Page 1

[ method guidelines ]
XBridge™ ost C18 Method guidelines
*ost: oligonuCleotide separation teChnology
XBridge™ OST C18 columns are based on Waters second generation of
hybrid-silica BEH Technology™ particles and can be effectively used
for the lab scale purification and analysis of detritylated synthetic
oligonucleotides using ion-pair, reversed-phase chromatography. This
document provides useful method guidelines for the effective use of
this column chemistry for this group of compounds.
Contents
I. PRINCIPLES OF OLIGONUCLEOTIDE SEPARATIONS
II. SAMPLE PREPARATION
III. RECOMMENDED MOBILE PHASES
IV. RECOMMENDED INJECTOR WASH SOLVENT
V. GENERAL CONSIDERATIONS IN DEVELOPING SEPARATIONS
VI. ANALYSIS OF MODIFIED OLIGONUCLEOTIDES
VII. PURIFICATION CONSIDERATIONS
Page 2
[ method guidelines ]
+
+
+
+
-
-
-
-
-
-
-
-
+
TEA
XBridge™ OST C
18
chain
PO group on Oligo chain
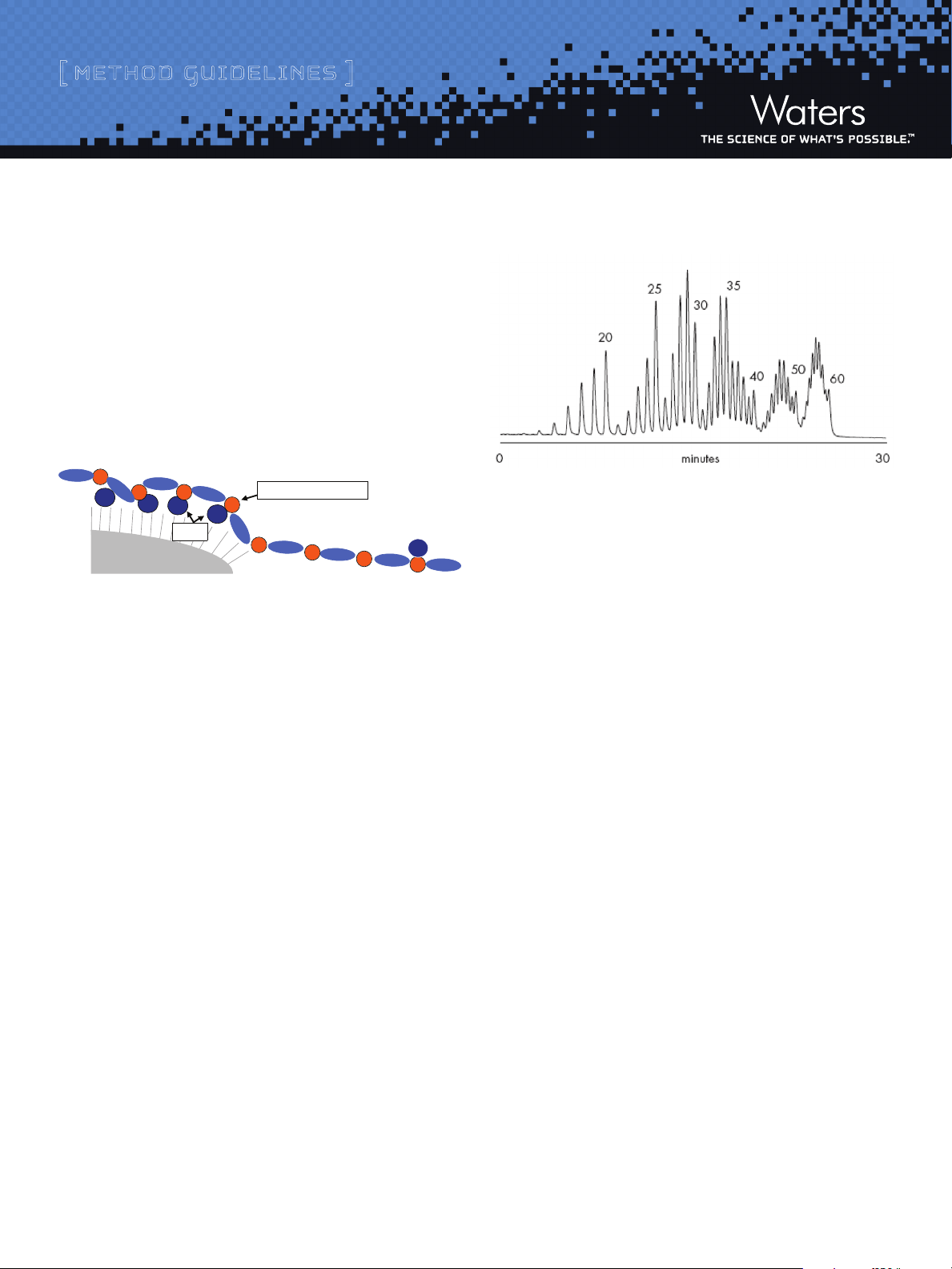
i. p rinCiples o F o l i gonu C l eotide s e parat ions
Separations of detritylated synthetic oligonucleotides on an XBridge™
O ST C18 column are based on ion-pair, reversed-phase chromatographic
principles (IP-RP-LC). As shown in Figure 1, the ion-pairing additive in
the mobile phase is adsorbed on a hydrophobic sorbent and provides
for charge-to-charge interactions with negative charges contained on
the oligonucleotide backbone (e.g., phosphate groups).
