Page 1

NanoDrop Micro-UV/Vis Spectrophotometer
NanoDrop One
User Guide
269-309102 NanoDrop One UG Revision C March 2021
Page 2

© 2020 Thermo Fisher Scientific Inc. All rights reserved.
Wi-Fi is either a trademark or a registered trademark of Wi-Fi Alliance in the United States and/or other countries.
Bluetooth is either a trademark or a registered trademark of Bluetooth Special Interest Group. Windows is either a
trademark or a registered trademark of Microsoft Corporation in the United States and/or other countries. All other
trademarks are the property of Thermo Fisher Scientific Inc. and its subsidiaries.
For U.S. Technical Support, please contact:
Thermo Fisher Scientific
3411 Silverside Road
Tatnall Building, Suite 100
Wilmington, DE 19810 U.S.A.
Telephone: 302 479 7707
Toll Free: 1 877 724 7690 (U.S. & Canada only)
For International Support, please contact:
http://www.thermofisher.com/
NanoDropSupport
Contact your local distributor. For contact
information go to:
http://www.thermofisher.com/
NanoDropDistributors
E-mail: nanodrop@thermofisher.com
Thermo Fisher Scientific Inc. provides this document to its customers with a product purchase to
use in the product operation. This document is copyright protected and any reproduction of the
whole or any part of this document is strictly prohibited, except with the written authorization of
Thermo Fisher Scientific Inc.
The contents of this document are subject to change without notice. All technical information in this
document is for reference purposes only. System configurations and specifications in this document
supersede all previous information received by the purchaser.
Thermo Fisher Scientific Inc. makes no representations that this document is complete, accurate or
error-free and assumes no responsibility and will not be liable for any errors, omissions, damage or
loss that might result from any use of this document, even if the information in the document is
followed properly.
This document is not part of any sales contract between Thermo Fisher Scientific Inc. and a
purchaser. This document shall in no way govern or modify any Terms and Conditions of Sale,
which Terms and Conditions of Sale shall govern all conflicting information between the two
documents.
For Research Use Only. This instrument or accessory is not a medical device and is not intended to
be used for the prevention, diagnosis, treatment or cure of disease.
WARNING Avoid an explosion or fire hazard. This instrument or accessory is not
designed for use in an explosive atmosphere.
Page 3

Contents
Chapter 1 About the Spectrophotometer . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .9
C
Features . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 10
Touchscreen . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .10
Cuvette Holder. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .11
USB-A port . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .11
Accessories. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 12
DYMO™ LabelWriter™ 450 USB Label Printer . . . . . . . . . . . . . . . . .12
PR-1 Pedestal Reconditioning Kit . . . . . . . . . . . . . . . . . . . . . . . . . . .12
PV-1 Performance Verification Solution . . . . . . . . . . . . . . . . . . . . . . . 12
Instrument Detection Limits . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .13
Chapter 2 Instrument Set up. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .15
Register Your Instrument . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 15
Update Software . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .15
Setting Up User Account Control (Optional). . . . . . . . . . . . . . . . . . . . . .16
User Account Control . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .16
Security Administration Policies . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 17
Technical Support . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .19
For U.S./Canada Support, please contact: . . . . . . . . . . . . . . . . . . . .19
For International Support, please contact: . . . . . . . . . . . . . . . . . . . . .19
Chapter 3 Application Measurement Ranges . . . . . . . . . . . . . . . . . . . . . . . . . .21
Detection Limits for All Applications . . . . . . . . . . . . . . . . . . . . . . . . . . . .21
Detection limits for standard applications . . . . . . . . . . . . . . . . . . . . . .21
Detection limits for pre-defined dyes . . . . . . . . . . . . . . . . . . . . . . . . .23
Chapter 4 Nucleic Acid Applications . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .25
Measure dsDNA, ssDNA or RNA. . . . . . . . . . . . . . . . . . . . . . . . . . . . . .25
Measure dsDNA, ssDNA or RNA . . . . . . . . . . . . . . . . . . . . . . . . . . . .25
Best practices for nucleic acid measurements . . . . . . . . . . . . . . . . . .27
Nucleic Acid Reported Results. . . . . . . . . . . . . . . . . . . . . . . . . . . . . .29
Settings for Nucleic Acid Measurements . . . . . . . . . . . . . . . . . . . . . . 32
Calculations for Nucleic Acid Measurements . . . . . . . . . . . . . . . . . . .32
Thermo Scientific NanoDrop One User Guide iii
Page 4

Contents
Measure Microarray. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 35
Measure Microarray Samples. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 35
Microarray Reported Results . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 38
Settings for Microarray Measurements. . . . . . . . . . . . . . . . . . . . . . . .41
Calculations for Microarray Measurements. . . . . . . . . . . . . . . . . . . . . 45
Measure using a Custom Factor . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .49
Measure Nucleic Acid using a Custom Factor . . . . . . . . . . . . . . . . . . 49
Custom Factor Reported Results. . . . . . . . . . . . . . . . . . . . . . . . . . . .51
Settings for Nucleic Acid Measurements using a Custom Factor . . . .52
Detection Limits for Nucleic Acid Measurements using a Custom
Factor . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .53
Measure Oligo DNA or Oligo RNA . . . . . . . . . . . . . . . . . . . . . . . . . . . . .55
Measure Oligo DNA or Oligo RNA . . . . . . . . . . . . . . . . . . . . . . . . . . .55
Oligo Reported Results . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 58
Settings for Oligo DNA and Oligo RNA Measurements. . . . . . . . . . . .60
Detection Limits for Oligo DNA and Oligo RNA Measurements. . . . . . 62
Calculations for Oligo DNA and Oligo RNA Measurements. . . . . . . . . 63
Chapter 5 Protein Applications. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 67
Measure Protein A280. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .67
Measure Protein Concentration at A280 . . . . . . . . . . . . . . . . . . . . . .67
Best practices for protein measurements. . . . . . . . . . . . . . . . . . . . . .69
Protein A280 Reported Results . . . . . . . . . . . . . . . . . . . . . . . . . . . . .71
Settings for Protein A280 Measurements . . . . . . . . . . . . . . . . . . . . . . 72
Protein editor . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .75
Detection Limits for Protein A280 Measurements. . . . . . . . . . . . . . . .78
Calculations for Protein A280 Measurements. . . . . . . . . . . . . . . . . . .79
Measure Protein A205. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .84
Measure Protein Concentration at A205 . . . . . . . . . . . . . . . . . . . . . .84
Protein A205 Reported Results . . . . . . . . . . . . . . . . . . . . . . . . . . . . .86
Settings for Protein A205 Measurements . . . . . . . . . . . . . . . . . . . . . . 89
Calculations for Protein A205 Measurements. . . . . . . . . . . . . . . . . . .91
Measure Proteins and Labels . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .93
Measure Labeled Protein Samples. . . . . . . . . . . . . . . . . . . . . . . . . . .93
Proteins & Labels Reported Results . . . . . . . . . . . . . . . . . . . . . . . . . . 96
Settings for Proteins and Labels Measurements. . . . . . . . . . . . . . . . .98
Detection Limits for Proteins and Labels Measurements . . . . . . . . .100
Calculations for Proteins and Labels Measurements . . . . . . . . . . . .100
Measure Protein BCA . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .103
Measure Total Protein Concentration . . . . . . . . . . . . . . . . . . . . . . . .103
Protein BCA Reported Results. . . . . . . . . . . . . . . . . . . . . . . . . . . . .111
Settings for Protein BCA Measurements . . . . . . . . . . . . . . . . . . . . .115
iv NanoDrop One User Guide Thermo Scientific
Page 5

Contents
Measure Protein Bradford . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .117
Measure Total Protein Concentration . . . . . . . . . . . . . . . . . . . . . . . .117
Protein Bradford Reported Results . . . . . . . . . . . . . . . . . . . . . . . . .121
Settings for Protein Bradford Measurements . . . . . . . . . . . . . . . . . . 124
Measure Protein Lowry . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 127
Measure Total Protein Concentration . . . . . . . . . . . . . . . . . . . . . . . .127
To measure Protein Lowry standards and samples . . . . . . . . . . . . .128
Protein Lowry Reported Results. . . . . . . . . . . . . . . . . . . . . . . . . . . . 130
Settings for Protein Lowry Measurements . . . . . . . . . . . . . . . . . . . .132
Measure Protein Pierce 660 . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .135
Measure Total Protein Concentration . . . . . . . . . . . . . . . . . . . . . . . .135
To measure Protein Pierce 660 standards and samples. . . . . . . . . . 136
Protein Pierce 660 Reported Results . . . . . . . . . . . . . . . . . . . . . . . .138
Settings for Protein Pierce 660 Measurements. . . . . . . . . . . . . . . . . 141
Chapter 6 Measure OD600 . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 143
Measure OD600 . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .143
To measure OD600 samples . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .145
OD600 Reported Results. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .147
Settings for OD600 Measurements . . . . . . . . . . . . . . . . . . . . . . . . .148
Calculations for OD600 Measurements . . . . . . . . . . . . . . . . . . . . . .151
Chapter 7 Custom Applications . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 153
Measure UV-Vis. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .154
Measure UV-Vis . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 154
Best practices for UV-Vis measurements . . . . . . . . . . . . . . . . . . . . .155
UV-Vis Reported Results . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .156
Settings for UV-Vis Measurements. . . . . . . . . . . . . . . . . . . . . . . . . .159
Measure Custom. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .161
Measure using a Custom Method . . . . . . . . . . . . . . . . . . . . . . . . . .161
Delete Custom Method . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .164
Custom Method Reported Results. . . . . . . . . . . . . . . . . . . . . . . . . .165
Manage Custom Methods . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .167
Chapter 8 Measure Kinetics . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .177
Measure Kinetics . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .177
Create Kinetics Method . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .181
Edit Kinetics Method . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .182
Kinetics Reported Results . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .183
Settings for Kinetic Measurements. . . . . . . . . . . . . . . . . . . . . . . . . .188
Chapter 9 Learning Center . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .191
Micro-Volume Sampling—How it Works . . . . . . . . . . . . . . . . . . . . . . .192
Thermo Scientific NanoDrop One User Guide v
Page 6

Contents
Set Up the Instrument . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .194
Connect Power . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .194
Connect an Accessory. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .194
Set Up Bluetooth Connections. . . . . . . . . . . . . . . . . . . . . . . . . . . . . 194
Set Up Ethernet Connection . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .199
Set up Wireless Connections . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 200
Assess Instrument Connectivity . . . . . . . . . . . . . . . . . . . . . . . . . . . .202
Operating Specifications . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 203
Measure a Micro-Volume Sample . . . . . . . . . . . . . . . . . . . . . . . . . . . .204
Best practices for micro-volume measurements. . . . . . . . . . . . . . . .205
Recommended sample volumes . . . . . . . . . . . . . . . . . . . . . . . . . . .206
Measure a Sample Using a Cuvette. . . . . . . . . . . . . . . . . . . . . . . . . . .209
Best practices for cuvette measurements . . . . . . . . . . . . . . . . . . . .210
Prepare Samples and Blanks . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .212
Preparing Samples . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .212
Run a Blanking Cycle. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .215
Basic Instrument Operations . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .217
NanoDrop One Home Screen . . . . . . . . . . . . . . . . . . . . . . . . . . . . .217
NanoDrop One Measurement Screens . . . . . . . . . . . . . . . . . . . . . .220
NanoDrop One General Operations . . . . . . . . . . . . . . . . . . . . . . . . .235
Acclaro Sample Intelligence. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .245
Activate Detection . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .245
View Acclaro Sample Intelligence Information. . . . . . . . . . . . . . . . . . 246
Contaminant Analysis . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .247
On-Demand Technical Support . . . . . . . . . . . . . . . . . . . . . . . . . . . .251
Invalid-Results Alerts . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .252
Instrument Settings . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .253
System Settings. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 253
Network Settings . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .255
Export Settings . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .256
General Settings . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 258
Data Deletion Settings . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .259
PC Control Software . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .261
PC Control Home Screen overview . . . . . . . . . . . . . . . . . . . . . . . . .261
Control options . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .262
Instrument Status . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .265
Measurement Screen Display Options . . . . . . . . . . . . . . . . . . . . . . .266
Chapter 10 Maintenance . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .267
Maintenance Schedule . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .268
Daily Maintenance . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 268
Periodic Maintenance . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .268
Every 6 Months . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .268
Cleaning the Touchscreen . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 269
vi NanoDrop One User Guide Thermo Scientific
Page 7

