
Ion AmpliSeq™ SARS‑CoV‑2 Research Panel
Instructions for use on the Genexus™ Integrated Sequencer
Pub. No. MAN0019278 Rev. B.0
QUICK REFERENCE
WARNING! Read the Safety Data Sheets (SDSs)
and follow the handling instructions. Wear appropriate
protective eyewear, clothing, and gloves. Safety Data
Sheets (SDSs) are available from thermofisher.com/
support.
This quick reference provides guidelines and instructions for using
the Ion AmpliSeq™ SARS‑CoV‑2 Research Panel to prepare Ion
AmpliSeq™ libraries from SARS-CoV-2 samples and sequence the
libraries on the Genexus™ Integrated Sequencer, then analyze the
sequencing results using Genexus™ Software.
Product description .............................. 1
■
Ordering instructions ............................. 1
■
Isolate and quantify viral RNA ...................... 1
■
Create samples in Genexus™ Software ............... 3
■
Plan a sample run with the Ion AmpliSeq™ SARS‑CoV‑2
■
Research Panel in Genexus™ Software ............... 3
Fill Genexus™ Primer Pool Tubes .................... 3
■
Load the sample plate ............................ 4
■
Start a sequencing run ............................ 4
■
Analyze SARS‑CoV‑2 sequencing results in Genexus
■
Software ....................................... 4
Guidelines for sample quality, viral copy number, and variant
■
calling ......................................... 5
Limited product warranty .......................... 5
■
™
Ordering instructions
To order the Ion AmpliSeq™ SARS‑CoV‑2 Research Panel, follow
these steps.
1. Go to AmpliSeq.com and sign in, or register for a new
account.
2. In the navigation bar, go to the Fixed Panels dropdown
menu, then select Community Panels.
3. In the Research Area navigation pane on the left side of the
screen, select the Infectious Disease checkbox to filter the
list. Find the Ion AmpliSeq™ SARS‑CoV‑2 Research Panel in
the filtered list, then click Preview Order.
Note: Alternatively, enter SARS-CoV-2 in the search field at
the top of the screen to find the panel page.
4. In the Order options window, select Genexus in the
Choose instrument section, then click Next.
5. In the Order summary window, review the order, then
select Proceed to cart. As an option, select the List
recommended consumables checkbox, then click Preview
to see a list of additional products that you may need. Select
the items, then click Add all to cart.
6. Click Proceed to checkout to complete the order at
thermofisher.com.
Unless otherwise indicated, all other materials listed in this
quick reference are available at thermofisher.com.
Isolate and quantify viral RNA
Product description
The Ion AmpliSeq™ SARS‑CoV‑2 Research Panel consists of
two 5X primer pools that target 237 amplicons specific to the
SARS‑CoV‑2 (the virus that causes COVID‑19), and 5 human
expression controls. With an amplicon length range of 125–
275 bp, the panel provides >99% coverage of the SARS‑CoV‑2
genome (~30 kb), and covers all potential serotypes. The panel
is a community Ion AmpliSeq™ panel available for order through
AmpliSeq.com.
When used in conjunction with the Genexus™ Integrated
Sequencer, the Ion AmpliSeq™ SARS‑CoV‑2 Research Panel
oers high sensitivity, high throughput (up to 16 samples per
sample-to-result sequencing run), fast turnaround time, and
minimal hands‑on time in SARS‑CoV‑2 research studies.
Guidelines for RNA isolation and sample normalization
For Research Use Only. Not for use in diagnostic procedures.
• A sample containing as little as 20 copies of viral RNA
(10 copies per target amplification reaction) can be used
to prepare an Ion AmpliSeq™ SARS‑CoV‑2 Research Panel
library. For optimal results, we recommend a viral copy
number in the 200 to 200,000 range, or an amount of
total RNA between 1–10 ng. For more information, see
“Guidelines for sample quality, viral copy number, and variant
calling” on page 5.
• The amount of viral RNA among samples should be
approximately equivalent so that the target amplification
conditions you select are optimal for all samples.
• See “Recommended materials for isolation and
quantification” on page 2 for recommended Thermo Fisher
Scientific kits and master mix.
