Page 1

[ CARE AND USE MANUAL ]
Hi3 STANDARDS
I. INTRODUCTION
The Hi3 Ecoli and Phos B standards are a specialized set of 6
synthetically prepared and highly purified peptides that provide
the top 6 ionizing peptides of both the E.coli ClpB and rabbit
Phosphorylase B proteins. These standards are quantitated via
AAA to equimolar ratios to provide greater accuracy to each
analysis. They are greater than 95% pure and have been verified
by mass identification. In addition, each vial is in a Waters Max
Recovery vial 186000327C which simplifies the sample
preparation process.
These standards are primarily intended for use with the Hi3
quantification method for MSE proteomics data processed with
ProteinLynx Global Server™. The E. coli standard is intended for
samples of animal origin, and the Phos B standard is intended for
samples of microbial origin. T he standards may also be of use in the
evaluation and benchmarking of proteomic LC/MS systems comprised
of nanoACQUITY UPLC® and Synapt® and Xevo® time-of-flight
mass spectrometers.
Table 1: Hi3 Phos B Standard (P/N 186006011)
Average
Peptide Sequence
Phos B
Sequence 1
Phos B
Sequence 2
Phos B
Sequence 3
Phos B
Sequence 4
Phos B
Sequence 5
Phos B
Sequence 6
The cysteine in sequence 2 is carbamidomethylated.
VLYPNDNFFEGK 1442.5913 1nmol/vial
TC*AYTNHTVLPEALER 1875.0931 1nmol/vial
IGEEYISDLDQLRK 1678.8602 1nmol/vial
LLSYVDDEAFIR 1440.6164 1nmol/vial
LITAIGDVVNHDPVVGDR 1890.1273 1nmol/vial
VFADYEEYVK 1262.3817 1nmol/vial
Molecular
Weight
Concentration
Table 2: Hi3 EColi Standard (P/N: 186006012)
Average
Peptide Sequence
Ecoli ClpB
Sequence 1
Ecoli ClpB
Sequence 2
Ecoli ClpB
Sequence 3
Ecoli ClpB
Sequence 4
Ecoli ClpB
Sequence 5
Ecoli ClpB
Sequence 6
VIGQNEAVDAVSNAIR 1655.8292 1nmol/vial
NNPVLIGEPGVGK 1293.4863 1nmol/vial
AIDLIDEAASSIR 1373.5266 1nmol/vial
VTDAEIAEVLAR 1286.4484 1nmol/vial
AIQQQIENPLAQQILSGELVPGK 2474.8412 1nmol/vial
LPQVEGTGGDVQPSQDLVR 1995.1776 1nmol/vial
Molecular
Weight
Concentration
II. STORAGE AND STABILITY
Lyophilized peptides generally have excellent stabilities, often
showing little or no degradation after a few years at -20 °C. Long
term storage (>1 year) should be at -80 °C desiccated, medium
term storage (1-12 months) should be at -20 °C desiccated, short
term storage (<1 month) may be at 4 °C. Once reconstituted the
solution should be used immediately to avoid degradation of
peptides which would compromise the quantitative benefit.
III. RECONSTITUTION OF THE
Hi3 DIGESTION STANDARDS
The Hi3 Digestion Standards contain 1 nmol of each of the peptides
listed in Table 1 or 2. It is recommended that 100 µL of 0.5%
trifluoroacetic acid in 3% acetonitrile/water be added to a vial of
standard to obtain a 10 pmol/µL stock solution. Because peptides can
adhere to glass and other surfaces resulting in significant losses from
highly dilute solutions, further dilutions should be made just prior to
addition to samples and excess diluted standard should be discarded.
Unused stock solutions may be stored in a freezer for a brief time.
Analysis of small amounts of standard (ca. 50 fmol) should only be
carried out in the presence of a sample matrix such as E. coli digest
of a serum digest (60 ng/µL or greater).
