Page 1

................................................................................................................................
Profinity
™
Epoxide Resin
Instruction Manual
Please read these instructions prior to using Profinity epoxide resin. If
you have any questions or comments regarding these instructions,
please contact your Bio-Rad Laboratories representative.
Page 2

Table of Contents
Section 1 Product Description ................................................1
Section 2 General Considerations for
Ligand Coupling.......................................................3
Section 3 Column Packing.......................................................5
Section 4 Protein Binding and Elution ..................................7
Section 5 Renaturation of Eluted Proteins...........................8
Section 6 Assessing Protein Purity........................................8
Section 7 Regeneration and Storage.....................................9
Section 8 References................................................................9
Section 9 Ordering Information ..............................................9
Page 3
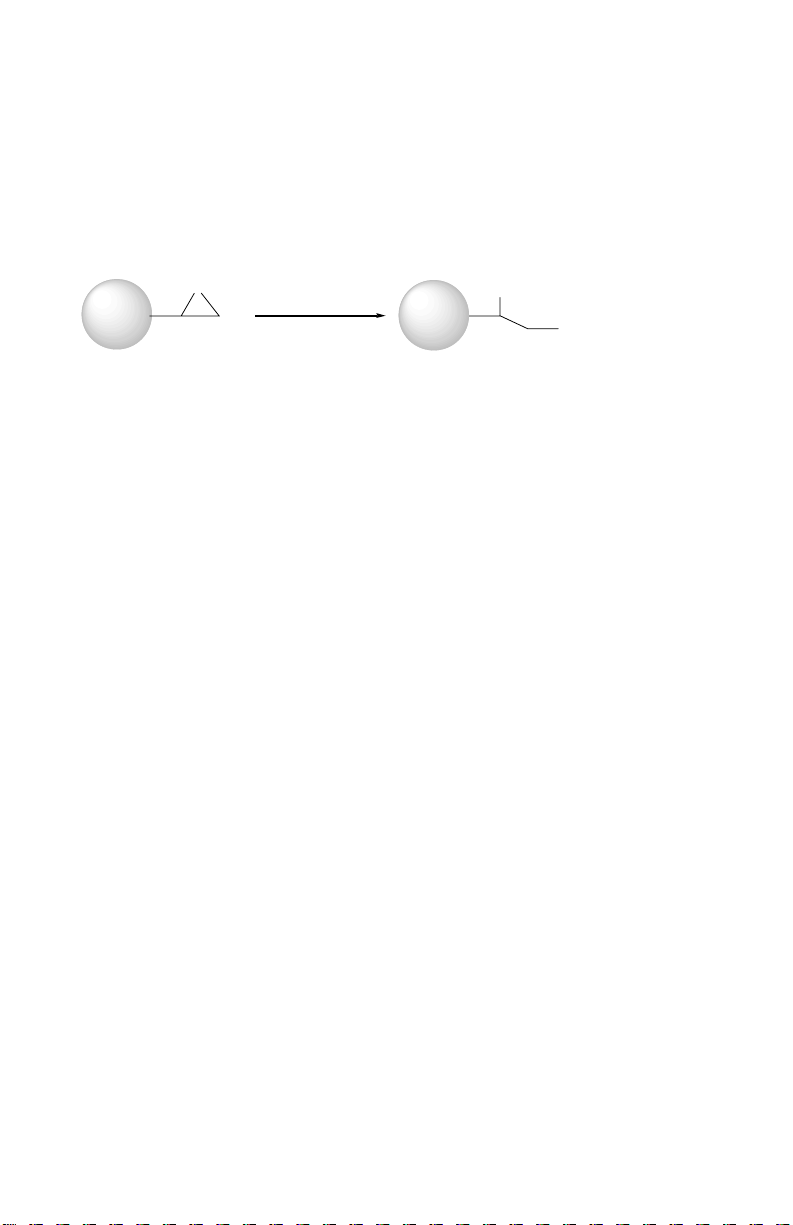
Section 1
UNOsphere
O
OH
NHR (-SR, -OR)
R-NH2, -SH, -OH
UNOsphere
Product Description
Profinity epoxide is an activated macroporous resin for the immobilization of
various ligands of interest. Ligands containing amino, thiol, or hydroxyl groups can
be coupled to Profinity epoxide through an epoxy ring-opening reaction under mild
conditions.
