Page 1
Leica MM Monopole Detection
powered by MetaMorph
Analysis Software Drop-in for Leica MM AF
• Quantitation of mitotic cells with monopolar or bipolar spindles
• Adaptive background correction for improved segmentation
• Field and Cell-by-Cell data logging
®
Proper formation of a bipolar spindle is vital for the segregation
of chromosomes during mitosis. In some serious diseases where
cells proliferate uncontrollably, such as cancer, progression
through mitosis can be stopped by simply disrupting the normal bipolar spindle formation. Several classical chemotherapy drugs act
on microtubules to disrupt the bipolar spindle formation. However,
these treatments have side effects in interphase cells.
Recently, a new compound named monastrol was found to disrupt
spindle formation by affecting centrosome separation. In comparison with microtubule drugs, this effect was specic to mitosis.
When the two centrosomes fail to replicate or separate, a monopolar spindle forms instead of a normal bipolar spindle. Other compounds that can produce monopolar spindles are actively being
investigated.
The Leica MM Monopole Detection for Leica MM AF software is
designed for the quantitation of mitotic cells with monopolar or bipolar spindles where cells are labeled with a DNA stain and a second probe for microtubules.
The module utilizes Adaptive Background Correction adapting the
cell detection algorithm to the local intensity ranges between and
within cells to provide the most robust segmentation available.
This technique enables probe detection even with highly variable
background uorescence within a single image.
The simple interface minimizes setup efforts and at the same time
enables users to customize the settings and measurements to obtain the best possible results specic to the type of cells used in
the experiments.
Page 2
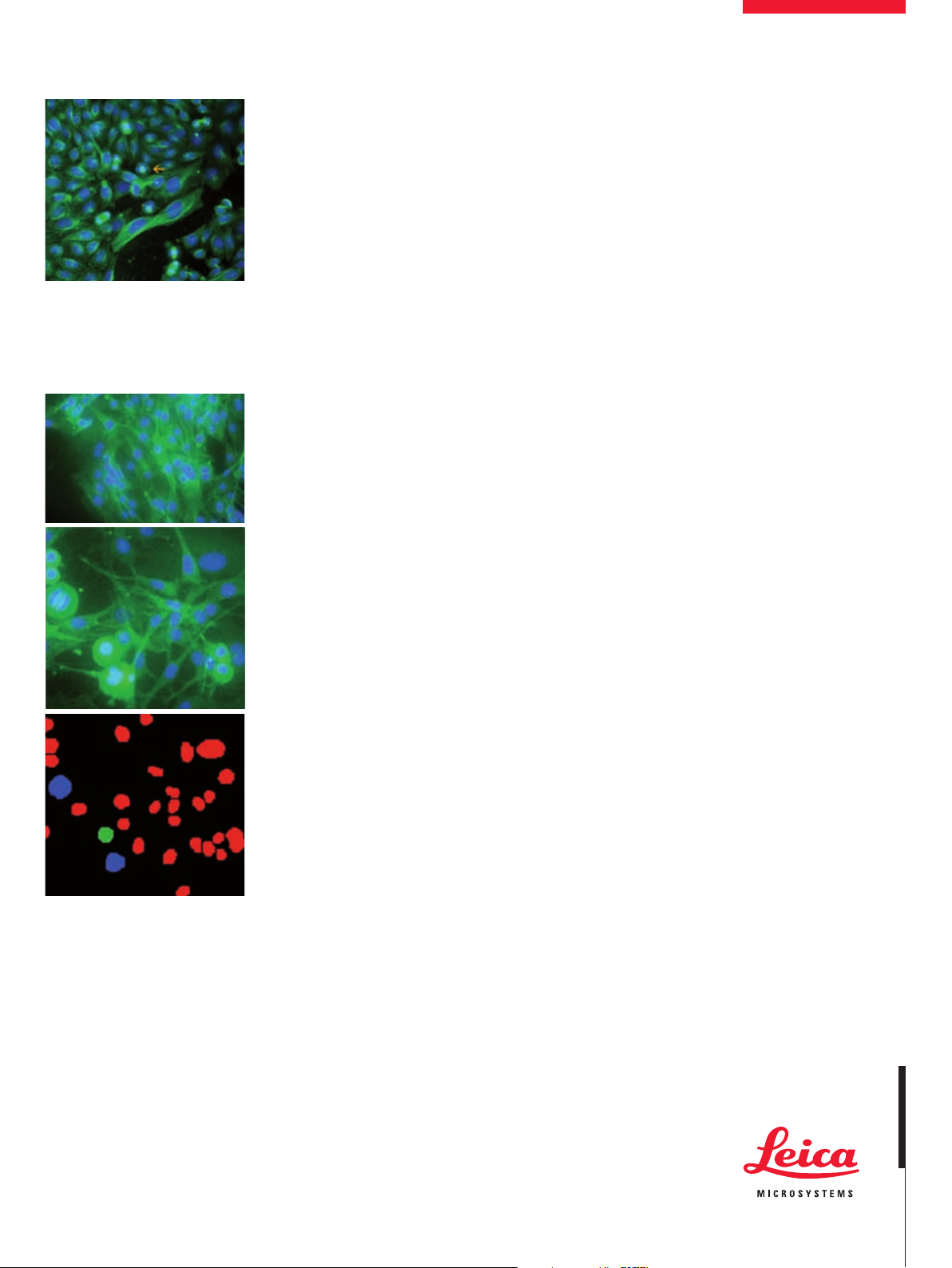
Multiple wavelength acquisition
CHO-K1 cells treated with monastrol and stained
with mouse anti-beta tubulin primary antibody
detected with a FITC conjugated goat antimouse secondary antibody. Nuclei are stained
with Hoeschst 33342. Orange arrow shows monopole.
Configuration for analysis
1. Select the DNA stained image
2. Specify the size range of DNA-stained cells
and intensity above local background
3. Select the microtubules image
4. Set cell classication limits based on
DNA/microtubule staining correlation
5. Optionally set reporting parameters
Interactive data display
Once the analysis is run, the Cellular Results table allows you to interactively view individual
cells’ data. Clicking one or multiple cells in the
image highlights the data for the selected cell(s)
in the table.
Customization through journaling
Journals are sophisticated and powerful macros
that record and perform a series of tasks without
the need for a programming language. The modules can be incorporated into a Leica MM AF
journal to increase the customization and automation of your analysis.
Multi-parameter analysis
The application module can generate a number
of eld or cell-by-cell parameters. Field measurements include:
• Count and percentage of monopoles, bipoles
and interphase cells
• Area of DNA structures, monopolar, bipolar and
interphase cells
• DNA and microtubule average intensities
Cell-by-cell measurements include:
• Cell classication
• Cell correlation coefcient (DNA versus
microtubule staining)
• Cell DNA structures area
• Integrated and average intensities of DNA
and microtubules
“MetaMorph® is a Registered Trademark of MDS Analyti-
cal Technologies”
Robust segmentation and analysis
3T3-L1 mouse fibroblast cells treated with monastrol and stained with mouse anti-beta tubulin
primary antibody detected with a FITC conjugated goat antimouse secondary antibody. Nuclei
are stained with Hoeschst 33342. Top: control,
middle: monastrol, bottom: segmented image
shows interphase cells (red), bipolar spindles
(blue) and monopole (green).
www.leica-microsystems.com
Powerful data export capabilities
All measurements can be directly exported to a
text le or Microsoft® Excel® for further analysis.
 Loading...
Loading...