Figure 1: Proposed Mechanism of IP-RP-LC for Synthetic Oligonucleotide
Separations
As a result, an efficient charge-based (length-based) oligonucleotide
separation is achieved (Figure 2). Gradient elution using an acetonitrile
or methanol eluent displaces both ion-pairing agent and the oligonucleotides from the sorbent surface.
Figure 2: Separation of a 15 - 60mer Deoxythymidine Ladder on
XBridge™ OST C
18
HPLC system: Waters BioAlliance™ 2796, PDA Detector with micro UV cell
Sample Injected: Approximately 100 pmoles of a detritylated 15 – 60mer
oligonucleotide ladder diluted in
0.1 M TEAA
Column: Waters X Bridge™ OST C18, 2.5 µm (2.1 x 50 mm)
Mobile P hases: A: 0.1 M TEAA,
B: Acetonitrile / 0.1M TEAA, 20/80, v/v
Flow rate: 0.2 mL/min
Column Temp.: 60 ˚C
Gradient delay: 0.45 mL
Gradient: 40 to 62.5% B in 30 minutes (8-12.5% acetonitrile, 0.15%
acetonitrile per minute)
Detection: 260 nm, 5 scans per second
Separation selectivity and resolution decreases with increasing
oligonucleotide length (Figure 2) making the separation of long
oligonucleotides challenging. Modified oligonucleotides such as phosphorothioates and 2-O alkyl modified species are also more difficult
to analyze. Special mobile phase may be required (see Section III,
Recommended Mobile Phases).
Two commonly used ion-pairing agents for oligonucleotide applications
are triethyl ammonium and dimethylbutyl ammonium ions. The final pH of
these mobile phases containing either of these ion-pairing reagents is
adjusted by the addition of Acetic Acid, or in some cases, Hexafluoroisopropanol (HFIP). These mobile phases are volatile making them
suitable for LC-MS applications.
The ability to adequately resolve synthetic oligonucleotide mixtures
by ion-pair, reversed-phase chromatography is significantly affected
by the particle size of the material contained in an efficiently packed
column (see Figure 3). Consequently, XBridge™ OST C18 columns are
efficiently packed with 2.5 micron material to maximize detritylated
oligonucleotide component resolution. In order to improve oligonucleotide
separation efficiency and speed, elevated separation temperature (e.g.
60 ˚C) is recommended. Elevated temperature will also reduce operating
LC System back pressure.