Contents
Maintaining the Pedestals . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .269
Cleaning the Pedestals . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .269
Reconditioning the Pedestals. . . . . . . . . . . . . . . . . . . . . . . . . . . . . .272
Decontaminating the Instrument . . . . . . . . . . . . . . . . . . . . . . . . . . . . .274
Maintaining the Cuvette Sampling System. . . . . . . . . . . . . . . . . . . . . .276
Instrument Diagnostics . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 276
Intensity Check . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .277
Performance Verification . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .279
Pedestal Image Check . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 284
Chapter 11 Safety and Operating Precautions . . . . . . . . . . . . . . . . . . . . . . . . .285
Operating Precautions. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .286
Safety Information . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .287
Safety and Special Notices . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .287
When the System Arrives . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 289
Lifting or Moving the Instrument . . . . . . . . . . . . . . . . . . . . . . . . . . . . 289
Electrical Requirements and Safety . . . . . . . . . . . . . . . . . . . . . . . . . 290
Power Cords . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .290
Fire Safety and Burn Hazards . . . . . . . . . . . . . . . . . . . . . . . . . . . . .291
Optical Safety . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .292
Hazardous Materials . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .292
Thermo Scientific NanoDrop One User Guide vii
Page 8

Contents
viii NanoDrop One User Guide Thermo Scientific
Page 9

About the Spectrophotometer
NanoDrop OneC Spectrophotometer
Arm
1
Pedestal
Cuvette holder
Note Locate the instrument away from air vents and exhaust fans to minimize
evaporation
The Thermo Scientific™ NanoDrop™ OneC is a compact, stand-alone
UV-Visible spectrophotometer developed for micro-volume analysis of a wide variety
of analytes. The patented sample retention system enables the measurement of
highly concentrated samples without the need for dilutions.
The NanoDrop One system comes with preloaded software and a touchscreen
display. NanoDrop One PC Control software can be installed on a local PC and used
to control the instrument and view data. The instrument can be connected to an
optional printer with a USB cable or to a remote printer through an Ethernet
connection or wireless network.
Note Before operating a NanoDrop One instrument, please read the safety and
operating precautions and then follow their recommendations when using the
instrument.
Thermo Scientific NanoDrop One User Guide 9
Page 10

1 About the Spectrophotometer
Features
Features
Touchscreen
TheNanoDrop OneC spectrophotometer features the patented micro-volume sample
retention system. The NanoDrop One
dilute samples using standard UV-Visible cuvettes.
TheNanoDrop OneC comes with a built-in, 7-inch high-resolution touchscreen
preloaded with easy-to-use instrument control software. The touchscreen can slide
left or right to accommodate personal preference, and tilt forward or back for optimal
viewing
C
also features a cuvette holder for analyzing
10 NanoDrop One User Guide Thermo Scientific
Page 11

Cuvette Holder
Instrument light path
1
About the Spectrophotometer
Cuvette holder
Features
USB-A port
The NanoDrop OneC includes a cuvette holder for measuring dilute samples,
colorimetric assays, cell cultures and kinetic studies. The cuvette system has these
features:
• extended lower detection limits
• 37 °C heater option for temperature-sensitive samples and analyses
• micro-stirring option to ensure sample homogeneity and support kinetic studies
For details, see Measure a Sample using a Cuvette.
One USB-A port is located on the front of the instrument and two more USB-A ports
are located on instrument back panel.
Thermo Scientific NanoDrop One User Guide 11
Page 12

1 About the Spectrophotometer
Accessories
Accessories
This section lists the accessories included for use with the NanoDrop OneC.
DYMO™ LabelWriter™ 450 USB Label Printer
Prints two 5/16-in x 4-in self-adhesive labels for transferring sample data directly into
laboratory notebooks or posting on bulletin boards. The software allows printing of
data from each sample measurement or from a group of samples logged and
measured together.
The printer connects to the instrument (front or back) via a USB cable (included).
PR-1 Pedestal Reconditioning Kit
Specially formulated conditioning compound
that can be applied to the pedestals to restore
them to a hydrophobic state (required to
achieve adequate surface tension for accurate
sample measurements). The kit includes
conditioning compound and applicators. For
more information, see Reconditioning the
Pedestals.
PV-1 Performance Verification Solution
Liquid photometric standard used to check instrument performance. For more
information, see Performance Verification.
12 NanoDrop One User Guide Thermo Scientific
Page 13
Instrument Detection Limits
1
About the Spectrophotometer
Instrument Detection Limits
Measurement
Location Pathlength (mm)
Upper Detection Limit
(10 mm Equivalent Absorbance)
Pedestal 1.0 12.5
0.2 62.5
0.1 150
0.05 300
0.03 550
Cuvette 10 1.5
53
2 7.5
115
Thermo Scientific NanoDrop One User Guide 13
Page 14