Hi3 Standards 1
Page 2
[ CARE AND USE MANUAL ]
IV. USING THE Hi 3 DIGESTION STANDARDS
FOR QUANTIFICATION
For a typical proteomic analysis carried out with 75 µ i.d. UPLC®
Columns, sufficient Hi3 standard should be added to each sample to
provide ca. 50 fmol of standard per injection. The injected quantity
should be scaled for other column diameters. LC/MSE data should be
obtained in the normal manner.
For data processing, first append the appropriate protein sequence
below to the databank to be searched for protein identification
using a text editor.
>CLPB_ECOLI (P03815) ClpB protein (Heat shock protein F84.1)
MRLDRLTNKFQLALADAQSLALGHDNQFIEPLHLMSALLNQEGGSVS
PLLTSAGINAGQLRTDINQALNRLPQVEGTGGDVQPSQDLVRVLNLCDK
LAQKRGDNFISSELFVLAALESRGTLADILKAAGATTANITQAIEQMRG
GESVNDQGAEDQRQALKKYTIDLTERAEQGKLDPVIGRDEEIRRTIQVL
QRRTKNNPVLIGEPGVGKTAIVEGLAQRIINGEVPEGLKGRRVLALDMG
ALVAGAKYRGEFEERLKGVLNDLAKQEGNVILFIDELHTMVGAGKADG
AMDAGNMLKPALARGELHCVGATTLDEYRQYIEKDAALERRFQKVFV
AEPSVEDTIAILRGLKERYELHHHVQITDPAIVAAATLSHRYIADRQLP