2
Page 3
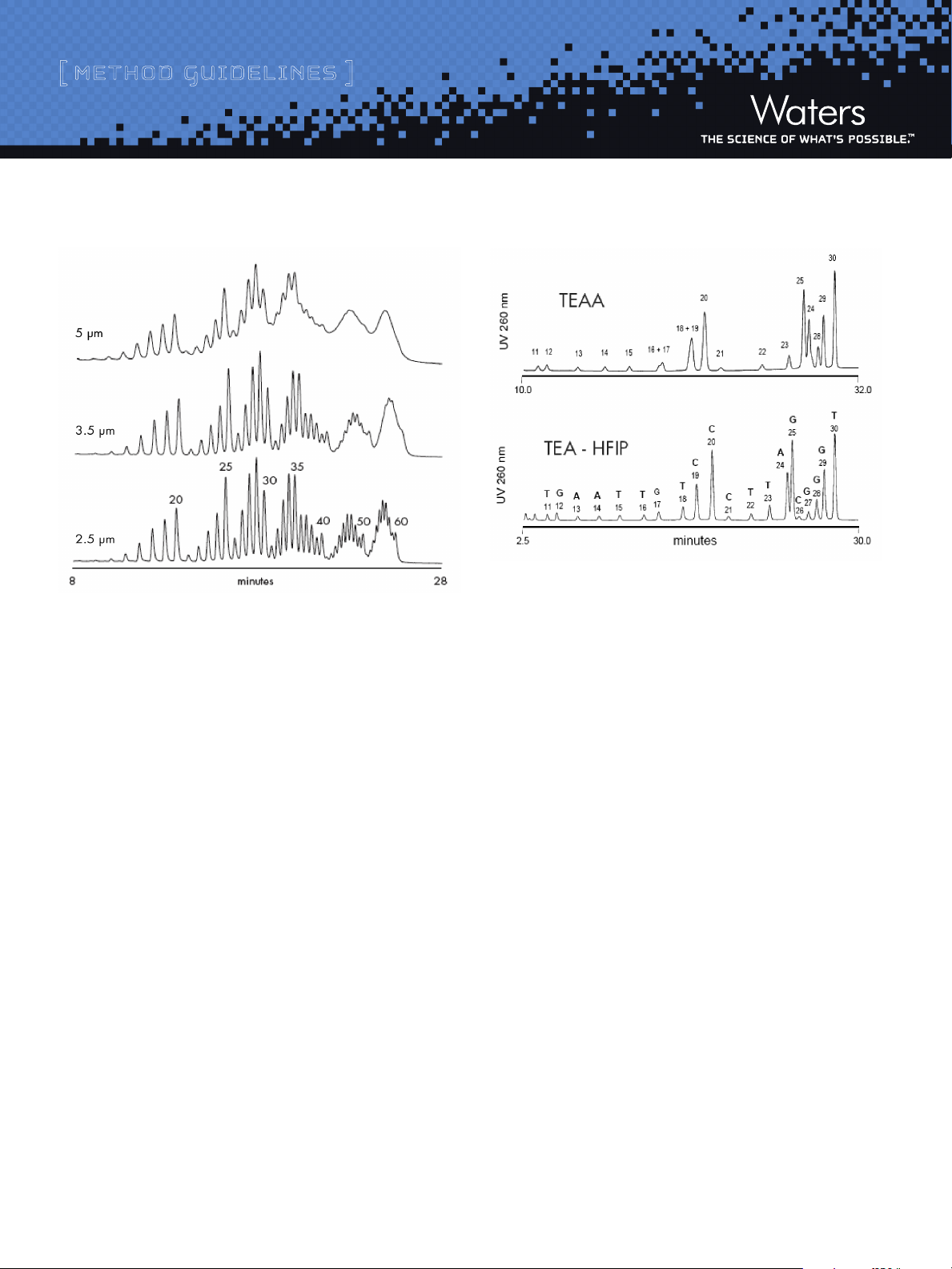
[ method guidelines ]
Figure 3: Effectiveness of Waters BEH Technology™ Hybrid-Silica C18
Particle Size on Deoxythymidine Ladder Separations
HPLC system: Waters BioAlliance™ 2796, PDA Detector with micro UV cell
Sample Injected: Approximately 100 pmoles of detritylated 15 – 60mer crude
oligonucleotide ladder diluted in 0.1 M TEAA
Column: Waters BEH Hybrid-Silica C18 particles (2.1 x 50 mm)
Mobile P hases: A: 0.1 M TEAA,
B: Acetonitrile / 0.1M TEAA, 20/80, v/v
Flow rate: 0.2 mL/min
Column Temp.: 60 ˚C
Gradient delay: 0.45 mL
Gradient: 40 to 62.5% B in 30 minutes (8-12.5% acetonitrile,
0.15% acetonitrile per minute)
Detection: 260 nm, 5 scans per second
In addition to ion-pairing, a hydrophobic reversed-phase mechanism
also takes place in the oligonucleotide separation. The residual interaction
of nucleobases has an impact on overall retention and separation
selectivity, especially when using Triethylammonium Acetate (TEAA)
ion-pairing mobile phases. Separation of N and N-1mers may be either
enhanced or suppressed by the sequence contribution. More potent
ion-pairing systems such as Triethylammonium ion with Hexafluoroisopropanol counter ion provide for more regular “charge-based”
separations (Figure 4).