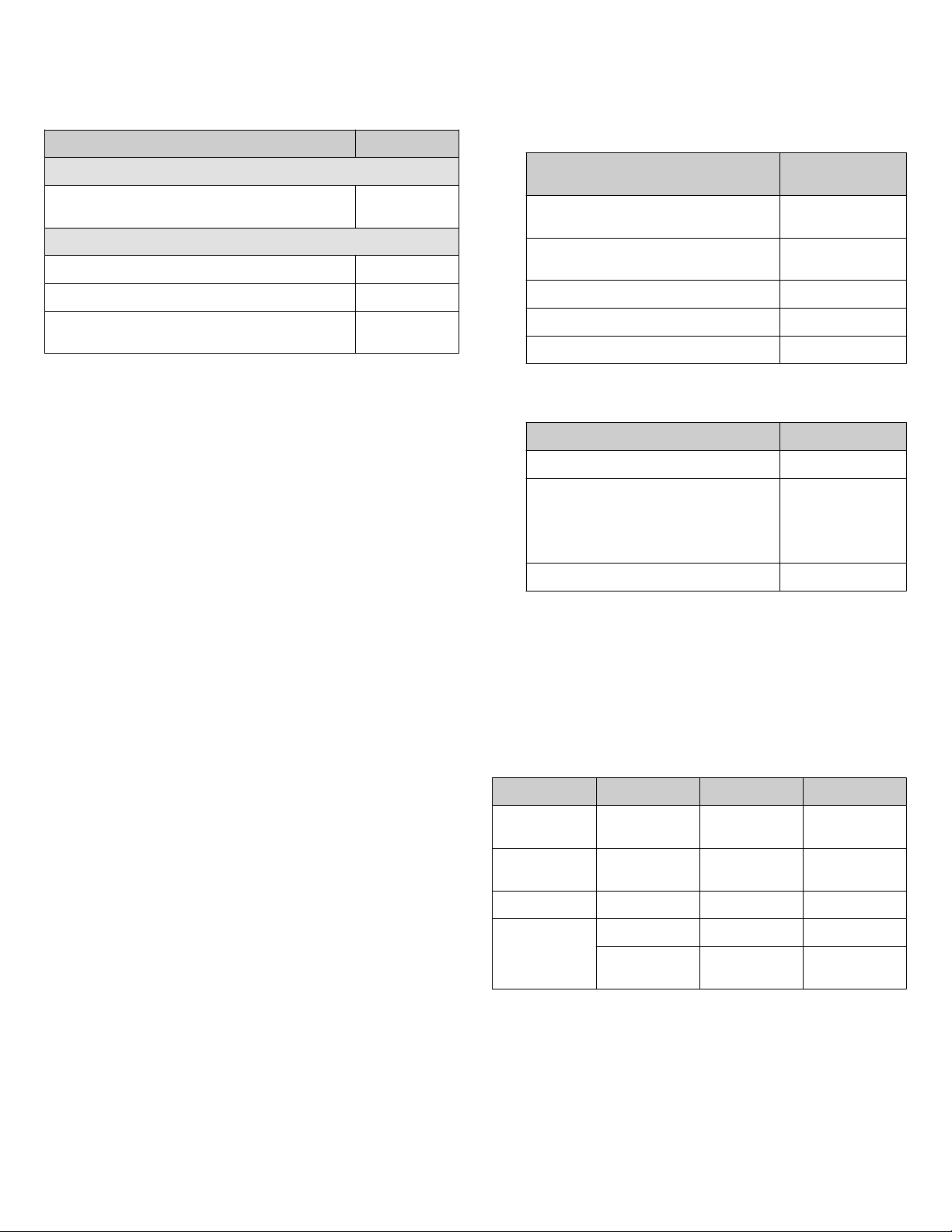
Recommended materials for isolation and quantification
We recommend the following Thermo Fisher Scientific kits and
master mix for the isolation and quantification of SARS‑CoV‑2
RNA.
Item Cat. No.
Isolation
MagMAX™ Viral/Pathogen Nucleic Acid Isolation Kit A42352 or
A48310
Quantification
TaqMan™ 2019-nCoV Assay Kit v1 A47532
TaqMan™ 2019-nCoV Control Kit v1 A47533
TaqPath™ 1-Step RT-qPCR Master Mix, CG A15299 or
A15300
Additional positive controls are available at the BEI Resources
Repository at https://www.beiresources.org, or through other
commercial providers.