Profinity epoxide is based on Bio-Rad’s proprietary and innovative UNOsphere
™
technology (US patent 6,423,666). Resins made using this technology have
properties of superb mechanical strength, open pore structure, optimized ligand
density, and low, nonspecific binding effects. These unique features of the
UNOsphere base matrix enable ligand-coupled Profinity resin to exhibit excellent
flow properties and to perform separations at very fast flow rates without
compromising protein binding, recovery, or purity. Profinity resin’s open-pore
structure is particularly useful for the purification of large biomolecules.
The base matrix of Profinity epoxide resin is stable across the entire pH range
(1–14) and is compatible with most reagents commonly used in protein
purification, such as denaturing agents, detergents, and reducing agents. It is
amenable to separations under native or denaturing conditions using liquid
chromatographic instrumentation, gravity flow columns, or sample preparation spin
columns.
The resin is supplied dry and is available in 5 g and 25 g quantities.
Note: UNOsphere support, from which Profinity epoxide is derived, was designed
to achieve the highest productivity possible (as measured in grams of target
molecule per operational hour per liter of support). UNOsphere media may be run
at the highest rates and loading capacities while staying within the pressure limits
of the column and chromatography system.
1
Page 4
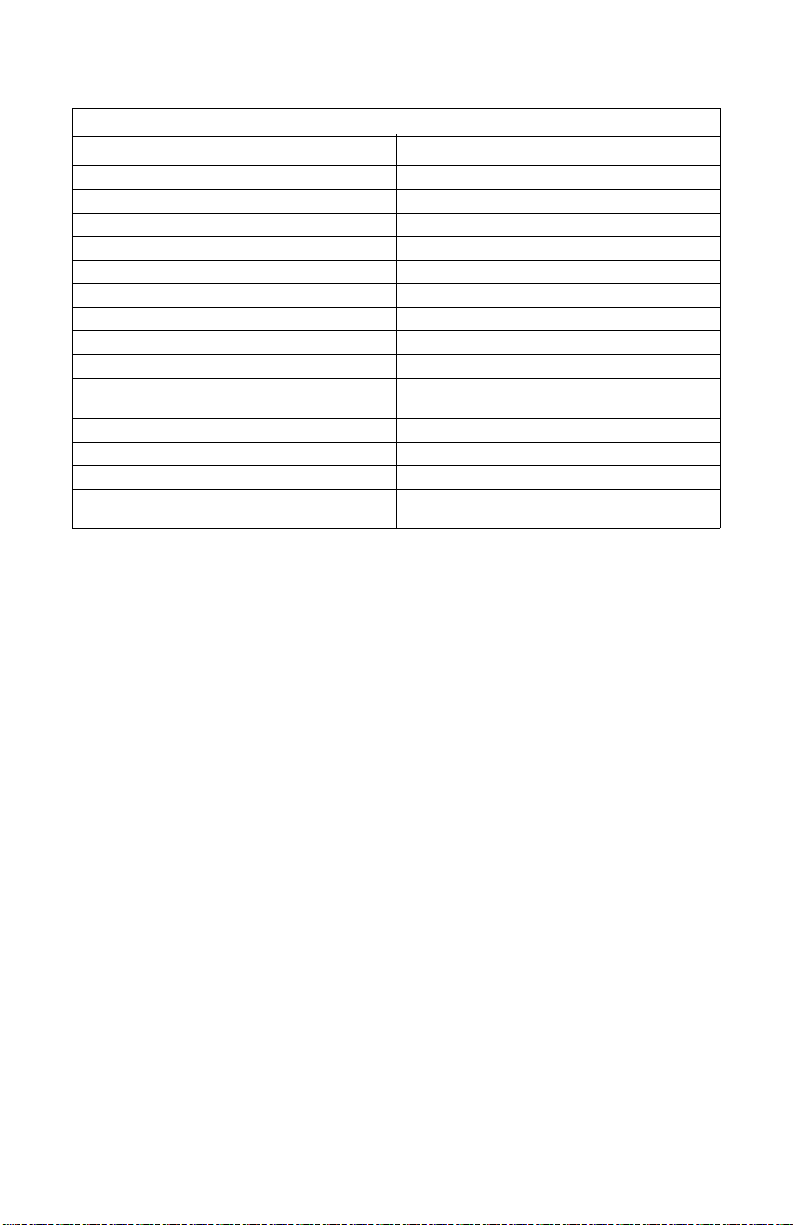
Table 1: Characteristics of Profinity epoxide resin
Profinity epoxide
Functional group Epoxy group
Base matrix UNOsphere base matrix
Form Dry powder
Particle size 45–90 µm
Mean particle size 60 µm
Functional group density 50–132 µmol/g UNOsphere epoxide resin
Swelling factor (ml drained resin/g resin) 5.5–8.0
Recommended linear flow rate <600 cm/hr at 25°C
Maximum operating pressure (net)*
pH stability (base matrix of coupled resin)** 1–14
Chemical compatibility (base matrix
of coupled resin)**
Storage 4°C ambient temperature
Shelf life > 1 year at ambient temperature
Operational temperature 4–40°C
Autoclavability (base matrix
of coupled resin)**
**
Maximum pressure test: Profinity epoxide resin packed in a 1.1 x 30 cm Amicon column to a bed height
of 20 cm with 20 mM sodium phosphate buffer up to 43 psi (3 bar). Flow rates were increased stepwise
to 200 cm/h and held for 2 min at each step. The pressure-low curve for Profinity epoxide becomes
nonlinear at pressures above 80 psi.
****
Refers to base matrix of coupled resin. The stability of coupled ligands may be a limitation of an affinity
resin’s stability.
≥ 80 psi
Compatible with common buffers
and aqueous solutions
0.1 M sodium acetate at 120°C for 30 min
2
Page 5

3
Section 2