Figure 4: Impact of Ion-pairing System on Separation of a 10-30mer
Heterooligonucleotide Ladder
HPLC system: Waters BioAlliance™ 2796, PDA Detector with micro UV cell
Sample: 20 mer: TCC C TA GCG T TG AAT TGT CC
25 mer: TC C CTA GCG TTG AAT TGT C CC TTA G
30 mer: TCC CTA GC G TTG AAT TGT CCC TTA GCG GGT
Ladder was prepared by hydrolyzing detritylated
20, 25, and 30mer oligonucleotides with a
3’-exonuclease
Column: Waters X Bridge™ OST C18, 2.5 µm (4.6 x 50 mm)
Mobile p hases: Upper chromatogram: 0.1 M TEAA with acetonitrile gradient; Lower
chromatogram: 16.3 mM T EA - 400 mM HFIP with methanol gradient
Flow rate: 1.0 mL/min
Column Temp.: 60 ˚C
Gradient delay: 0.45 mL
Detection: 260 nm, 5 scans per second
ii. saMple preparation
1. Dissolve the detritylated synthetic oligonucleotide sample in Mobile
Phase A (e.g., 0.1 M TEAA). For example, a 0.05 - 0.2 µmole scale
synthesis can be prepared in 0.1 mL of 0.1 M TEAA. Proportionately
larger or smaller volumes of 0.1M TEAA are required when dissolving
samples from different scale syntheses. Due to the nature of gradient
separations, relatively large volumes of sample (in low organic
strength eluent) can be injected and concentrated onto the head of
the column before beginning the gradient elution program.
2. Samples must be completely in solution and free of particulates
before injecting onto the column. Remove all particles from the
sample (Controlled Pore Glass Synthesis Support, etc.), which may
block the inlet column frit, increase the operating pressure, and
shorten the column life time. Sample contamination with high concentration of salts and/or detergents may also interfere with analysis.
3
Page 4

[ method guidelines ]
3. To remove particulates the sample may be filtered with a 0.2 μm
membrane. Be sure that the selected membrane is compatible and
does not dissolve with the selected Mobile Phase diluent. Contact
the membrane manufacturer with solvent compatibility questions.
An alternative method of particulate removal involves centrifugation
for 20 minutes at 8,000 rpm, followed by the transfer of the supernatant
liquid to an appropriate vial.
iii . reCo M M ended M o Bile p h ases
The most common ion-pair mobile phase for synthetic oligonucleotide
separations is based on Triethylammonium Acetate (TEAA). This
mobile phase can be prepared by titrating Glacial Acetic Acid aqueous
solution with Triethylamine (TEA).
Note: To maximize column life, it is ESSENTIAL that all prepared OST
Mobile Phases be ltered through a solvent compatible, 0.45 µm membrane and contained in bottles that are clean and particulate free.
TEAA
1L of 0.1 M TEAA may be prepared as follows:
advisable to use aqueous buffer consisting of 15 mM TEA and 400
mM HFIP (pH 7.9).
TEA-HFIP System 1
1L of 8.6 mM TEA / 100 mM HFIP is prepared as follows:
1) Perform work in a hood
2) Add 10.4 mL of HFIP (16.8 g) into 988.4 g of water and mix well.
3) Slowly add 1.2 mL of TEA.
4) The pH is approximately 8.3 +/- 0.1.
TEA-HFIP System 2
1 L of 15 mM TEA / 400 mM HFIP is prepared as follows:
1) Perform work in a hood
2) Add 41.56 mL (67.17 g) of HFIP into 956.36 g of water and mix well.
3) Slowly add 2.08 mL (1.52 g) of TEA.
4) The pH of final buffer is approximately 7.9 +/- 0.1.
1) Perform work in a hood.