1 About the Spectrophotometer
This page is intentionally blank.
14 NanoDrop One User Guide Thermo Scientific
Page 15

Instrument Set up
Register Your Instrument
Register your instrument to receive e-mail updates on software and accessories for
the NanoDrop One instrument. An Internet connection is required for registration.
To register your instrument
1. Do one of the following:
– From any PC that is connected to the Internet, use any web browser to
navigate to our website.
2
On the website, locate NanoDrop One Registration and follow the instructions to
register the instrument.
Update Software
Quickly and easily download and install the latest NanoDrop One software and
release notes from our website. Follow the steps to update or upgrade the software
on your local instrument and/or install or update the NanoDrop One software on a
personal computer (PC). An Internet connection is required to download software.
To install or update NanoDrop One software on a PC
1. Insert the USB flash drive containing the installer software into an available USB
port on your PC, or open the installation folder downloaded from the internet.
2. Launch Start.exe and click Install. The software installer will run.
To install or update NanoDrop One software on the instrument
1. Copy the .zip file with the new software from your computer to a USB storage
device. Do not attempt to unzip the folder.
2. Insert the USB device into any USB port on the NanoDrop One instrument.
3. From the instrument Home screen, tap Settings > System > Update
Software and choose the latest version of software.
Thermo Scientific NanoDrop One User Guide 15
Page 16
2 Instrument Set up
Setting Up User Account Control (Optional)
Setting Up User Account Control (Optional)
User account control is managed using the Security Administration application. The
Thermo Scientific Security Administration software for NanoDrop One may be
purchased for instruments used in labs requiring 21 CFR Part 11 compliance. When
you launch Security Administration, you will need to enter your Windows log-in
information.
User Account Control
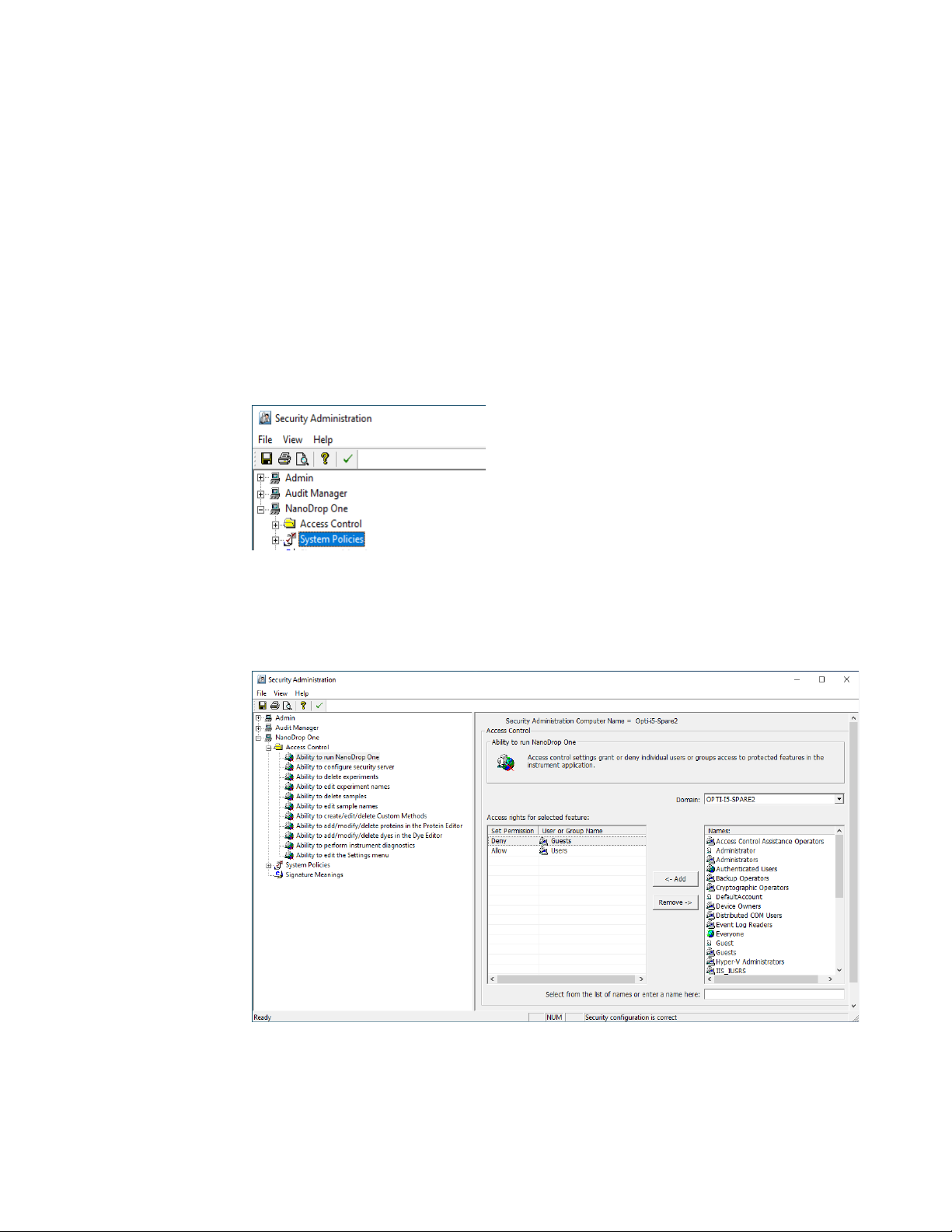
Launch the Security Administration application and select NanoDrop One from the
directory on the left to reveal Access Control and System Policies.
Access control
Access control is used to grant or deny individual users or groups access to
protected features in the instrument application. Add and remove users and groups
to the access list and set access rights using the drop-down for each entity.
16 NanoDrop One User Guide Thermo Scientific
Page 17
System policies
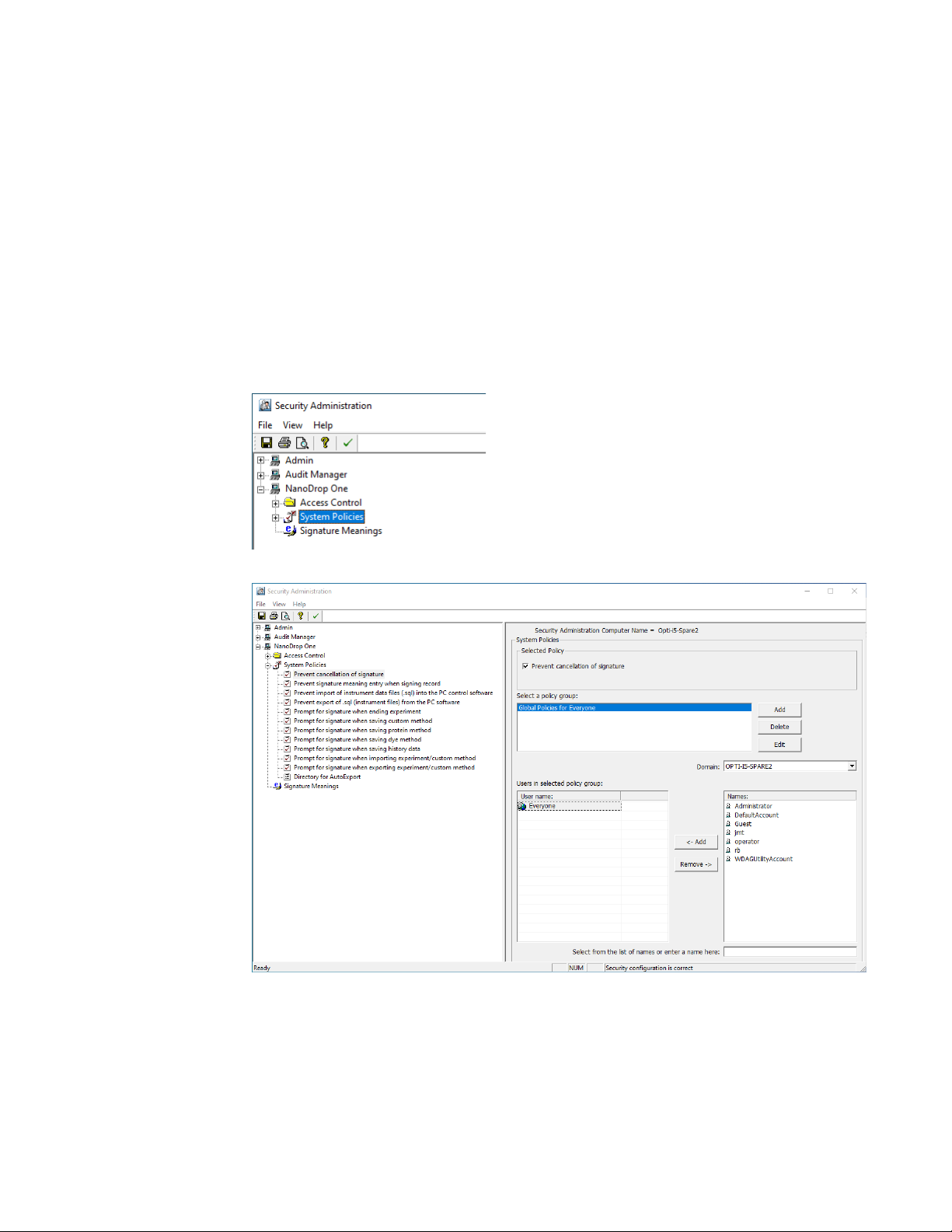
System Policies is used to set options that define the behavior of the client
application. See “Security Administration Policies.”
Security Administration Policies
System policies allow you to assign data and method creation and deletion and
editing privileges for users and groups.
Launch the Security Administration application and select NanoDrop One->
System Policies
2
Setting Up User Account Control (Optional)
Instrument Set up
Thermo Scientific NanoDrop One User Guide 17
Page 18
2 Instrument Set up
Setting Up User Account Control (Optional)
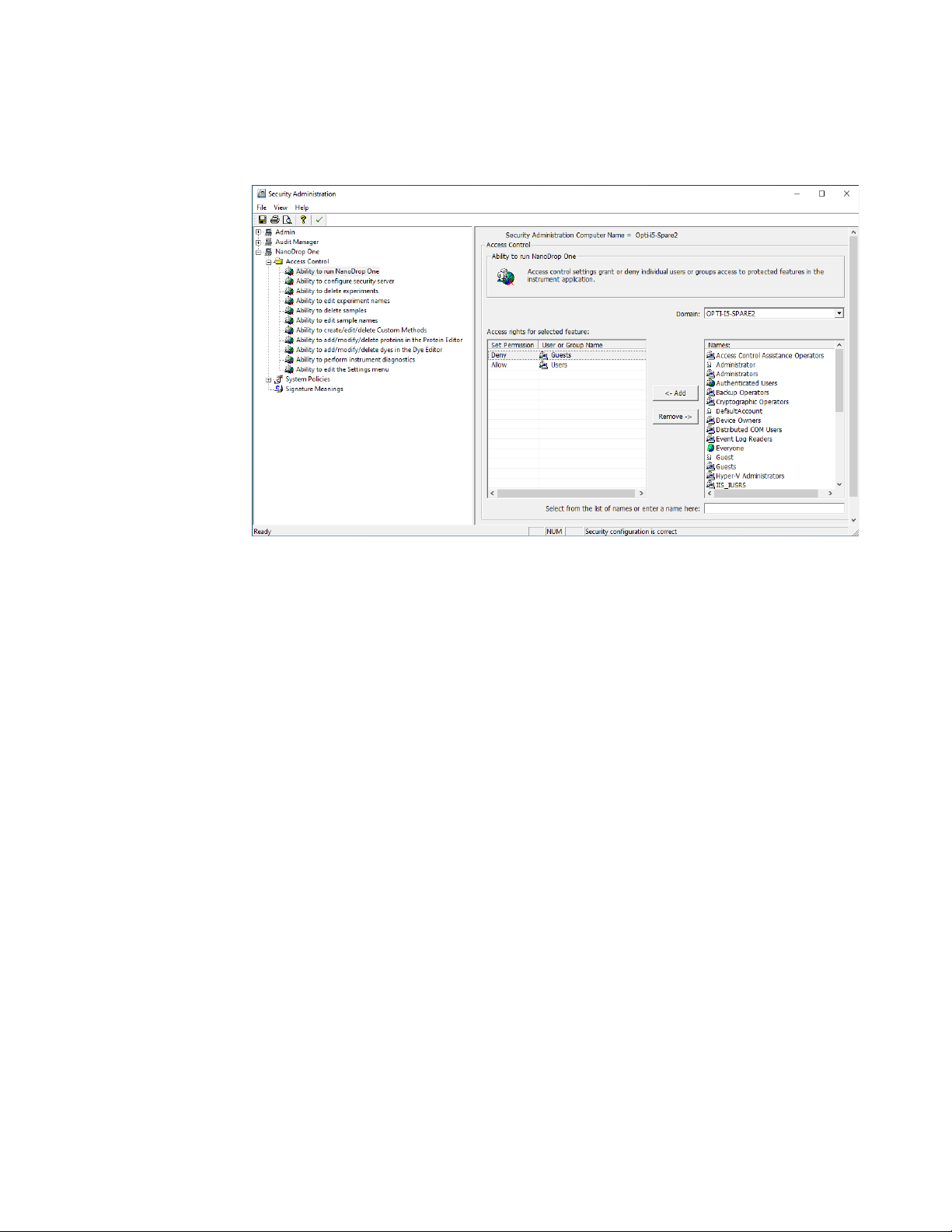
You can add, delete, or edit policy groups and enable or disable the group’s users
permission to delete data. Select NanoDrop One-> Access Control.
When you are finished, select Save. Changes will take effect the next time
NanoDrop One is launched.
18 NanoDrop One User Guide Thermo Scientific
Page 19

Technical Support
For U.S./Canada Support, please contact:
Thermo Fisher Scientific
3411 Silverside Road
Tatnall Building, Suite 100
Wilmington, DE 19810 U.S.A.
Telephone: 302 479 7707
Toll Free: 1 877 724 7690 (U.S. & Canada only)
Fax: 302 792 7155
E-mail: nanodrop@thermofisher.com
Website: www.thermofisher.com/nanodrop
2
Instrument Set up
Technical Support
For International Support, please contact:
Contact your local distributor. For contact information go to:
http://www.thermofisher.com/NanoDropDistributors
If you are experiencing an issue with your system, refer to the troubleshooting
information. If the issue persists, contact us. If you are outside the U.S.A. and
Canada, please contact your local distributor.
If your instrument requires maintenance or repair, contact us or your local distributor.
Thermo Scientific NanoDrop One User Guide 19
Page 20

2
Instrument Set up
This page is intentionally blank.
20 NanoDrop One User Guide Thermo Scientific
Page 21
Application Measurement Ranges
Detection Limits for All Applications
3
Note Detection limits provided in the tables below are approximate and apply to
micro-volume measurements only; they are based on the instrument’s
photometric absorbance range (10 mm equivalent) of 0–550 A. For
measurements with 10 mm pathlength cuvettes, the photometric absorbance
range is 0–1.5 A.
Detection limits for standard applications
Sample Type
dsDNA 2.0 ng/µL (pedestal)
ssDNA 1.3 ng/µL (pedestal)
Lower Detection
Limit
0.20 ng/µL (cuvette)
0.13 ng/µL (cuvette)
Upper Detection
Limit
27,500 ng/µL
(pedestal)
75 ng/µL (cuvette)
18,150 ng/µL
(pedestal)
49.5 ng/µL (cuvette)
Typical Reproducibility
±2.0 ng/µL for sample
concentrations between 2.0 and
100 ng/µL samples;
±2% for samples >100 ng/µL
±2.0 ng/µL for sample
concentrations between 2.0 and
100 ng/µL samples;
±2% for samples >100 ng/µL
a
Thermo Scientific NanoDrop One User Guide 21
Page 22