General Considerations for Ligand Coupling
Removal of fines
Fine particles in resin may clog the column screen or filter membrane and increase
the column backpressure. Before Profinity epoxide resin is bottled, fines in the
medium have already been removed, so removal of fines is not necessary for most
applications. If necessary for a particular application, the very small amount of
remaining fines may be removed by decanting. Weigh out required amount of dry
resin in a hood (1 g of dry resin gives 5.5–8.0 ml of settled resin bed). Slurry resin
in 3 column volumes (CV) distilled water or buffer by agitation or mixing with a
paddle. Do not use magnetic stirrers, since magnetic stirbars will grind the resin
and more fines will be generated. Let resin settle for 25–40 min, and then carefully
decant the supernatant. The decanting process may be repeated a couple of
times if needed.
General protocols
Profinity epoxide resin can be used for coupling of a variety of ligands, such as
protein A, StrepTactin, subtilisin, and immunoglobulins. Since Profinity epoxide is
based on the UNOsphere platform, it has large pores. The medium’s open pore
structure is particularly useful for coupling large ligands and for purifying large targets. The general ligand coupling protocol is as follows:
1. Weigh out appropriate amount of dry resin (1 g dry resin swells to 5.5–8.0 ml
of settled resin in distilled water or buffers). Swell and wash resin with distilled
water or buffer. Do not use buffers containing Tris, glycine, or thiols since
these nucleophiles will compete with ligands for the resin’s active groups.
Removal of fines is not necessary for most applications. See Removal of fines
above.
2. Add protein ligand (5–20 mg per ml settled resin) to coupling solution. A
buffer: resin ratio of 1:1 to 2:1 is suitable for coupling. Salts such as
ammonium or potassium sulfate may be added to facilitate coupling.
3. Rotate the stoppered vessel containing the ligand and resin at ambient
temperature overnight (avoid using magnetic stirbars for mixing since the
resin’s physical properties will be damaged). Wash away unreacted ligand
using an ample amount of coupling buffer.
4. Collect ligand-coupled resin on a frit. Deactivate or block remaining active
groups by mixing resin cake with 1 M ethanolamine, pH 8–9 for at least
4 hours.
5. Wash the product thoroughly with coupling buffer. Additional wash cycles
alternating acidic and basic buffers are recommended. Each cycle should
consist of a wash of a pH 4.0 buffer (100 mM acetate, 500 mM NaCl)
Page 6

followed by a wash of a pH 8.0 buffer (100 mM phosphate, 500 mM NaCl).
The product can now be used for intended applications or stored in the
presence of a bacteriostat at 4–8°C for future use.