2) Add 5.6 mL of glacial Acetic Acid into 950 mL of water and mix well.
3) Slowly add 13.86 mL of TEA.
4) The pH should be adjusted to pH 7 +/- 0.5 by careful addition
of Acetic Acid.
5) Adjust final volume to 1 L with water.
Alternatively, premixed TEAA can be used [(e.g., Sigma 1 M TEAA
(part no. 90357)]. Mix 100 mL with 900 mL of water to prepare 1 L of
0.1 M TEAA mobile phase.
Alternative ion-pairing reagents are recommended for improved separation of phosphorothioates or when performing LC-MS analyses. An
ion-pairing mobile phase based on Triethylamine (TEA) and Hexafluoroisopropanol (HFIP) as the buffering acid produces an efficient eluent
system for improved separations involving these application types.
As indicated below, two ion-pairing systems are useful.
For routine detritylated oligonucleotide applications, aqueous buffer
consisting of 8.6 mM TEA and 100 mM HFIP is effective. For applications
such as those involving the separation of G-rich oligonucleotides, it is
iV. reCoMMended inJeCtor Wash solVents
Between analyses, the HPLC system injector seals should be washed. A
90% Water / 10% Acetonitrile injector wash solvent is recommended.
V. general Consideration in deVeloping separations
Separation of detritylated synthetic oligonucleotides by ion-pair,
reversed-phase chromatography uses very shallow gradients. With
both TEAA and TEA-HFIP ion-pairing systems, a rate of strong eluent
change between 0.1-0.25 % Acetonitrile (or Methanol) per minute is
recommended. However, the formation of shallow gradients can place
performance demands on LC pumps and mixers that can compromise
the quality of the separation. Consequently, it is strongly advised
that Mobile Phase B formulation contain a premix blend of aqueous
and organic solvents (e.g., Mobile Phase A= 0.1 M TEAA and Mobile
Phase B = Acetonitrile / 0.1M TEAA, 20/80, v/v) to minimize potentially inadequate solvent mixing that can compromise component
resolution.
4
Page 5

[ method guidelines ]
As illustrated in Figures 5 through 7, these analyses were performed
with the following mobile phases:
Mobile Phase A: 0.1 M TEAA
Mobile Phase B: Acetonitrile (ACN) containing 0.1 M TEAA, 20:80 (v:v)
The 0.1% ACN / min gradient change from an initial 5 to 10% Acetonitrile
concentration over 50 minutes was programmed as specified in Table 1:
Table 1
Time % A % B Actual Acetonitrile (ACN) Concentration
0 min 75 25 5%
50 min 50 50 10%
Example:
For the initial 5% Acetonitrile concentration:
Initial %B = desired ACN % / Volume Fraction of ACN in Mobile Phase
B. So, initial %B = 5% / 0.2 = 25%
For the final 10% Acetonitrile concentration:
The organic solvent concentration at initial sample loading conditions
has to be well chosen. If the initial organic solvent strength is too
high, then some desired oligonucleotide sequences may be unretained.
In the other extreme, when the gradient starts with too low an organic
concentration, the analysis is excessively long without the benefit
of enhanced component resolution. A suitable gradient separation
method can be approximated from the oligonucleotide base (C, G, A,
and T) composition. The initial gradient is typically adjusted while
keeping the gradient slope constant.
Table 2: Suggested Gradient Conditions for Non-Standard Detritylated
Synthetic Oligonucleotide Sequences
Gradient 1
[Stan dard
oligonucleotides (1)]
Initial % ACN 7.0 0 % 5.25% 7.5 0 %
Final % ACN 10.75% 9.00% 12. 50 %
Gradient Length(4) 15 min 15m in 20min
Gradient 2
[Hig h GC content or s hort
oligonucleotides (2)]
Gradient 3
[Hig h AT content or long
oligonucleotides (3)]
1: Standard oligonucleotides: 10 – 30mers
Final %B = desired ACN % / Volume Fraction of ACN in Mobile Phase
B. So, final %B = 10% / 0.2 = 50%
With TEAA mobile phases, the unmodified oligonucleotides elute
within a 7-10 % ACN gradient window. However, C and G rich oligonucleotide sequences are generally less retained (i.e., elute within a
5-8% ACN gradient window) than A and T rich sequences (i.e., elute
within a 8-11% ACN gradient span). When using a shallow gradient,
the total length of analysis for an unknown sample sequence may be
excessive. Use of a fast scouting gradient with a 1% ACN per minute
change is recommended in such cases. Information gathered from this
scouting analysis can then be used to create a more appropriate and
time efficient set of gradient conditions for the particular sample.
