creates a single FCS file for every well.
(ignoring the smallest clip
i16 export (FlowJo). When using i16 export and importing with FlowJo, FlowJo uses the
gain to rescale the data but is then not able to go back to the logscale. Therefore it is recommended to use raw mode with i16 and FlowJo. To switch to logscale use then the
FlowJo function: Add derived ... and convert the required parameters to log. Doubles and
Multi-dataset FCS Files can not be read by FlowJo. Please note, that if you have data
within a X,Y logscale and you rescale the X,Y data with different scaling factors (as might
happen with the automated scaling during export because of range differences) there might
be a shape change.
An alternative to FlowJo is FCS Express (http://www.denovosoftware.com/site/Download.shtml)
which is supposed to stick more strictly to ISAC FCS 3.0 standard.
Page 83

Analysis Software Manual Chapter 6 – Appendix 77
6.3 Libraries
6.3.1 Object Analyzers Library (OAL)
Page 84

78 Chapter 6 – Appendix
The password-protected Object Analyzers Library can be accessed through ModulesObject Analyzers Library.
It allows managing the set of parameters selectable in the Edit Assay tab Parameters; see Chapter 4.6,
Measurement Parameters
Add. Click here to add a new entry to the list.
Parameter. Select the desired parameter from the drop down menu to add it to the list.
.
6.3.2 Object Finders Library (OFL)
The password protected Object Finders Library can be accessed through ModulesObject Finders
Library. It lists and allows administrating the available Object Finder Modules. Each module may have
different settings lists and may thus appear several times.
Add/Remove. Click here to add or delete a new entry to the list.
6.3.3 Image Processing Library
The password-protected Image Processing Library can be accessed through ModulesImage Processors…
It allows managing the set of available Image Processing algorithms in the Image Processing modules.
Add Module. Click here to add a new entry to the list.
Remove Module. Click here remove an entry from the list.
Page 85

Analysis Software Manual Chapter 6 – Appendix 79
6.3.4 Virtual Channel Library
The password-protected Virtual Channel Library can be accessed through ModulesVC Processors…
Page 86

80 Chapter 6 – Appendix
It allows managing the set of available Virtual Channel algorithms in the module selector on the Edit
Assay Virtual Channels tab.
Add Module. Click here to add a new entry to the list.
Remove Module. Click here remove an entry from the list.
6.4 Valid functions for derived parameters
abs(x) Absolute Value Returns the absolute value of x.
x
) Inverse Cosine Computes the inverse cosine of x in radians.
acos(
x
) Inverse Hyperbolic Cosine Computes the inverse hyperbolic cosine of x.
acosh(
x
) Inverse Sine Computes the inverse sine of x in radians.
asin(
x
) Inverse Hyperbolic Sine Computes the inverse hyperbolic sine of x.
asinh(
x
) Inverse Tangent Computes the inverse tangent of x in radians.
atan(
y,x
atan2(
atanh(
ceil(
ci(
cos(
cosh(
cot(
csc(
exp(
expm1(
floor(
getexp(
gamma(
getman(
int(
intrz(
ln(
lnp1(x) Natural Logarithm (Arg +1) Computes the natural logarithm of (x + 1).
log(x) Logarithm Base 10 Computes the logarithm of x (to the base of 10).
log2(x) Logarithm Base 2 Computes the logarithm of x (to the base of 2).
max(x,y) Max & Min Compares x and y and returns the larger value.
min(x,y) Max & Min Compares x and y and returns the smaller value.
mod(x,y) Quotient & Remainder Computes the remainder of x/y, when the quotient is rounded toward –
pow(x,y) Power of X Computes x raised to the y power.
) Inverse Tangent (2 Input) Computes the arctangent of y/x in radians.
x
) Inverse Hyperbolic Tangent Computes the inverse hyperbolic tangent of x.
x
) Round Toward +Infinity Rounds
x
) Cosine Integral Evaluates the cosine integral for any real nonnegative number x.
x
) Cosine Computes the cosine of x, where x is in radians.
x
) Hyperbolic Cosine Computes the hyperbolic cosine of x.
x
) Cotangent Computes the cotangent of x (1/tan(x)), where x is in radians.
x
) Cosecant Computes the cosecant of x (1/sin(x)), where x is in radians.
x
) Exponential Computes the value of e raised to the x power.
x
) Exponential (Arg) – 1 Computes one less than the value of e raised to the x power ((e^x) – 1).
x
) Round To –Infinity Truncates x to the next lower integer (largest integer
x
) Mantissa & Exponent Returns the exponent of x.
x
) Gamma Evaluates the gamma function or incomplete gamma function for x.
x
) Mantissa & Exponent Returns the mantissa of x.
x
) Round To Nearest Rounds x to the nearest integer.
x
) — Rounds
x
) Natural Logarithm Computes the natural logarithm of x (to the base of e).
Infinity.
x
to the next higher integer (smallest integer
x
to the nearest integer between x and zero.
>= x
<= x
).
).
Page 87

Analysis Software Manual Chapter 6 – Appendix 81
rand( ) Random Number (0 – 1) Produces a floating-point number between 0 and 1 exclusively.
x,y
) Quotient & Remainder Computes the remainder of x/y, when the quotient is rounded to the nearest
rem(
x
) Sine Integral Evaluates the sine integral for any real number x.
si(
x
) Secant Computes the secant of x, where x is in radians (1/cos(x)).
sec(
x
) Sign Returns 1 if x is greater than 0, returns 0 if x is equal to 0, and returns –1 if x
sign(
x
) Sine Computes the sine of x, where x is in radians.
sin(
x
) Sinc Computes the sine of x divided by x (sin(x)/x), where x is in radians.
sinc(
x
) Hyperbolic Sine Computes the hyperbolic sine of x.
sinh(
x
) Spike Generates the spike function for any real number x.
spike(
x
) Square Root Computes the square root of x.
sqrt(
x
) Step Generates the step function for any real number x.
step(
x
) Tangent Computes the tangent of x, where x is in radians.
tan(
x
) Hyperbolic Tangent Computes the hyperbolic tangent of x.
tanh(
integer.
is less than 0.
Page 88

OLYMPUS LIFE SCIENCE EUROPA GMBH
Postfach 10 49 08, 20034, Hamburg, Germany
®
Printed in Germany 2008 12 M 1.1.0.6
 Loading...
Loading...