General coupling efficiency considerations
Amount of Ligand Used and Monitoring Amount Coupled
While the amount of ligand coupled is, to a certain extent, proportional to the
amount of ligand added to the coupling solution, the efficiency of ligand coupled
(varying with the ligand and conditions of coupling) will generally taper off at a
certain ligand concentration. In most cases, 5–20 mg of ligand per ml settled resin
is a good starting point to study optimal ligand concentration in a coupling
solution.
Soluble (unbound) ligand remaining in the coupling and wash buffers may be
monitored by measuring OD 280 or by using Bio-Rad protein assay kit II (catalog
#500-0002) or DC™ protein assay kit II (catalog #500-0112).
Coupling Buffers and pH
In order to maintain pH control, a minimum buffer strength of 10 mM is
recommended. Suitable buffers include carbonate, borate, and phosphate. Do not
use buffers such as Tris or glycine. They contain primary amino groups that will
couple to the resin, as will any other compounds containing nucleophiles.
Profinity epoxide couples ligands best at a pH range of 9–13. Often the choice of
coupling pH is limited by the stability of ligands in basic buffers. Profinity epoxide
resin’s epoxy groups react faster with ligands at a higher pH; however, competing
hydrolysis reactions of the resin’s epoxy groups also occur more frequently at
higher pH. Coupling of ligands through their hydroxyl groups requires a pH
around 13.
Coupling Temperature and Time
Coupling at 20°C is recommended for most applications. Carrying out the
coupling reaction at a higher temperature, up to 40°C, is normally faster if the
ligand is stable at the selected temperature. A water bath should be used to raise
the reaction temperature to the desired level.
Coupling ligands at ~20°C overnight is recommended for most applications. A
shorter coupling time is needed if both the pH of the coupling buffer and the
reaction temperature are high. A prolonged coupling reaction sometimes leads to
degradation of ligands and should be avoided.
Deactivation (Blocking) of Remaining Active Groups and Washing of
Ligand-Coupled Resin
Active groups remaining on the resin after coupling need to be deactivated or
blocked to avoid undesirable reaction with proteins of interest during affinity
chromatography. They can be deactivated or blocked by mixing the resin cake
with 1 M ethanolamine, pH 8–9 for at least 4 hr.
4
Page 7

After coupling, ligand-coupled resin needs to be washed thoroughly with coupling
buffer to remove excess ligand, but sometimes small amounts of ligand are still
trapped on the resin through ionic interactions. Additional wash cycles alternating
acidic and basic buffers are recommended. Each cycle should consist of a wash
with a pH 4.0 buffer (100 mM acetate, 500 mM NaCl) followed by a wash with a
pH 8.0 buffer (100 mM phosphate, 500 mM NaCl).
Section 3
Column Packing
General handling
Profinity base resin is a rigid support that can operate under high flow rates and
pressures. However, bead damage due to excessive physical force is possible.
Magnetic stirrers or excessive stirring may cause mechanical damage and fracture
of some beads. Fine particles generated in this manner may clog the column and
increase the column backpressure; in addition, they may enter the pores of intact
beads and reduce their binding capacity.
Small column packing — slurry packing
Slurry packing is the preferred packing method for small columns. Use this
method for packing ligand-coupled Profinity epoxide resin into 5–15 mm ID
columns. For best results, a bed height of 5 to 30 cm should be used. Since the
slurry volume may exceed the column volume, a packing reservoir may be
necessary.
1. Make a 50–70% slurry of ligand-coupled Profinity epoxide resin in degassed
binding buffer of choice. Resuspend the slurry by gently swirling or stirring
with a glass or plastic rod. Do not use a magnetic stirrer.
2. Connect the packing reservoir to the column with the outlet valve closed.
3. Fill the column to about 10% of its volume with buffer. Ensure that the bed
support is fully hydrated and free of any bubbles.
4. Pour the slurry into the reservoir and insert the flow adaptor. Allow buffer to
back-flow out of the adaptor to remove trapped air.
5. To pack the column bed, open the column outlet and pump 5–10 CV of
buffer through the column at 125–200% of the selected operating flow rate.
For optimal performance, pack the column at the maximum flow rate allowed
by the column hardware and resin.
6. Turn off the pump and close the column outlet. Reposition the flow adaptor
firmly against the bed. Additional pumping may be required for the final
adjustment of the flow adaptor.