Gradient slope has a direct impact on the achievable oligonucleotide
component resolution (along with the type of ion-pairing agent,
sequence, and oligonucleotide modification). Steeper gradients (e.g.,
1% ACN change per minute on a 4.6 x 50mm column at a 1.0 mL/min
flow) are recommended for labeled oligonucleotides or for short, 5-15
mer sequences. Separation of longer sequences are typically performed
using more shallow gradient slopes (e.g. 0.15% ACN change per minute
on a 4.6 x 50mm column at a 1.0 mL/min flow).
2: Short oligonucleotides: Less than 10mer
3: Long oligonucleotides: 30 – 60mers
4: Assuming use of a 2.1 x 50mm XBridge™ OST C18 column at a flow
of 0.2 mL/min and a separation temperature of 60 ˚C.
The retention of single and dual dye-labeled oligonucleotides is dictated
by the nature of label. For example, the retention of 25 mer oligonucleotide increases according to the type of label attached as follows:
no label<6FAM< <TAMRA<TET<HEX<Cy3.
Vi. analysis oF ModiFied oligonuCleotides
XBridge™ OST C18 columns are suitable for analysis of unmodified as
well as modified detritylated oligodeoxyribonucleotides and oligoribonucleotides. Phosphorothioate and 2’-O -alkyl modified oligonucleotides
can also be analyzed with IP-RP-HPLC method. However, these full
length oligonucleotide products are usually more difficult to resolve
from their shorter length failure sequences. The recommended ion-pair
system for phosphorothioate oligonucleotide analysis is TEA-HFIP (see
Recommended mobile phases). An example of a 25mer phosphorothioate
oligonucleotide analysis is shown in Figure 5.
5
Page 6

[ method guidelines ]
Figure 5: Analysis of a Digested 25mer Phosphorothioate Oligonucleotide
HPLC system: Waters BioAlliance™ 2796, PDA Detector with micro UV cell
Sample: Detritylated 25mer phosphorot hioate oligonucleotide mix
(CTC TCG CAC CCA TC T CTC TCC TTC T ) digested with 3’ exonuclease
Column: XBridge™ OST C18, 2.5 µm (2.1 x 50 mm)
Mobile p hase: A: 15 mM TEA with 400 mM HFIP
B: methanol
Flow rate: 0.2 mL/min
Column Temp.: 60 ˚C
Gradient delay: 0 mL
Gradient: 15 to 20% B in 20 minutes (0.25% methanol per minute)
Detection: 260 nm, 2 scans per second
Figure 6: Analysis of a 20mer Oligodeoxythymidine Crude Synthesis Mixture
HPLC system: Waters BioAlliance™ 2796, PDA Detector with micro UV cell
Sample: ~600 pmol of a detritylated 20mer, ~18 pmol of 19mer, ~4.5 pmol
of 17mer was injected on column.
Column: XBridge™ OST C18, 2.5 µm (2.1 x 50mm)
Mobile p hase: A: 0.1 M TE AA,
B: Acetonitrile / 0.1M TEAA, 20/80 (v/v)
Flow rate: 0.2 mL/min
Column Temp: 60 ˚C
Gradient Delay: 0 mL
Gradient: 35 to 50% B in 30 minutes (7-10% acetonitrile)
Detection: 260 nm, 2 scans per second
Peptide nucleic acids (PNA) can also be analyzed using XBridge™ OST C18
columns. The ion-pairing system recommended for analysis of PNA is
similar to those used for peptide analysis (0.1% Trifluoroacetic Acid
or Formic Acid).
UV detection of eluted oligonucleotide peaks is often performed at
260 nm. Injection of 50 pmol of detritylated oligonucleotide sample
on a 2.1 x 50mm XBridge™ OST C18 column yields relatively abundant
peaks. Limits of quantitation (LOQ) vary with the type of oligonucleotide,
LC system and detector; LOQ generated on Waters 2996 PDA detector
equipped with micro UV cell is approximately 1 pmol (2.1 x 50mm
XBridge™ OST C18). The Limit of detection (LOD) estimate is shown in
Figure 6.