3 Application Measurement Ranges
Detection Limits for All Applications
Sample Type
Lower Detection
Limit
RNA 1.6 ng/µL (pedestal)
0.16 ng/µL (cuvette)
DNA Microarray
1.3 ng/µL (pedestal)
(ssDNA)
0.13 ng/µL (cuvette)
Purified BSA by
Protein A280
IgG by Protein
A280
Purified BSA by
0.06 mg/mL (pedestal)
0.006 mg/mL (cuvette)
0.03 mg/mL (pedestal)
0.003 mg/mL (cuvette)
0.06 mg/mL (pedestal)
Proteins &
Labels
0.006 mg/mL (cuvette)
Protein BCA 0.2 mg/mL (20:1
reagent/sample
volume)
Upper Detection
Limit
22,000 ng/µL
(pedestal)
Typical Reproducibility
±2.0 ng/µL for sample
concentrations between 2.0 and
a
100 ng/µL samples;
60 ng/µL (cuvette)
495 ng/µL (pedestal)
±2% for samples >100 ng/µL
±2.0 ng/µL for sample
concentrations between 2.0 and
49.5 ng/µL (cuvette)
100 ng/µL samples;
±2% for samples >100 ng/µL
825 mg/mL (pedestal)
±0.10 mg/mL (for 0.10–10 mg/mL
samples);
±2% for samples >10 mg/mL
402 mg/mL (pedestal)
19 mg/mL (pedestal) ±0.10 mg/mL for 0.10–10 mg/mL
samples
8.0 mg/mL (pedestal)
0.20 mg/mL (cuvette)
2% over entire range
0.01 mg/mL over entire range
0.01 mg/mL (1:1
reagent/sample
volume)
Protein Lowry 0.2 mg/mL (pedestal) 4.0 mg/mL (pedestal) 2% over entire range
Protein Bradford 100 µg/mL (50:1
reagent/sample
volume)
15 µg/mL (1:1
reagent/sample
8000 µg/mL
100 µg/µL
±25 µg/mL for 100–500 µg/mL
samples
±5% for 500–8000 µg/mL samples
±4 µg/mL for 15–50 µg/mL samples
±5% for 50–125 µg/mL samples
volume)
Protein Pierce
660
50 µg/mL (15:1
reagent/sample
2000 µg/mL
±3 µg/mL for 50–125 µg/mL samples
±2% for samples > 125 µg/mL
volume)
±3 µg/mL for 25–125 µg/mL samples
25 µg/mL (7.5:1
1000 µg/mL
±2% for samples >125 µg/mL
reagent/sample
volume)
a
Based on five replicates (SD=ng/µL; CV=%)
22 NanoDrop One User Guide Thermo Scientific
Page 23

Note To minimize instrument error with highly concentrated samples, make
dilutions to ensure that measurements are made within these absorbance limits:
• For micro-volume measurements, maximum absorbance at 260 nm (for
nucleic acids) or 280 nm (for proteins) should be less than 62.5 A.
• For measurements with 10 mm pathlength cuvettes, maximum absorbance
at 260 nm (or 280 nm for proteins) should be less than 1.5 A, which is
approximately 75 ng/µL dsDNA.
Detection limits for pre-defined dyes
3
Application Measurement Ranges
Detection Limits for All Applications
Sample Type
Cy3, Cy3.5, Alexa
Fluor 555, Alexa Fluor
Lower Detection
Limit
0.2 pmol/µL
(pedestal)
660
Cy5, Cy5.5, Alexa
Fluor 647
Alexa Fluor 488, Alexa
Fluor 594
0.12 pmol/µL
(pedestal)
0.4 pmol/µL
(pedestal)
Alexa Fluor 546 0.3 pmol/µL
(pedestal)
a
Values are approximate
b
Based on five replicates (SD=ng/µL; CV=%)
Upper Detection
a
Limit
100 pmol/µL
(pedestal)
60 pmol/µL
(pedestal)
215 pmol/µL
(pedestal)
145 pmol/µL
(pedestal)
Typical Reproducibility
b
±0.20 pmol/µL for sample
concentrations between 0.20 and
4.0 pmol/µL;
±2% for samples >4.0 pmol/µL
±0.12 pmol/µL for sample
concentrations between 0.12 and
2.4 pmol/µL;
±2% for samples >2.4 pmol/µL
±0.40 pmol/µL for sample
concentrations between 0.40 and
8.0 pmol/µL;
±2% for samples >8.0 pmol/µL
±0.30 pmol/µL for sample
concentrations between 0.30 and
6.0 pmol/µL;
±2% for samples >6.0 pmol/µL
Thermo Scientific NanoDrop One User Guide 23
Page 24

3
Application Measurement Ranges
This page is intentionally blank.
24 NanoDrop One User Guide Thermo Scientific
Page 25

Nucleic Acid Applications
Measure dsDNA, ssDNA or RNA
Measures the concentration of purified
dsDNA, ssDNA or RNA samples that
absorb at 260 nm.
Measure dsDNA, ssDNA or RNA
Reported Results
Settings
4
Detection Limits
Calculations
Measure dsDNA, ssDNA or RNA
Use the dsDNA, ssDNA and RNA applications to quantify purified double-stranded
(ds) or single-stranded (ss) DNA or RNA samples. These applications report nucleic
acid concentration and two absorbance ratios (A260/A280 and A260/A230). A
single-point baseline correction can also be used.
To measure dsDNA, ssDNA or RNA samples
NOTICE
• Do not use a squirt or spray bottle on or near the instrument as liquids will
flow into the instrument and may cause permanent damage.
• Do not use hydrofluoric acid (HF) on the pedestals. Fluoride ions will
permanently damage the quartz fiber optic cables.
Thermo Scientific NanoDrop One User Guide 25
Page 26

4 Nucleic Acid Applications
Measure dsDNA, ssDNA or RNA
Before you begin...
Before taking pedestal measurements with the NanoDrop One instrument, lift the
instrument arm and clean the upper and lower pedestals. At a minimum, wipe the
pedestals with a new laboratory wipe. For more information, see Cleaning the
Pedestals.
To measure nucleic acid
1. From the Home screen, select the Nucleic Acids tab and select dsDNA,
2. Specify a baseline correction if desired.
3. Pipette 1–2 µL blanking solution onto the lower pedestal and lower the arm, or
ssDNA or RNA, depending on the samples to be measured.
insert the blanking cuvette into the cuvette holder.
Tip: If using a cuvette, make sure to align the cuvette light path with the
instrument light path.
4. Tap Blank and wait for the measurement to complete.
Tip: If Auto-Blank is On, the blank measurement starts automatically after you
lower the arm. (This option is not available for cuvette measurements.)
5. Lift the arm and clean both pedestals with a new laboratory wipe, or remove the
blanking cuvette.
6. Pipette 1-2 µL sample solution onto the pedestal and lower the arm, or insert the
sample cuvette into the cuvette holder.
7. Start the sample measurement:
– Pedestal: If Auto-Measure is On, lower arm; if Auto-Measure is off, lower arm
and tap Measure.
– Cuvette: Tap Measure.
When the sample measurement is completed, the spectrum and reported values
are displayed (see the next section).
8. When you are finished measuring samples, tap End Experiment.
9. Lift the arm and clean both pedestals with a new wipe, or remove the sample
cuvette.
26 NanoDrop One User Guide Thermo Scientific
Page 27
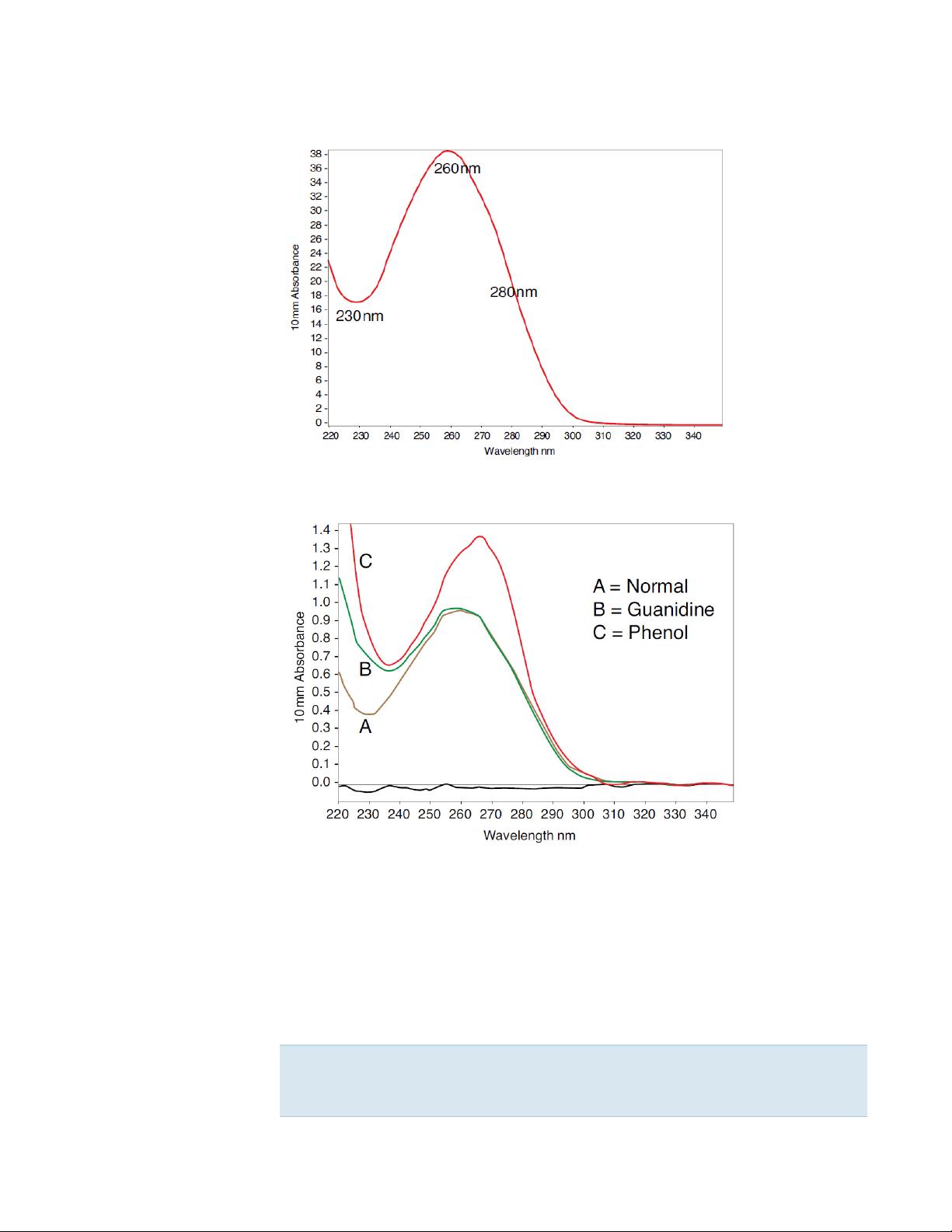
Typical nucleic acid spectrum
4
Nucleic Acid Applications
Measure dsDNA, ssDNA or RNA
Comparison of nucleic acid spectra with and without two
common contaminants
Best practices for nucleic acid measurements
• Isolate and purify nucleic acid samples before measurement to remove
impurities. Depending on the sample, impurities could include DNA, RNA, free
nucleotides, proteins, some buffer components and dyes. See Preparing
Samples for more information.
Note Extraction reagents such as guanidine, phenol, and EDTA contribute
absorbance between 230 nm and 280 nm and will affect measurement
results if present in samples (even residual amounts).
Thermo Scientific NanoDrop One User Guide 27
Page 28