DKAIDLIDEAASSIRMQIDSKPEELDRLDRRIIQLKLEQQALMKESDEAS
KKRLDMLNEELSDKERQYSELEEEWKAEKASLSGTQTIKAELEQAKIAI
EQARRVGDLARMSELQYGKIPELEKQLEAATQLEGKTMRLLRNKVTDA
EIAEVLARWTGIPVSRMMESEREKLLRMEQELHHRVIGQNEAVDAVSN
AIRRSRAGLADPNRPIGSFLFLGPTGVGKTELCKALANFMFDSDEAMV
RIDMSEFMEKHSVSRLVGAPPGYVGYEEGGYLTEAVRRRPYSVILLDEV
EKAHPDVFNILLQVLDDGRLTDGQGRTVDFRNT VVIMTSNLGSDLIQER
FGELDYAHMKELVLGVVSHNFRPEFINRIDEVVVFHPLGEQHIASIAQIQ
LKRLYKRLEERGYEIHISDEALKLLSENGYDPVYGARPLKRAIQQQIENP
LAQQILSGELVPGKVIRLEVNEDRIVAVQ
ELEPHKFQNKTNGITPRRWLVLCNPGLAEIIAERIGEEYISDLDQLRKLLS
YVDDEAFIRDVAKVKQENKLKFAAYLEREYKVHINPNSLFDVQVKRIH
EYKRQLLNCLHVITLYNRIKKEPNKFVVPRTVMIGGKAAPGYHMAKMII
KLITAIGDVVNHDPVVGDRLRVIFLENYRVSLAEKVIPAADLSEQISTAG
TEASGTGNMKFMLNGALTIGTMDGANVEMAEEAGEENFFIFGMRVEDV
DRLDQRGYNAQEYYDRIPELRQIIEQLSSGFFSPKQPDLFKDIVNMLMH
HDRFKVFADYEEYVKCQERVSALYKNPREWTRMVIRNIATSGKFSSDR
TIAQYAREIWGVEPSRQRLPAPDEKIP
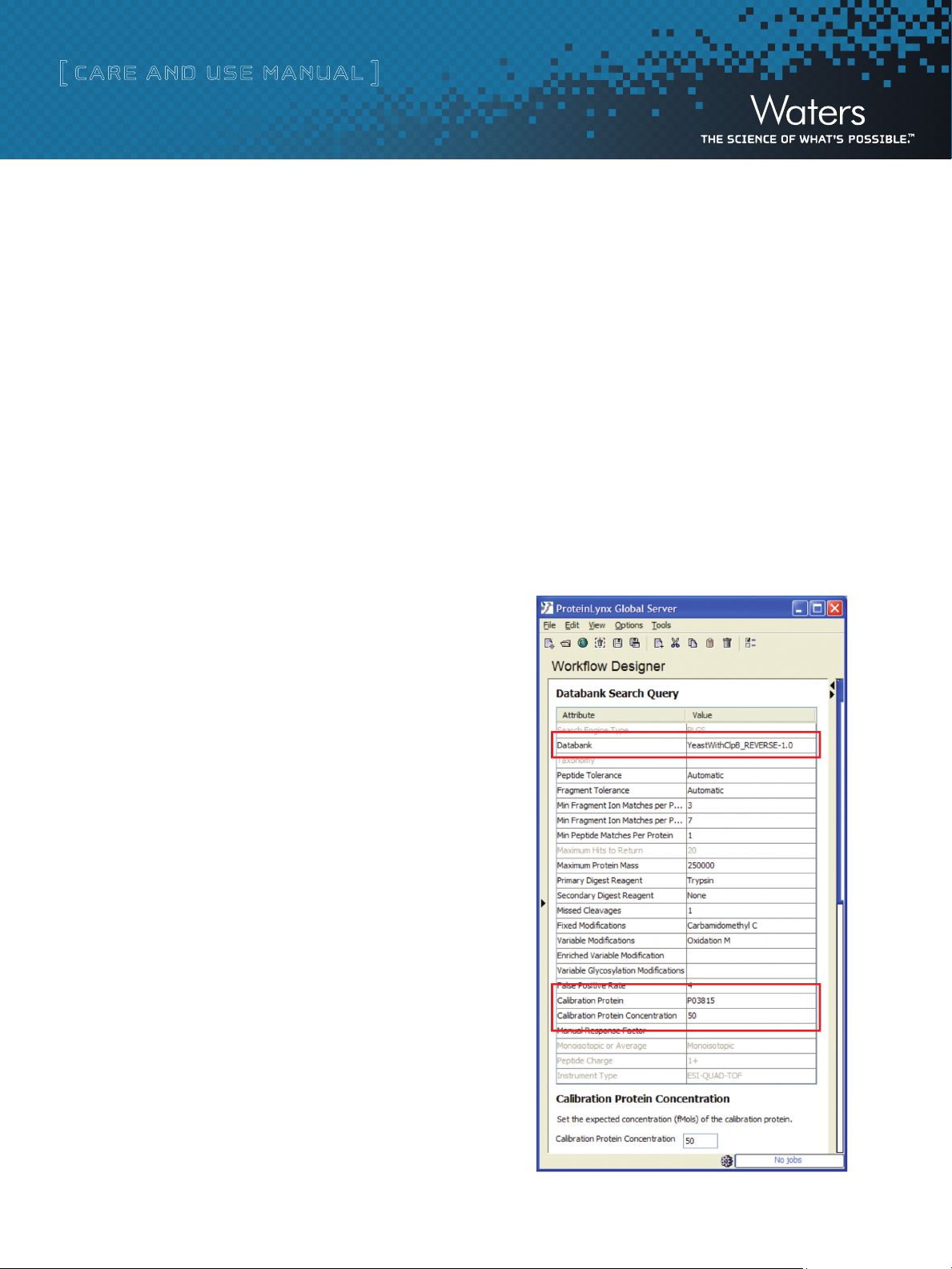
Configure the modified databank with the PLGS Databank Admin Tool.