Vii. puriFiCation Considerations
XBridge™ OST C18 columns are designed for laboratory scale oligonucleotide purifications and analyses. Sufficient amount of isolated
material suitable for molecular biology and other experiments can
be prepared in a single injection. For example, a 4.6 X 50 mm XBridge™
OST C18 column can suitably purify approximately 20-200 nmoles of
sample in a single injection. It is important to understand that column
overloading results in a peak broadening and that some earlier eluting
impurities may co-elute with the component of interest. With a proper
heart-cutting technique, a good purity of the target oligonucleotide
can be obtained without significant yield sacrifice (Figure 7).
Chromatographers frequently develop a separation on the analytical
scale before moving to preparative work. The steps required to optimize the analytical separation involve:
1) Selecting the appropriate column packing material and mobile phase.
2) Determining the optimal flow rate, gradient during and separation
temperature.
3) Determining the amount of material that can be satisfactorily
loaded and separated on the analytical scale column.
6
Page 7

[ method guidelines ]
Once the separation has been optimized, one begins preparing for the
preparative separation. The steps to successfully scale a separation
from an analytical to a preparative column, containing the same packing
material composition, are detailed below
Step A: Calculate the flow rate for use on the Preparative column.
Prep arative Column Flow Rate =
Analytic al Column Flow Rate x (Diameter of Prep Column)
(Diam eter of Anal Column)
2
2
Step B: To get similar chromatography, the gradient elution profile
should be created on both columns using the same number of column
volumes. When the analytical and preparative columns are of the
same length, as is recommended for this application, then the gradient
duration should be the same.
Note: This assumes use of the same ow rate linear velocity for both
runs as calculated above. For preparative runs, it is also important to
note that an initial gradient delay is required to allow the entire sample
to load onto the head of the column prior to beginning chromatography.
* XBridge™ OST C18 Custom Column
** Values are only approximate and vary depending on detritylated
oligonucleotide length, base composition, and “heart-cutting” fraction
collection method used
Figure 7 shows the separation of 90 nmoles of a detritylated 30 mer
deoxythymidine crude reaction mixture on a 4.6 x 50mm XBridge
OST C18 column. The collection interval is suggested by the lines. Due
to partial column overloading, the N -1, N -2... impurities are partially
displaced and elute earlier than expected. With the proper hearth-cutting
technique, 95-98% purity is typically achieved for 15-35 mer oligonucleotides at this purification scale.
Figure 7: Purication of a Detritylated 30mer Deoxythymidine Sample
™
Step C: The last calculation involves determining how much sample
can be loaded on the preparative column. This calculation compares
the relative volumes of the two columns assuming that both columns
are the same length as recommended for this application.
Preparative Column Sample Load =
Previously Determined Analytical Column Sample Load x (Diameter of Prep Column)2
(Diameter of Anal Column)
2
Table 3: XBridge™ OST C18 Column Selection Guide for Detritylated
Oligonucleotide Purication
Column (mm) Approx Mass Load (µmoles)** Flow Rate (mL/min)
2.1 x 50 0.04 0.2
4.6 x 50 0.20 1.0
10.0 x 50 1.0 0 4.5
19.0 x 50* 4.00 16. 0
30.0 x 50* 9.00 40.0
50.0 x 50* 25.00 110 . 0
HPLC system: Waters BioAlliance™ 2796, PDA Detector with micro UV cell
Sample: Crude detritylated 30mer oligothymidine, 200 nmole dissolved in
100 µl of mobile p hase A, 45 µl was injected on column
Column: XBridge™ OST C18, 2.5 µm (4.6 x 50mm)
Mobile p hase: A: 0.1M TEAA with 400 mM HFIP
B: Acetonitrile/0.1M TEAA, 20/80 (v/v)
Flow rate: 1.0 mL/min
Column Temp.: 60 ˚C
Gradient delay: 0 mL (compensated)
Gradient: 35 to 65% B in 24 minutes (7-13% ACN, 0.25% ACN per minute)
Detection: 260 nm, 2 scans per second
Table 4: Ordering Information
Description Particle Size Pore Size Dimension Part No.