5
Page 8

Recommended Columns
Bio-Rad’s Bio-Scale
™
MT high-resolution columns may be used for the above
column packing procedure. These columns are empty but may be packed with
the support of choice. Bio-Scale MT columns are convenient for use with
Bio-Rad’s BioLogic
™
system or with any medium- or high-pressure system.
• Bio-Scale MT2 column (7 x 52 mm) for bed volumes up to 2 ml
• Bio-Scale MT5 column (10 x 64 mm) for bed volumes up to 5 ml
• Bio-Scale MT10 column (12 x 88 mm) for bed volumes up to 10 ml
• Bio-Scale MT20 column (15 x 113 mm) for bed volumes up to 20 ml
Column packing — sample preparation sized columns
Use this method for packing Profinity resin into small micro spin columns for
sample preparation (such as Bio-Rad’s Micro Bio-Spin
columns, empty, 100, catalog #732-6204).
1. Make a 50% slurry of ligand-coupled Profinity epoxide resin in degassed
binding buffer of choice. Resuspend the slurry by gently swirling or stirring
with a glass or plastic rod. Do not use a magnetic stirrer.
2. Place the column into an appropriate collection vessel (for example, a 2 ml
capless collection tube), and spin.
3. Using a pipet, transfer enough Profinity resin to a microcentrifuge tube. If
using Bio-Rad’s Micro Bio-Spin column, transfer ~0.2 ml slurried Profinity
resin to a Micro Bio-Spin column. This is equivalent to ~100 µl of a packed
resin bed.
™
chromatography
4. Remove excess binding buffer by centrifugation. Centrifuge at 1,000 x g for
15 sec to pack resin. The column is now ready for separation.
Recommended columns for gravity-flow chromatography
The following Bio-Rad columns may be used for gravity-flow chromatography.
They are empty and can be filled with Profinity resin.
• Poly-Prep
sample
• Econo-Pac
• Glass Econo-Column
diameter
®
columns, for up to 2 ml chromatography support and 10 ml
®
columns, for up to 20 ml
®
columns, from 5–170 cm long and 0.5–5.0 cm in
6
Page 9

Section 4
Protein Binding and Elution
General strategies are listed below. Refer to textbooks (Gagnon 1996, Hermanson
et al. 1992, Matejtschuk 1997) and literature articles for additional information and
guidance.
Binding
Use only the required amount of Profinity support. If excess support is used,
sample elution becomes more difficult because the sample continues to bind and
elute as it passes down the column. Stronger elution conditions become
necessary, residence time is longer, the eluted peak is broader, and there is a
greater risk of denaturation and poor recovery. One method to ensure that only the
required amount of Profinity resin is used is to apply the sample to the top of the
column and elute using reverse flow. Another method is to titrate the resin with
sample, checking the supernatant for unbound sample after each addition.
Continue until the resin is saturated. This method can be used with a small
amount of resin and sample to determine the resin capacity and the amount of
resin required for the purification.
Removal of unbound solutes
Proteins or other solutes that are not bound, or are weakly bound by nonspecific
interactions, must be washed off prior to elution. This can be done by washing
with binding or equilibration buffers, with salts (1 M NaCl), or with detergents
(0.5% Triton X-100). In many cases, the elution buffer can be used, but at a lower
concentration. This frequently neglected wash step eliminates proteins that may
complicate final elution and helps yield a more highly purified product.
Elution
Elution is usually the most demanding step in affinity chromatography. Often the
objective is to obtain high purity and high recovery of a stable and active product.
In attempting to maximize yields, elution conditions that denature the proteins are
often chosen.
Antigens and antibodies are bound to each other by a combination of ionic
bonding, hydrogen bonding, and hydrophobic interactions (Frost et al. 1981). The
strength of different antigen-antibody complexes varies widely. Other parameters
such as ligand density, steric orientation, and nonspecific interactions can be
important.
Acid elution is the most commonly employed desorption method and is
frequently very effective. Eluants such as glycine-HCl (pH 2.5), 20 mM HCl, and
sodium citrate (pH 2.5) can be used to disrupt antigen-antibody interactions. Acid
elution can give low recoveries due to hydrophobic interactions between the
antigen and the antibody. An eluant such as 1 M propionic acid, or the addition of
10% ethylene glycol to an acidic eluant, is more effective in dissociating such
complexes.