Create a search workflow using the PLGS Workflow Designer,
being sure that the modified databank is selected, the proper
identifier for the standard protein is entered on the Calibration
Protein line and the number of femtomoles injected is entered
on the Calibration Protein Concentration line, as below:
>PHS2_RABIT (P00489) GLYCOGEN PHOSPHORYLASE, MUSCLE
FORM (ECSRPLSDQEKRKQISVRGLAGVENVTELKKNFNRHLHFTLVK
DRNVATPRDYYFALAHTVRDHLVGRWIRTQQHYYEKDPKRIYYLSLE
FYMGRTLQNTMVNLALENACDEATYQLGLDMEELEEIEEDAGLGNGGL
GRLAACFLDSMATLGLAAYGYGIRYEFGIFNQKICGGWQMEEADDWL
RYGNPWEKARPEFTLPVHFYGRVEHTSQGAKWVDTQVVLAMPYDTP
VPGYRNNVVNTMRLWSAKAPNDFNLKDFNVGGYIQAVLDRNLAENIS
RVLYPNDNFFEGKELRLKQEYFVVAATLQDIIRRFKSSKFGCRDPVRTN
FDAFPDKVAIQLNDTHPSLAIPELMRVLVDLERLDWDKAWEVTVKTCA
YTNHTVLPEALERWPVHLLETLLPRHLQIIYEINQRFLNRVAAAFPGDV
DRLRRMSLVEEGAVKRINMAHLCIAGSHAVNGVARIHSEILKKTIFKDFY
Hi3 Standards 2
Page 3
[ CARE AND USE MANUAL ]
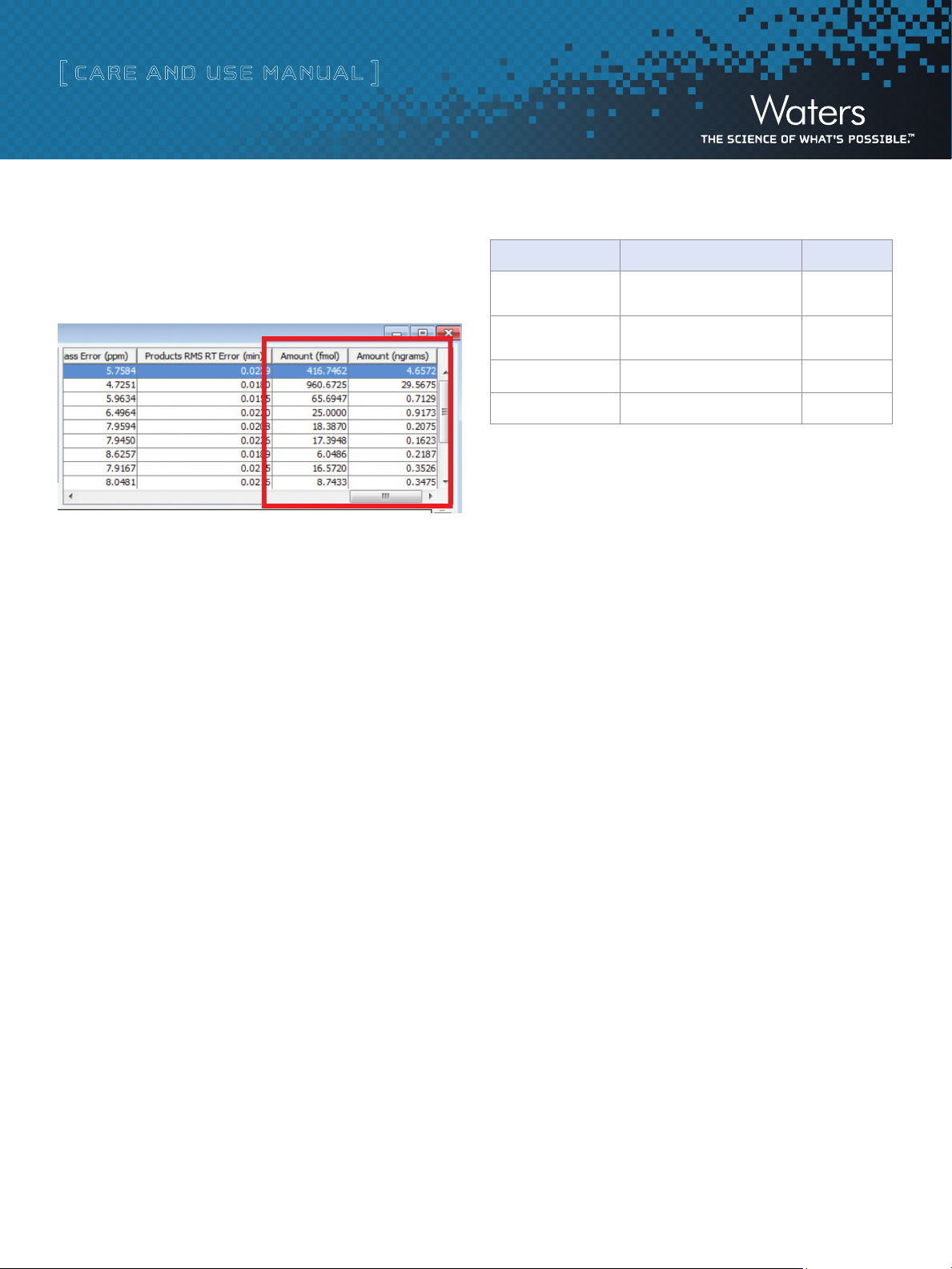
Process and search the data in the normal fashion. Calculated
quantities for each identified protein (in fmol and ng) will be
found in the rightmost two columns of the protein results table
in the search result window (below):
Refer to IdentityE and ProteinLynx Global Server documentation
for further information.
MSDS information: The Hi3 standards contain various non-
hazardous peptides in powder form. They are for laboratory use only;
solely for purposes of scientific experimentation, analysis, research
or development. T hese standards should be used only as directed
and in accordance with good laboratory practice. T hese standards
are non hazardous as defined by OSHA’s Hazard Communication
Standard, 29 CFR 1910.1200(c) and Appendix A and B, and are
thus exempt from the requirements of the Hazard Communication