4 Nucleic Acid Applications
Measure dsDNA, ssDNA or RNA
• Ensure the sample absorbance is within the instrument’s absorbance detection
• Blank with the same buffer solution used to resuspend the analyte of interest.
• Run a blanking cycle to assess the absorbance contribution of your buffer
• For micro-volume measurements:
limits.
The blanking solution should be a similar pH and ionic strength as the analyte
solution.
solution. If the buffer exhibits strong absorbance at or near the analysis
wavelength (typically 260 nm), you may need to choose a different buffer or
application. See Choosing and Measuring a Blank for more information.
– Ensure pedestal surfaces are properly cleaned and conditioned.
– If possible, heat highly concentrated or large molecule samples, such as
genomic or lambda DNA, to 63 °C (145 °F) and gently (but thoroughly) vortex
before taking a measurement. Avoid introducing bubbles when mixing and
pipetting.
– Follow best practices for micro-volume measurements.
– Use a 1-2 µL sample volume. See Recommended Sample Volumes for more
information.
C
• For cuvette measurements (NanoDrop One
instruments only), use compatible
cuvettes and follow best practices for cuvette measurements.
Related Topics
• Measure a Micro-Volume Sample
• Measure a Sample Using a Cuvette
• Best Practices for Micro-Volume Measurements
• Best Practices for Cuvette Measurements
• Prepare Samples and Blanks
• Basic Instrument Operations
28 NanoDrop One User Guide Thermo Scientific
Page 29
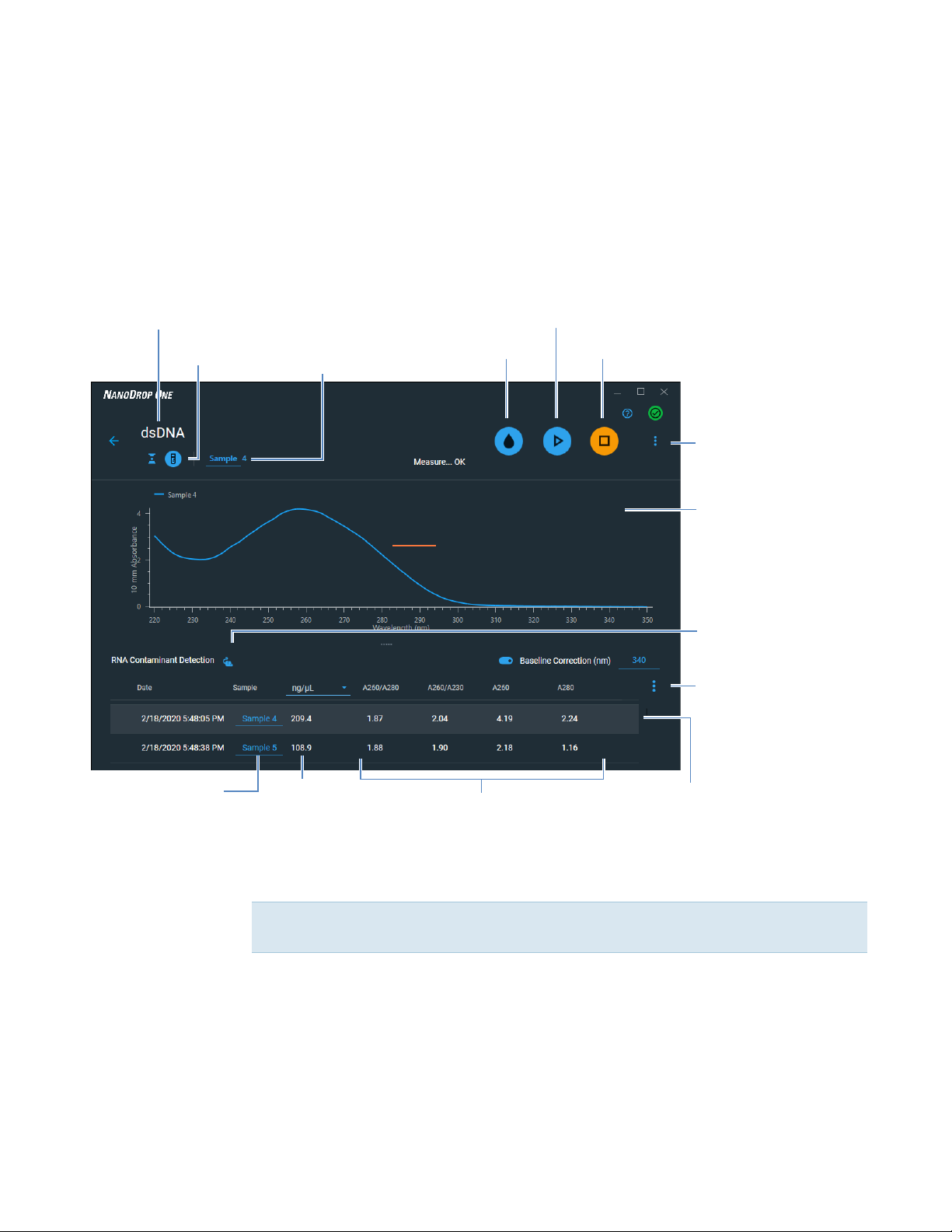
Nucleic Acid Reported Results
dsDNA measurement screen
For each measured sample, the dsDNA, ssDNA and RNA applications show the UV
absorbance spectrum and a summary of the results. Below is an example of the
measurement screen of the PC control software:
4
Nucleic Acid Applications
Measure dsDNA, ssDNA or RNA
Application
Sampling
method
Sample name of
next measurement;
select to edit
Measure sample
Run Blank
UV spectrum
End experiment
Menu of options;
click to open
Right click graph
area to view
options
Mammalian
display
.
Contaminant
Detection
mouse icon to
toggle on and off
Menu of table options;
click to choose which
columns to report
; select
Sample name;
select to edit
Nucleic acid
concentration
Purity ratios
Click row to select sample
and update spectrum.
Measurement screen of PC Control software
Note Micro-volume absorbance measurements and measurements taken with
nonstandard cuvettes are normalized to a 10.0 mm pathlength equivalent.
Thermo Scientific NanoDrop One User Guide 29
Page 30
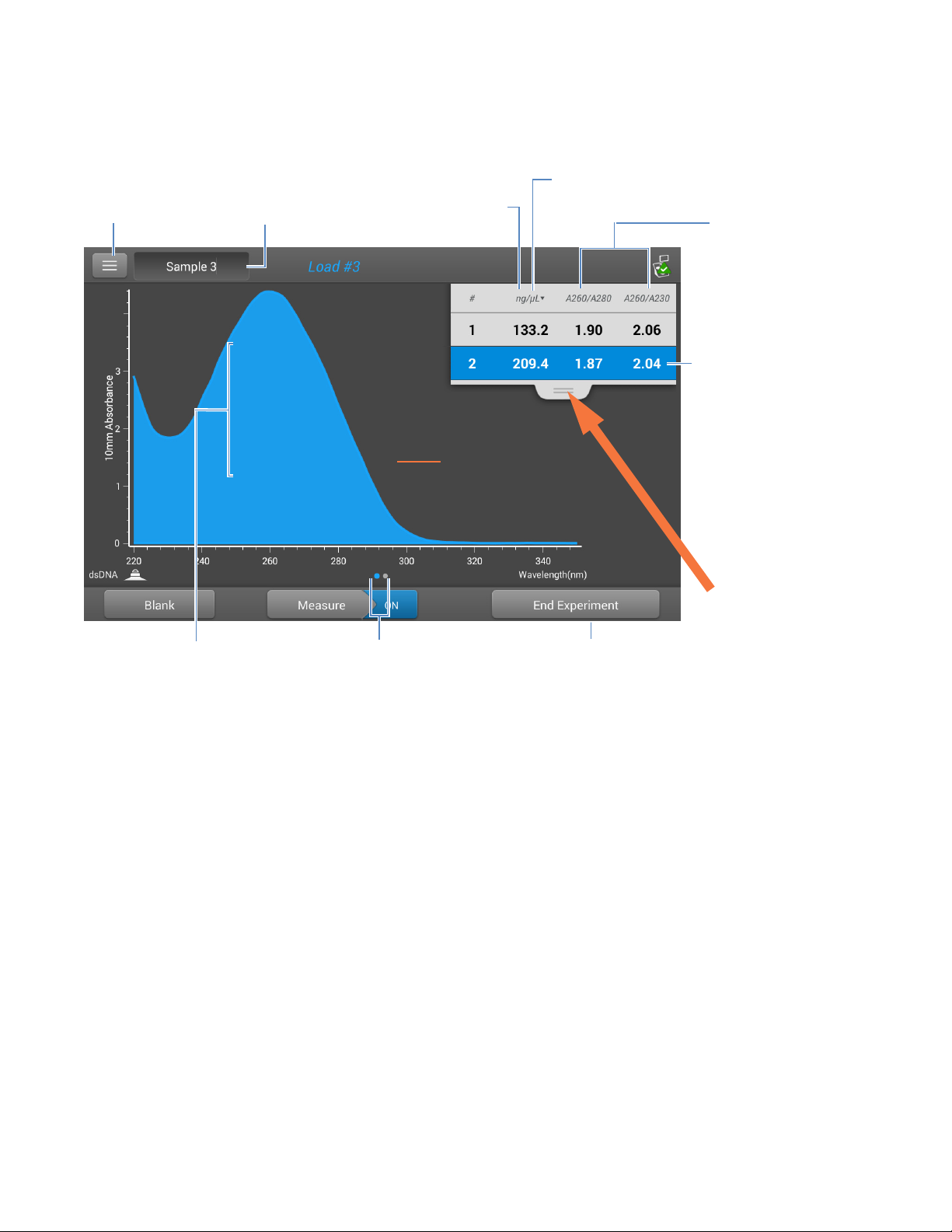
4 Nucleic Acid Applications
Measure dsDNA, ssDNA or RNA
The following is an example of the measurement screen of the local control:
Menu of options;
tap to open
Pinch and zoom to
adjust axes; double-tap
to reset
Sample name;
tap to edit
Nucleic acid
concentration
UV spectrum
Swipe screen left to view
table with more
measurement results
Tap to select unit
Tap to end
experiment and
export data
Purity ratios
Tap r ow to select
sample and update
spectrum; tap
more rows to
overlay up to five
spectra. Press and
hold sample row to
view measurement
details.
Drag tab down/up
to see more/less
sample data
Measurement screen of NanoDrop One local control software
30 NanoDrop One User Guide Thermo Scientific
Page 31

dsDNA, ssDNA and RNA reported values
The initial screen that appears after each measurement (see previous image) shows
a summary of the reported values. To view all reported values, press and hold the
sample row. Here is an example:
4
Nucleic Acid Applications
Measure dsDNA, ssDNA or RNA
Application
Sampling
method
Sample name;
tap to edit
Factor Baseline Correction
(shown here if applied)
Date/time
measured
nucleic acid
concentration
A260/A280
A260/A230
purity ratio
A260
absorbance
A280
absorbance
• sample details (application and sampling method used, that is, pedestal or
cuvette)
• sample name
• created on (date sample measurement was taken)
• nucleic acid concentration
• A260/A280
• A260/A230
• A260
• A280
• factor
• baseline correction
Thermo Scientific NanoDrop One User Guide 31
Page 32