XBridge™ OST C18 2.5 μm 135 Å 2.1 x 50 mm 186003952
XBridge™ OST C18 2.5 μm 135 Å 4.6 x 50 mm 186003953
XBridge™ OST C18 2.5 μm 135 Å 10.0 x 50 mm 186003954
Custom
XBridge™ OST C
18
-- -- -- 186003955
7
Page 8

[ method guidelines ]
sales oFFiC es
Australia
Waters Australia Pty. Limited
Tel: 2 9933 1777
Fax: 2 9898 1455
Austria and European Export
(Central South Eastern Europe,
CIS and Middle East)
Tel: 431 877 18 07
Fax: 431 877 18 08
Belgium
Waters S.A.-N.V.
Tel: 32 2 726 1000
Fax: 32 2 726 1100
Brazil
Waters Technology
Tel: 55 11 5094 3788
Fax: 55 11 5093 6413
Canada
Waters Limited
Tel: 800 252 4752
Fax: 905 678 9237
China
Waters Technologies
(Shanghai) Limited
Tel: 8621 6495 6999
Fax: 8621 6495 1999
CIS/Russia
Tel: +7 495 727 4490
Fax: +7 495 336 7000
Czech Republic
Tel: 42 02 617 11384
Fax: 42 02 617 11386
Denmark
Waters A/S
Tel: 45 46 59 8080
Fax: 45 46 59 8585
Finland
Waters Finland
Tel: (09) 5659 6288
Fax: (09) 5659 6282
France
Waters S.A
Tel: (33) 1 30 48 72 00
Fax: (33) 1 30 48 72 01
Germany
Waters GmbH
Tel: 49 6196 400600
Fax: 49 6196 4006010
Hungary
Tel: 36 1 350 5086
Fax: 36 1 350 5087
Hong Kong
Waters China Ltd.
Tel: 852 29 64 1800
Fax: 852 25 49 6802
India
Waters India Pvt. Ltd.
Tel: 91 80 2 837 1900
Fax: 91 80 2 839 2157
Ireland
Waters Chromatography
Ireland Ltd.
Tel: 353 1 448 1500
Fax: 353 1 448 1510
Italy
Waters S.P.A.
Tel: 39 02 274 211
Fax: 39 02 250 1827
Japan
Nihon Waters K.K.
Tel: (81) 3 3471 7191
Fax: (81) 3 3471 7118
Korea
Waters Korea Limited,
Tel: (82) 2 820 2700
Fax: (82) 2 820 2730
Mexico
Waters S.A. DE C.V.
Tel: 5255 5200 1860
Fax: 5255 5524 9376
The Netherlands
Waters Chromatography B.V.
Tel: +31 (0)76-50 87 200
Fax: +31 (0)76-50 87 280
Norway
Waters AS
Tel: 47 63 84 60 50
Fax: 47 63 84 60 51
Poland
Tel: (48) 22 833 4400
Fax: (48) 22 833 0987
Puerto Rico
Waters Technologies Corporation
Tel: 787 747 8445
Fax: 787 747 8448
Singapore
Waters Asia Ltd.
Tel: 65 6273 1221
Fax: 65 6273 6116
Spain
Waters Cromatografía, S.A.
Tel: 34 93 600 93 00
Fax: 34 93 600 93 60
Sweden
Waters Sverige AB
Tel: 46 8 555 11500
Fax: 46 8 555 11550
Switzerland
Waters AG
Tel: 41 62 889 2030
Fax: 41 62 889 2059
Taiwan
Tel: 886 2 2543 1898
Fax: 886 2 2543 1918
United Kingdom
Waters U.K. Ltd.
Tel: 44 208 238 6100
Fax: 44 208 207 7070
All Other Countries
Waters Corporation
34 Maple Street
Milford, MA 01757 USA
Tel: 508 478 2000
800 252 4752
Fax: 508 872 1990
http://www.waters.com
Waters, XBridge, BEH Technology and BioAlliance are trademarks of
Waters Corporation.
©2008 Waters Corporation.
April 2008 715001476 Rev B SC
Waters Corporation
34 Maple Street
Milford, MA 01757 U.S.A.
T: 1 508 478 2000
F: 1 508 872 1990
www.waters.com
 Loading...
Loading...