Standard, including the requirement to supply MSDSs.
V. ORDERING INFORMATION
Part Number Product Description Qty
186002326
186003196
186006011 Hi3 Phos B Standard 1/pk
186006012 Hi3 Ecoli Standard 1/pk
MassPREP™ Digestion Standard,
Phosphorylase B
MassPREP Ecoli Digestion
Standard
1/pk
1/pk
Hi3 Standards 3
Page 4

[ CARE AND USE MANUAL ]
Austria and European Export
(Central South Eastern Europe, CIS
and Middle East) 43 1 877 18 07
Australia 61 2 9933 1777
Belgium 32 2 726 1000
Brazil 55 11 5094-3788
Canada 1 800 252 4752 x2205
China 86 21 6879 5888
CIS/Russia +497 727 4490/290 9737
Czech Republic 420 2 617 1 1384
Denmark 45 46 59 8080
Finland 09 5659 6288
France 33 1 30 48 72 00
Germany 49 6196 400600
Hong Kong 852 2964 1800
The Netherlands 31 76 508 7200
Norway 47 6 384 60 50
Poland 48 22 6393000
Puerto Rico 1 787 747 8445
Singapore 86 21 6879 5888
Spain 34 936 009 300
Sweden 46 8 555 11 500
Switzerland 41 56 676 70 00
Taiwan 886 2 2543 1898
United Kingdom 44 208 238 6100
All other countries:
Waters Corporation U.S.A.
1 508 478 2000
1 800 252 4752
www.waters.com
Hungary 36 1 350 5086
India and India Subcontinent
91 80 2837 1900
Ireland 353 1 448 1500
Italy 39 02 265 0983
Japan 81 3 3471 7191
Korea 82 2 6300 4800
Mexico 52 55 5524 7636
©2012 Waters C orporation. Waters, UP LC, nanoAC QUITY
UPLC, Synapt, and Xevo are registered trademarks of Waters
Corporation. The Science of What's Possible, MassP rep and
ProteinLynx G lobal Server are trademarks of Waters Corporation.
All other trademarks are the property of their respective owners.
March 2012 720004332EN I H-PDF
Waters Corporation
34 Maple Street
Milford, MA 01757 U.S.A.
T: 1 508 478 2000
F: 1 508 872 1990
www.waters.com
Hi3 Standards 4
 Loading...
Loading...