4 Nucleic Acid Applications
Measure dsDNA, ssDNA or RNA
Settings for Nucleic Acid Measurements
To show the dsDNA, ssDNA or RNA settings, from the dsDNA, ssDNA or RNA
measurement screen, tap > Nucleic Acid Setup.
Setting Available Options Description
Baseline Correction On or off
Enter baseline
correction wavelength
in nm or use default
value (340 nm)
Optional user-defined baseline correction. Can
be used to correct for any offset caused by light scattering
particulates by subtracting measured absorbance at
specified baseline correction wavelength from absorbance
values at all wavelengths in sample spectrum. As a result,
absorbance of sample spectrum is zero at specified
baseline correction wavelength.
Calculations for Nucleic Acid Measurements
The Nucleic Acid applications use a
modification of the Beer-Lambert equation
(shown at right) to calculate sample
concentration where the extinction
coefficient and pathlength are combined
and referred to as a “factor.”
Extinction Coefficients vs Factors
Using the terms in the Beer-Lambert equation, factor (f) is
defined as:
factor (f) = 1/( * b)
where:
= wavelength-dependent molar extinction coefficient in
ng-cm/µL
b = sample pathlength in cm
As a result, analyte concentration (c) is calculated as:
c = A * [1/( * b)]
or
c = A * f
where:
c = analyte concentration in ng/µL
A = absorbance in absorbance units (A)
f = factor in ng-cm/µL (see below)
32 NanoDrop One User Guide Thermo Scientific
Page 33

4
Nucleic Acid Applications
Measure dsDNA, ssDNA or RNA
For the dsDNA, ssDNA and RNA
applications, the generally accepted factors
for nucleic acids are used in conjunction
with Beer’s Law to calculate sample
concentration. For the Custom Factor
application, the user-specified factor is
used.
Calculated nucleic acid concentrations are
based on the absorbance value at 260 nm,
the factor used and the sample pathlength.
A single-point baseline correction (or
analysis correction) may also be applied.
Concentration is reported in mass units.
Calculators are available on the Internet to
convert concentration from mass to molar
units based on sample sequence.
Absorbance values at 260 nm, 280 nm and
sometimes 230 nm are used to calculate
purity ratios for the measured nucleic acid
samples. Purity ratios are sensitive to the
presence of contaminants in the sample,
such as residual solvents and reagents
typically used during sample purification.
Factors Used
• dsDNA (factor = 50 ng-cm/µL)
• ssDNA (factor = 33 ng-cm/µL)
• RNA (factor = 40 ng-cm/µL)
• Custom Factor (user entered factor between
15 ng-cm/µL and 150 ng-cm/µL
Measured Values
Note: For micro-volume absorbance measurements and
measurements taken with nonstandard (other than 10 mm)
cuvettes, the spectra are normalized to a 10 mm pathlength
equivalent.
A260 absorbance
• Nucleic acid absorbance values are measured at 260 nm
using the normalized spectrum. This is the reported A260
value if Baseline Correction is not selected.
• If Baseline Correction is selected, the absorbance value
at the correction wavelength is subtracted from the
absorbance at 260 nm. The corrected absorbance at
260 nm is reported and used to calculate nucleic acid
concentration.
A230 and A280 absorbance
• Normalized and baseline-corrected (if selected)
absorbance values at 230 nm and 280 nm are used to
calculate A260/A230 and A260/A280 ratios.
Sample Pathlength
• For micro-volume measurements, the software selects
the optimal pathlength (between 1.0 mm and 0.03 mm)
based on sample absorbance at the analysis wavelength.
• For cuvette measurements, pathlength is determined by
the cuvette Pathlength setting in the software (see
General Settings).
• Displayed spectra and absorbance values are normalized
to a 10 mm pathlength equivalent.
Thermo Scientific NanoDrop One User Guide 33
Page 34

Reported Values
• Nucleic acid concentration. Reported in selected
unit (i.e., ng/µL, µg/uL or µg/mL). Calculations are based
on modified Beer’s Law equation using corrected nucleic
acid absorbance value.
• A260/A280 purity ratio. Ratio of corrected
absorbance at 260 nm to corrected absorbance at
280 nm. An A260/A280 purity ratio of ~1.8 is generally
accepted as “pure” for DNA (~2.0 for RNA). Acidic
solutions may under represent the reported value by
0.2-0.3; the opposite is true for basic solutions.
• A260/A230 purity ratio. Ratio of corrected
absorbance at 260 nm to corrected absorbance at
230 nm. An A260/A230 purity ratio between 1.8 and 2.2
is generally accepted as “pure” for DNA and RNA.
Note: Although purity ratios are important indicators of
sample quality, the best quality indicator quality is
functionality in the downstream application of interest (e.g.,
real-time PCR).
• Factor. Used in conjunction with Beer’s Law to calculate
sample concentration
• Contaminant - If a contaminant was identified by the
Acclaro software, the contaminant will be displayed in this
column.
• A260 absorbance.
• A280 absorbance.
• Baseline correction.
Page 35

Measure Microarray
Measures the concentration of purified
nucleic acids that have been labeled with
up to two fluorescent dyes for use in
downstream microarray applications.
Measure Microarray Samples
Reported Results
Settings
Detection Limits
Calculations
Measure Microarray Samples
4
Nucleic Acid Applications
Measure Microarray
Use the Microarray application to quantify nucleic acids that have been labeled with
up to two fluorescent dyes. The application reports nucleic acid concentration, an
A260/A280 ratio and the concentrations and measured absorbance values of the
dye(s), allowing detection of dye concentrations as low as 0.2 picomole per
microliter.
To measure microarray samples
NOTICE
• Do not use a squirt or spray bottle on or near the instrument as liquids will
flow into the instrument and may cause permanent damage.
• Do not use hydrofluoric acid (HF) on the pedestals. Fluoride ions will
permanently damage the quartz fiber optic cables.
Before you begin...
Before taking pedestal measurements with the NanoDrop One instrument, lift the
instrument arm and clean the upper and lower pedestals. At a minimum, wipe the
pedestals with a new laboratory wipe. For more information, see Cleaning the
Pedestals.
Thermo Scientific NanoDrop One User Guide 35
Page 36

4 Nucleic Acid Applications
Measure Microarray
To measure a microarray sample
1. From the Home screen, select the Nucleic Acids tab and select Microarray.
2. Specify the sample type and factor and the type of dye(s) used.
3. Pipette 1–2 µL blanking solution onto the lower pedestal and lower the arm, or
4. Select Blank and wait for the measurement to complete.
5. Lift the arm and clean both pedestals with a new laboratory wipe, or remove the
Tip: Select a dye from the pre-defined list or add a custom dye using the
Dye/Chromophore Editor.
insert the blanking cuvette into the cuvette holder.
Tip: If using a cuvette, make sure to align the cuvette light path with the
instrument light path.
Tip: If Auto-Blank is On, the blank measurement starts automatically after you
lower the arm. (This option is not available for cuvette measurements.)
blanking cuvette.
6. Pipette 1-2 µL sample solution onto the pedestal and lower the arm, or insert the
sample cuvette into the cuvette holder.
7. Start the sample measurement:
– Pedestal: If Auto-Measure is On, lower arm; if Auto-Measure is off, lower arm
and select Measure.
– Cuvette: Select Measure.
When the sample measurement is completed, the spectrum and reported values
are displayed (see the next section).
8. When you are finished measuring samples, select End Experiment.
9. Lift the arm and clean both pedestals with a new wipe, or remove the sample
cuvette.
36 NanoDrop One User Guide Thermo Scientific
Page 37

A260 absorbance peak used
to calculate nucleic acid
concentration
4
Nucleic Acid Applications
Measure Microarray
Dye absorbance peak used to
calculate dye concentration
Typical microarray spectrum
Related Topics
• Best Practices for Nucleic Acid Measurements
• Measure a Micro-Volume Sample
• Measure a Sample Using a Cuvette
• Best Practices for Micro-Volume Measurements
• Best Practices for Cuvette Measurements
• Prepare Samples and Blanks
• Basic Instrument Operations
Thermo Scientific NanoDrop One User Guide 37
Page 38

4 Nucleic Acid Applications
Measure Microarray
Microarray Reported Results
Microarray measurement screen (Local Control)
For each measured sample, this application shows the absorbance spectrum and a
summary of the results. Here is an example:
Menu of options;
tap to open
UV-visible spectrum
Pinch and zoom to
adjust axes; double-tap
to reset
Sample name;
tap to edit
Swipe screen left to view
table with more
measurement results
Nucleic acid
concentration
Tap to select unit
Tap to end
experiment and
export data
Dye
concentration(s)
Tap r ow to select
sample and update
spectrum; tap
more rows to
overlay up to five
spectra. Press and
hold sample row to
view measurement
details.
Drag tab
down/up to see
more/less sample
data
Note
• A baseline correction is performed at 850 nm (absorbance value at 850 nm is
subtracted from absorbance values at all wavelengths in sample spectrum).
• Micro-volume absorbance measurements and measurements taken with
nonstandard cuvettes are normalized to a 10.0 mm pathlength equivalent.
38 NanoDrop One User Guide Thermo Scientific
Page 39

Microarray reported values
The initial screen that appears after each measurement (see previous image) shows
a summary of the reported values. To view all reported values, press and hold the
sample row. Here is an example:
• sample details (application used and pedestal or cuvette)
• sample name
• created on (date sample measurement was taken)
• nucleic acid concentration
• A260
• A260/A280
• dye 1/dye 2 concentration
• sample type
4
Nucleic Acid Applications
Measure Microarray
• analysis correction
• factor
Thermo Scientific NanoDrop One User Guide 39
Page 40

4 Nucleic Acid Applications
Measure Microarray
Microarray measurement screen (PC Control)
For each measured sample, this application shows the absorbance spectrum and a
summary of the results. Here is an example:
Click to select cuvette or
pedestal measurement
Nucleic acid
concentration
Sample name;
Click to edit
UV-Vis Spectrum
Microarray
Setup
Run
Blank
Measure
sample
Dye
concentration(s)
Click to end
experiment and
export data
Menu of options;
click to open
Click and Drag
to adjust axes;
double-click to
reset
Click to
select unit
Click to
select data
columns
Click row to
select sample
and update
spectrum; click
more rows to
overlay spectra
Note
• A baseline correction is performed at 850 nm (absorbance value at 850 nm is
subtracted from absorbance values at all wavelengths in sample spectrum).
• Micro-volume absorbance measurements and measurements taken with
nonstandard cuvettes are normalized to a 10.0 mm pathlength equivalent.
40 NanoDrop One User Guide Thermo Scientific
Page 41

Settings for Microarray Measurements
Microarray settings
The Microarray Setup screen appears after you select the Microarray application
from the Nucleic Acids tab on the Home screen. To show the Microarry settings from
the Microarray measurement screen, tap > Microarray Setup.
Setting Available Options Description
4
Nucleic Acid Applications
Measure Microarray
Sample type and
Factor
dsDNA (with non-editable factor of
50 ng-cm/µL)
ssDNA (with non-editable factor of
33 ng-cm/µL)
RNA (with non-editable factor of
40 ng-cm/µL)
Oligo DNA with non-editable
calculated factor in ng-cm/µL
Oligo RNA with non-editable
calculated factor in ng-cm/µL
Custom (with user-specified factor in
ng-cm/µL)
Widely accepted value for double-stranded
DNA
Widely accepted value for single-stranded DNA
Widely accepted value for RNA
Factor calculated from user-defined DNA base
sequence. When selected, available DNA base
units (i.e., G, A, T, C) appear as keys. Define
sequence by tapping appropriate keys. Factor
is calculated automatically based on widely
accepted value for each base unit.
Factor calculated from user-defined RNA base
sequence. When selected, available RNA base
units (i.e., G, A, U, C) appear as keys. Define
sequence by tapping appropriate keys. Factor
is calculated automatically based on widely
accepted value for each base unit.
Enter factor between 15 ng-cm/µL and
150 ng-cm/µL
Dye 1/Dye 2
a
Type
Cy3, 5, 3.5, or 5.5,
Alexa Fluor 488, 546, 555, 594, 647,
or 660
Thermo Scientific NanoDrop One User Guide 41
Select pre-defined dye(s) used to label sample
material, or one that has been added using Dye
Editor.
Page 42