7
Page 10

Base elution is less frequently used than acid elution, but, in some cases, it is
more effective. Elution with 1 M NH
effective with membrane glycoproteins and certain antigens that precipitate in acid
but are stable in base (Izuta and Saneyoshi 1988). Organic solvents can also be
added to basic eluants as described above with acid elution.
Chaotropic agents disrupt the tertiary structure of proteins and, therefore, can be
used to dissociate antigen-antibody complexes. Chaotropic salts disrupt ionic
interactions, hydrogen bonding, and sometimes hydrophobic interactions.
Chaotropic anions are effective in the order SCN->ClO
cations are effective in the order guanidine>Mg
(up to 8 M), guanidine-HCl (up to 6 M), and NaSCN (up to 6 M) are effective in
disrupting most protein-protein interactions. However, these strong chaotropic
agents may destroy the activity of the antigen, the antibody, or both. Conditions as
mild as possible should always be used.
It is important to remove the eluted antigen or antibody from eluant as quickly as
possible to minimize the chance of denaturation. If acid or base is used, the
samples should be neutralized immediately following elution. If a chaotropic agent
is used for elution, it can be rapidly removed by desalting (Econo-Pac desalting
columns, Econo-Pac P6 desalting cartridges, Bio-Gel
very small volumes, Micro Bio-Spin columns).
OH or with 50 mM diethylamine, pH 11.5, is
4
->I>Br->Cl-. Chaotropic
4
+>K+
2
>Na+. Eluants such as urea
®
P-6 desalting gel, or for
Section 5
Renaturation of Eluted Proteins
Proteins that have been denatured during elution can often be renatured by the
addition of a chaotropic agent such as guanidine-HCl, followed by stepwise
dialysis against decreasing concentrations of the chaotrope. The high
concentration of guanidine-HCl puts the protein into a random coil configuration.
As the chaotrope is slowly removed, the protein will return to its native form.
Section 6
Assessing Protein Purity
Check the integrity of purified protein by SDS-PAGE analysis (such as on
Criterion
cannot be run directly on these gels due to precipitation of protein. First remove
the guanidine by desalting using Bio-Gel P-6 (or -30) spin columns.
™
or Criterion™XT precast midi gels). Samples containing guanidine-HCl
8
Page 11

9
Section 7
Regeneration and Storage
Regeneration conditions depend on which type of ligand or protein is used in the
coupling and chromatography process. Refer to textbooks and literature articles
for guidance. In general, 2–4 wash cycles alternating acidic and basic buffers are
useful. Each cycle should consist of a wash of a pH 4.0 buffer (100 mM acetate,
500 mM NaCl) followed by a wash of a pH 8.0 buffer (100 mM phosphate,
500 mM NaCl).
Profinity epoxide resin should be stored dry between 4°C and ambient temperature. Ligand-coupled Profinity resin should be stored at 4–8°C in an appropriate
buffer that contains a bacteriostat.
Section 8
References
Frost RG et al., Covalent immobilization of proteins to N-hydroxysuccinimide ester
derivative of agarose. Effect of protein charge on immobilization, Biochim Biophys
Acta 670, 163–169 (1981)
Gagnon P, Purification Tools for Monoclonal Antibodies, Validated Biosystems,
Inc., Tuscon (1996)
Hermanson GT et al., Immobilized Affinity Ligand Techniques, Academic Press,
New York (1992)
Izuta S and Saneyoshi M, AraUTP-Affi-Gel 10: a novel affinity absorbent for the
specific purification of DNA polymerase alpha-primase, Anal Biochem 174,
318–324 (1988)
Matejtschuk P (ed), Affinity Separations — A Practical Approach, Oxford University
Press, Oxford (1997)
Section 9
Ordering Information
Ordering Information
Catalog # Description
156-0200 Profinity Epoxide Resin, 5 g
156-0201 Profinity Epoxide Resin, 25 g
Streptactin is a trademark of Institut für Bioanalytik GmbH. Triton is a trademark of Union Carbide.
Page 12

Bio-Rad Laboratories, Inc.
2000 Alfred Nobel Dr., Hercules, CA 94547 USA
510-741-1000
10003245 Rev A
 Loading...
Loading...