Isolate viral RNA
The MagMAX™ Viral/Pathogen Nucleic Acid Isolation Kit can be
used in either a manual or a high-throughput automated mode
using the MagMAX™ Express Magnetic Particle Processor or
KingFisher™ Purification System. Follow these basic steps to
isolate SARS-CoV-2 RNA using the MagMAX™ Viral/Pathogen
Nucleic Acid Isolation Kit (manual extraction). For detailed
information on how to use the kit, and required materials not
supplied, see the following user guides, which are available for
download at thermofisher.com.
• MagMAX™ Viral/Pathogen Nucleic Acid Isolation Kit (manual
extraction) User Guide (Pub. No. MAN0018072) or the
• MagMAX™ Viral/Pathogen Nucleic Acid Isolation Kit
(automated extraction) User Guide (Pub. No. MAN0018073)
1. Digest 200–400 μL of each sample with Proteinase K in
a deep-well 96-well plate, then bind RNA to Nucleic Acid
Binding Beads.
2. Wash the Nucleic Acid Binding Beads.
3. Elute the RNA from the Nucleic Acid Binding Beads.
Use 1–10 ng total RNA in library target amplification reactions.
We recommend quantifying viral copy number by real-time PCR,
described in “Quantify by real-time qPCR”.
Quantify by real-time qPCR
To determine the optimal system-installed SARS‑CoV‑2 assay
parameter set to use for run planning in Genexus™ Software,
quantify viral RNA copy number in samples following these
steps and using the kits and mastermix listed in “Recommended
materials for isolation and quantification”. For more information
about reaction set up, see the TaqMan™ 2019-nCoV Assay Kit v1
Product Information Sheet (Pub. No. MAN0019096).
After you quantify RNA viral copy number, or if you do
not quantify copy number, follow the guidelines for selecting
an assay in step 3 of “Plan a sample run with the Ion
™
AmpliSeq
SARS‑CoV‑2 Research Panel in Genexus™ Software”
on page 3.
1. For each 2019-nCoV qPCR assay (N Protein, S Protein, and
ORF1ab), combine the following components per reaction to
make a reaction mix for the total number of reactions, plus
10% overage.
Component
TaqPath™ 1-Step RT-qPCR Master Mix,
CG (4X)
2019-nCoV assay (20X; N Protein,
S Protein, or ORF1ab)
RNAse P assay (20X) 1.25 µL
RT-PCR Grade Water 11.25 µL
Total reaction mix volume 20.0 µL
Volume per
reaction
6.25 µL
1.25 µL
2. For each reaction, combine the following components in a
MicroAmp™ Optical 96-Well Reaction Plate 0.2‑mL well.
Component
Reaction mix (from step 1) 20.0 µL
• Nucleic acid research sample or
• 1 µL 2019-nCoV Control v1 + 4 µL
RT-PCR Grade Water or
• NTC
Total reaction volume 25.0 µL
Volume per well
5.0 µL
3. Set up and run the reactions on a real-time PCR instrument
using the following settings:
• Analysis method: Comparative C
t
Note: You must use Comparative Ct to analyze 2019-
nCoV assay data using QuantStudio™ Design and
Analysis Software v2 and ExpressionSuite™ Software.
• Cycling mode: Standard
• Thermal cycling protocol:
Stage
Hold
Hold
Hold Activation
Cycling
(40 cycles)
[1]
Heat-labile UNG in TaqPath™ 1-Step RT-qPCR Master Mix, CG is
completely inactivated during the first ramp to 95°C.
[2]
Required for RT inactivation, first denaturation, and activation of the DNA
polymerase.
Step Temperature Time
UNG
incubation
Reverse
transcription
Denaturation 95°C 3 seconds
Extension
[1]
[2]
Anneal/
25°C 2 minutes
50°C 15 minutes
95°C 2 minutes
60°C 30 seconds
Use the Ct result for each 2019-nCoV qPCR assay to estimate
copy number in your sample. See “Copy number determination by
qPCR” on page 3 for example data.
2 Ion AmpliSeq
™
SARS‑CoV‑2 Research Panel on the Genexus™ Integrated Sequencer Quick Reference
 Loading...
Loading...