4 Nucleic Acid Applications
Measure Microarray
Setting Available Options Description
Dye 1/Dye 2 Unit picomoles/microliter (pmol/uL),
Select unit for reporting dye concentrations
micromoles (uM), or millimoles (mM)
Analysis
Correction
b
On or off
Enter analysis correction wavelength
in nm or use default value (340 nm)
Corrects sample absorbance measurement for
any offset caused by light scattering particulates
by subtracting absorbance value at specified
analysis correction wavelength from
absorbance value at analysis wavelength.
Corrected value is used to calculate sample
concentration.
Tip: If the sample has a modification that
absorbs light at 340 nm, select a different
correction wavelength or turn off Analysis
Correction.
a
To add a custom dye or edit the list of available dyes, use the Dye/Chromophore Editor.
b
The Analysis Correction affects the calculation for nucleic acid concentration only.
Dye/chromophore editor
Use the Dye/Chromophore Editor to add a custom dye to the list of available dyes in
Microarray Setup or Proteins & Labels Setup. You can also specify which dyes are
available in that list.
To access the Dye/Chromophore Editor:
• from the Home screen, select Settings > Dye Editor
• from the Microarray or Proteins & Labels measurement screen, tap >
Settings > Dye Editor
42 NanoDrop One User Guide Thermo Scientific
Page 43

Dye Editor
4
Nucleic Acid Applications
Measure Microarray
Locked dye (pre-defined;
cannot be edited or deleted)
Tap to add
custom dye
Tap to edit
selected
custom dye
Tap to
delete
selected
custom
dye
Selected dyes (will appear in Dye1 and
Custom dye (user-defined;
can be edited or deleted)
Dye2 lists in Microarray Setup or Proteins
& Labels Setup)
Tap to close Dye
Editor
These operations are available from the Dye/Chromophore Editor:
Add or remove a dye
To add or remove a dye from the Dye1 or Dye2 drop-down list in Microarray Setup or
Proteins & Labels Setup:
– select or deselect corresponding checkbox
Add custom dye
– tap to show New Dye box
– enter unique Name for new dye (tap field to display keyboard, tap Done key
to close keyboard)
Thermo Scientific NanoDrop One User Guide 43
Page 44

4 Nucleic Acid Applications
Measure Microarray
– select default Unit that will be used to display dye concentration
– enter dye’s Extinction Coefficient (or molar absorptivity constant) in
L/mole-cm (typically provided by dye manufacturer)
– specify Wavelength in nm (between 350 nm and 850 nm) that will be used
to measure dye’s absorbance
– specify dye’s correction values at 260 nm and 280 nm
– tap Add Dye
Note To determine dye correction values (if not available from dye
manufacturer):
– use instrument to measure pure dye and note absorbance at 260 nm,
280 nm and at analysis wavelength for dye (see above)
– calculate ratio of A
260/Adye wavelength
and enter that value for 260 nm
Correction
– calculate ratio of A
280/Adye wavelength
and enter that value for 280 nm
Correction
When a custom dye is selected before a measurement, the dye’s absorbance
and concentration values are reported and the corrections are applied to the
measured sample absorbance values, and to the resulting sample
concentrations and purity ratios.
Edit custom dye
Tip Dyes pre-defined in the software cannot be edited.
– tap to select custom dye
– tap
– edit any entries or settings
– tap Save Dye
Delete custom dye
Tip Dyes pre-defined in the software cannot be deleted.
– tap to select custom dye
– tap
NOTICE Deleting a custom dye permanently removes the dye and all
associated information from the software.
44 NanoDrop One User Guide Thermo Scientific
Page 45

Calculations for Microarray Measurements
4
Nucleic Acid Applications
Measure Microarray
As with the other nucleic acid applications,
the Microarray application uses a
modification of the Beer-Lambert equation
to calculate sample concentration where
the extinction coefficient and pathlength
are combined and referred to as a “factor.”
The Microarray application offers six
options (shown at right) for selecting an
appropriate factor for each measured
sample, to be used in conjunction with
Beer’s Law to calculate sample
concentration.
If the factor is known, choose the Custom
Factor option and enter the factor in
ng-cm/µL. Otherwise, choose the option
that best matches the sample solution.
Tip: Ideally, the factor or extinction
coefficient should be determined
empirically using a solution of the study
nucleic acid at a known concentration
using the same buffer.
Available Options for Factors
• dsDNA (factor = 50 ng-cm/µL)
• ssDNA (factor = 33 ng-cm/µL)
• RNA (factor = 40 ng-cm/µL)
• Oligo DNA (calculated from user entered DNA
nucleotide sequence)
• Oligo RNA (calculated from user entered RNA
nucleotide sequence)
• Custom Factor (user entered factor between
15 ng-cm/µL and 150 ng-cm/µL
Note: See Sample Type for more information.
Thermo Scientific NanoDrop One User Guide 45
Page 46

4 Nucleic Acid Applications
Measure Microarray
Calculated nucleic acid concentrations are
based on the absorbance value at 260 nm,
the factor used and the sample pathlength.
A single-point baseline correction (or
analysis correction) may also be applied.
Concentration is reported in mass units.
Calculators are available on the Internet to
convert concentration from mass to molar
units based on sample sequence.
Absorbance values at 260 nm, 280 nm and
sometimes 230 nm are used to calculate
purity ratios for the measured nucleic acid
samples. Purity ratios are sensitive to the
presence of contaminants in the sample,
such as residual solvents and reagents
typically used during sample purification.
Measured Values
A260 absorbance
Note: The absorbance value at 850 nm is subtracted from all
wavelengths in the spectrum. As a result, the absorbance at
850 nm is zero in the displayed spectra. Also, for
micro-volume absorbance measurements and
measurements taken with nonstandard (other than 10 mm)
cuvettes, the spectra are normalized to a 10 mm pathlength
equivalent.
• Nucleic acid absorbance values for all Microarray sample
types are measured at 260 nm using the 850-corrected
and normalized spectrum.
• If Analysis Correction is selected, the absorbance value at
the correction wavelength is subtracted from the
absorbance at 260 nm.
• If one or more dyes are selected, the dye correction
values at 260 nm are also subtracted from the
absorbance at 260 nm.
• The final corrected absorbance at 260 nm is reported
and used to calculate sample concentration.
A280 absorbance
• 850-corrected and normalized absorbance value at
280 nm (minus the A280 dye correction) is used to
calculate an A260/A280 ratio.
46 NanoDrop One User Guide Thermo Scientific
Page 47

4
Nucleic Acid Applications
Measure Microarray
Dye concentrations are calculated from the
absorbance value at the dye’s analysis
wavelength, the dye’s extinction coefficient,
and the sample pathlength. A sloped-line
dye correction may also be used.
Dye absorbance
• Dye absorbance values are measured at specific
wavelengths. See Dye/Chromophore Editor for analysis
wavelengths used.
• If Sloping Dye Correction is selected, a linear baseline is
drawn between 400 nm and 850 nm and, for each dye,
the absorbance value of the sloping baseline is
subtracted from the absorbance value at each dye’s
analysis wavelength. Baseline-corrected dye absorbance
values are reported and used to calculate dye
concentrations.
Dye correction
• Pre-defined dyes have known correction values for A260
and A280. See Dye/Chromophore Editor for correction
values used.
• A260 dye corrections are subtracted from the A260
absorbance value used to calculate nucleic acid
concentration, and from the A260 absorbance value
used to calculate the A260/A280 purity ratio.
Sample Pathlength
• For micro-volume measurements, the software selects
the optimal pathlength (between 1.0 mm and 0.03 mm)
based on sample absorbance at the analysis wavelength.
• For cuvette measurements, pathlength is determined by
the cuvette Pathlength setting in the software (see
General Settings).
• Displayed spectra and absorbance values are normalized
to a 10 mm pathlength equivalent.
Thermo Scientific NanoDrop One User Guide 47
Page 48

4 Nucleic Acid Applications
Measure Microarray
Reported Values
• Nucleic acid concentration. Reported in selected
unit (i.e., ng/µL, µg/uL or µg/mL). Calculations are based
on modified Beer’s Law equation using corrected nucleic
acid absorbance value.
• A260/A280 purity ratio. Ratio of corrected
absorbance at 260 nm to corrected absorbance at
280 nm. An A260/A280 purity ratio of ~1.8 is generally
accepted as “pure” for DNA (~2.0 for RNA). Acidic
solutions may under represent the reported value by
0.2-0.3; the opposite is true for basic solutions.
• Dye1/Dye2 concentration. Reported in pmol/µL.
Calculations are based on Beer’s Law equation using
(sloping) baseline-corrected dye absorbance value(s).
Note: Although purity ratios are important indicators of
sample quality, the best indicator of DNA or RNA quality is
functionality in the downstream application of interest (e.g.,
microarray).
Related Topics
• Calculations for Nucleic Acid Measurements
48 NanoDrop One User Guide Thermo Scientific
Page 49

Measure using a Custom Factor
Measures the concentration of
purified nucleic acids using a
custom factor for the
calculations.
Measure using Custom Factor
Reported Results
Settings
Detection Limits
Calculations
Measure Nucleic Acid using a Custom Factor
4
Nucleic Acid Applications
Measure using a Custom Factor
Use the Custom Factor application to quantify purified DNA or RNA samples that
absorb at 260 nm with a user-defined extinction coefficient or factor. The application
reports nucleic acid concentration and two absorbance ratios (A260/A280 and
A260/A230). A single-point baseline correction can also be used.
To measure nucleic acid samples using a custom factor
NOTICE
• Do not use a squirt or spray bottle on or near the instrument as liquids will
flow into the instrument and may cause permanent damage.
• Do not use hydrofluoric acid (HF) on the pedestals. Fluoride ions will
permanently damage the quartz fiber optic cables.
Before you begin...
Before taking pedestal measurements with the NanoDrop One instrument, lift the
instrument arm and clean the upper and lower pedestals. At a minimum, wipe the
pedestals with a new laboratory wipe. For more information, see Cleaning the
Pedestals.
To measure using a custom factor
1. From the Home screen, select the Nucleic Acids tab and select Custom
Factor.
Thermo Scientific NanoDrop One User Guide 49
Page 50

4 Nucleic Acid Applications
Measure using a Custom Factor
2. Enter the factor to be used for the calculations and specify a baseline correction
3. Pipette 1–2 µL blanking solution onto the lower pedestal and lower the arm, or
4. Select Blank and wait for the measurement to complete.
5. Lift the arm and clean both pedestals with a new laboratory wipe, or remove the
6. Pipette 1-2 µL sample solution onto the pedestal and lower the arm, or insert the
7. Start the sample measurement:
if desired.
insert the blanking cuvette into the cuvette holder.
Tip: If using a cuvette, make sure to align the cuvette light path with the
instrument light path.
Tip: If Auto-Blank is On, the blank measurement starts automatically after you
lower the arm. (This option is not available for cuvette measurements.)
blanking cuvette.
sample cuvette into the cuvette holder.
– Pedestal: If Auto-Measure is On, lower arm; if Auto-Measure is off, lower arm
and tap Measure.
– Cuvette: Tap Measure.
When the sample measurement is completed, the spectrum and reported values
are displayed (see the next section).
8. When you are finished measuring samples, tap End Experiment.
9. Lift the arm and clean both pedestals with a new wipe, or remove the sample
cuvette.
Typical nucleic acid spectrum
50 NanoDrop One User Guide Thermo Scientific
Page 51

Related Topics
• Measure a Micro-Volume Sample
• Measure a Sample Using a Cuvette
• Best Practices for Micro-Volume Measurements
• Best Practices for Cuvette Measurements
• Prepare Samples and Blanks
• Basic Instrument Operations
Custom Factor Reported Results
For each measured sample, this application shows the absorbance spectrum and a
summary of the results. Here is an example:
4
Nucleic Acid Applications
Measure using a Custom Factor
Note The Custom Factor measurement screen is identical to the measurement
screen for the other nucleic acid applications except the Custom Factor is
reported in the lower left corner (see image below).
Custom factor used to calculate
nucleic acid concentration
Thermo Scientific NanoDrop One User Guide 51
Page 52

4 Nucleic Acid Applications
Measure using a Custom Factor
Related Topics
• Basic Instrument Operations
• Nucleic Acid Reported Results
• Nucleic Acid Calculations
Settings for Nucleic Acid Measurements using a Custom Factor
To show the Custom Factor settings, from the local control, tap > Custom
Factor Setup.
When using the PC Control software, from the Custom Factor measurement screen,
select the settings icon to view the Custom Factor Setup.
Setting Available Options Description
Custom Factor Enter an integer value
between 15 ng-cm/µL
and 150 ng-cm/µL
Baseline Correction On or off
Enter baseline
correction wavelength
in nm or use default
value (340 nm)
Constant used to calculate nucleic acid concentration in
modified Beer’s Law equation. Based on extinction
coefficient and pathlength:
f = 1/(
where:
f= factor
260
* b))
= molar extinction coefficient at 260 nm in ng-cm/µL
b = sample pathlength in cm (1 cm for nucleic acids
measured with the NanoDrop One instruments)
Optional user-defined baseline correction. Can
be used to correct for any offset caused by light scattering
particulates by subtracting measured absorbance at
specified baseline correction wavelength from absorbance
values at all wavelengths in sample spectrum. As a result,
absorbance of sample spectrum is zero at specified
baseline correction wavelength.
NOTE: Baseline correction is selected from the
measurement screen of the PC control software and is not
shown in the Custom Factor Setup.
Related Topics
• Instrument Settings
52 NanoDrop One User Guide Thermo Scientific
Page 53

4
Nucleic Acid Applications
Measure using a Custom Factor
Detection Limits for Nucleic Acid Measurements using a Custom Factor
The lower detection limits and reproducibility specifications for nucleic acids are
provided here. The upper detection limits are dependent on the upper absorbance
limit of the instrument and the user-defined extinction coefficients.
To calculate upper detection limits for nucleic acid samples
To calculate upper detection limits in ng/µL, use the following equation:
(upper absorbance limit
For example, for a sample measurement using an extinction coefficient of 55, the
equation looks like this:
(550 AU * 55 ng-cm/µL) = 30,250 ng/µL
Note For measurements with 10 mm pathlength cuvettes, the upper
absorbance limit is 1.5 AU, which is approximately 75 ng/µL for dsDNA.
instrumen
* extinction coefficient
t
sample
)
Related Topics
• Detection Limits for All Applications
Thermo Scientific NanoDrop One User Guide 53
Page 54

4
This page is intentionally blank.
54 NanoDrop One User Guide Thermo Scientific
Page 55

Measure Oligo DNA or Oligo RNA
Measures the concentration of purified
ssDNA or RNA oligonucleotides that
absorb at 260 nm.
Measure Oligo DNA or RNA
Reported Results
Settings
Detection Limits
Calculations
Measure Oligo DNA or Oligo RNA
4
Nucleic Acid Applications
Measure Oligo DNA or Oligo RNA
Use the Oligo DNA and Oligo RNA applications to quantify oligonucleotides that
absorb at 260 nm. Molar extinction coefficients are calculated automatically based
on the user-defined base sequence of the sample. These applications report nucleic
acid concentration and two absorbance ratios (A260/A280 and A260/A230). A
single-point baseline correction can also be used.
Note If the oligonucleotide has been modified, for example with a fluorophore
dye, check with the oligo manufacturer to determine if the modification
contributes absorbance at 260 nm. If it does, we recommend using the
Microarray application to quantify nucleic acid concentration. The Microarray
application includes a correction to remove any absorbance contribution due to
the dye from the oligo quantification result.
To measure Oligo DNA or Oligo RNA samples
NOTICE
• Do not use a squirt or spray bottle on or near the instrument as liquids will
flow into the instrument and may cause permanent damage.
• Do not use hydrofluoric acid (HF) on the pedestals. Fluoride ions will
permanently damage the quartz fiber optic cables.
Before you begin...
Before taking pedestal measurements with the NanoDrop One instrument, lift the
instrument arm and clean the upper and lower pedestals. At a minimum, wipe the
pedestals with a new laboratory wipe. For more information, see Cleaning the
Pedestals.
Thermo Scientific NanoDrop One User Guide 55
Page 56

4 Nucleic Acid Applications
Measure Oligo DNA or Oligo RNA
To measure an oligonucleotide sample
1. From the Home screen, select the Nucleic Acids tab and select either Oligo
DNA or Oligo RNA, as needed.
2. Specify the Oligo base sequence and a baseline correction if desired.
3. Pipette 1–2 µL blanking solution onto the lower pedestal and lower the arm, or
insert the blanking cuvette into the cuvette holder.
Tip: If using a cuvette, make sure to align the cuvette light path with the
instrument light path.
4. Tap Blank and wait for the measurement to complete.
Tip: If Auto-Blank is On, the blank measurement starts automatically after you
lower the arm. (This option is not available for cuvette measurements.)
5. Lift the arm and clean both pedestals with a new laboratory wipe, or remove the
blanking cuvette.
6. Pipette 1-2 µL sample solution onto the pedestal and lower the arm, or insert the
sample cuvette into the cuvette holder.
7. Start the sample measurement:
– Pedestal: If Auto-Measure is On, lower arm; if Auto-Measure is off, lower arm
and tap Measure.
– Cuvette: Tap Measure.
When the sample measurement is completed, the spectrum and reported values
are displayed (see the next section).
8. When you are finished measuring samples, tap End Experiment.
9. Lift the arm and clean both pedestals with a new wipe, or remove the sample
cuvette.
56 NanoDrop One User Guide Thermo Scientific
Page 57

4
Measure Oligo DNA or Oligo RNA
Example Oligo DNA spectrum
Nucleic Acid Applications
Related Topics
• Best Practices for Nucleic Acid Measurements
• Measure a Micro-Volume Sample
• Measure a Sample Using a Cuvette
• Best Practices for Micro-Volume Measurements
• Best Practices for Cuvette Measurements
• Prepare Samples and Blanks
• Basic Instrument Operations
Thermo Scientific NanoDrop One User Guide 57
Page 58

4 Nucleic Acid Applications
Measure Oligo DNA or Oligo RNA
Oligo Reported Results
Oligo DNA measurement screen (Local Control)
For each measured sample, the Oligo DNA and Oligo RNA applications show the UV
absorbance spectrum and a summary of the results. Here is an example:
Menu of options;
tap to open
tap to edit
Nucleic acid
concentration
UV spectrum
Tap to select unitSample name;
Purity ratios
Tap row
to select
sample
and
update
spectrum;
tap more
rows to
overlay up
to five
spectra.
Press
and hold
sample
1
row to
view
measure
ment
details.
Pinch and zoom to
adjust axes; double-tap
to reset
1
Measured oligo: TTT TTT TTT TTT TTT TTT TTT TTT
Swipe screen left to view
table with more
measurement results
Tap to end
experiment and
export data
Drag tab down/up
to see more/less
sample data
Note Micro-volume absorbance measurements and measurements taken with
nonstandard cuvettes are normalized to a 10.0 mm pathlength equivalent.
58 NanoDrop One User Guide Thermo Scientific
Page 59

Oligo DNA and Oligo RNA reported values
The initial screen that appears after each measurement (see previous image) shows
a summary of the reported values. To view all reported values, press and hold the
sample row. Here is an example:
• sample details (application and sampling method used, i.e., pedestal or cuvette)
• sample name
• created on (date sample measurement was taken)
• nucleic acid concentration
• A260/A280
• A260/A230
• A260
• A280
4
Nucleic Acid Applications
Measure Oligo DNA or Oligo RNA
• factor
• oligo sequence
• baseline correction
• stirrer status
Note The five nucleotides that comprise DNA and RNA exhibit widely varying
A260/A280 ratios. See Oligo Purity Ratios for more information.
Thermo Scientific NanoDrop One User Guide 59
Page 60

4 Nucleic Acid Applications
Measure Oligo DNA or Oligo RNA
Oligo DNA and RNA measurement screen (PC Control)
For each measured sample, the Oligo DNA and Oligo RNA applications show the UV
absorbance spectrum reported results. Here is an example:
Click to select cuvette or
pedestal measurement
Sample name;
Click to edit
Oligo
RNA/RNA
Setup
Run
Blank
Measure
sample
Click to end
experiment and
export data
Menu of options;
click to open
Nucleic acid
concentration
UV spectrum
Purity ratios
Related Topics
• Basic Instrument Operations
• Oligo Calculations
Click and Drag
to adjust axes;
double-click to
reset
Click to
select unit
Click row to
select sample
and update
spectrum; click
more rows to
overlay spectra
Settings for Oligo DNA and Oligo RNA Measurements
The Oligo setup screen appears after you select the Oligo DNA or Oligo RNA
application from the Nucleic Acids tab on the Home screen.
In local instrument control, from the Oligo measurement screen, tap >
Oligo DNA Setup (or Oligo RNA Setup).
60 NanoDrop One User Guide Thermo Scientific
Page 61

From the PC control software, from the Oligo DNA or RNA measurement screen,
select the settings icon to view Oligo DNA Setup or Oligo RNA Setup.
Setting Available Options Description
4
Nucleic Acid Applications
Measure Oligo DNA or Oligo RNA
Oligo Base
Sequence
for DNA: Use the
G, A, T and C keys to
specify the DNA base
sequence
for RNA: Use the
G, A, U and C keys to
specify the RNA base
sequence
Specify your DNA or RNA base sequence.
Tap or click the corresponding keys:
Add guanine
Add adenine
Add thymine (DNA) or
uracil (RNA)
From the PC control software, you can also enter base
sequence using the keyboard, or by copy and pasting a
sequence from another application.
Each time a base is added to the sequence, the software
calculates the following:
• Factor. Constant used to calculate oligonucleotide
concentration in modified Beer’s Law equation. Based on
extinction coefficient and pathlength:
Add cytosine
Remove most
recent base
(seen in local
instrument
control)
f = 1/(
where:
f= factor
260
* b)
= molar extinction coefficient at 260 nm in ng-cm/µL
b = sample pathlength in cm (0.1 cm for nucleic acids
measured with the NanoDrop One instrument)
Thermo Scientific NanoDrop One User Guide 61
Page 62

4 Nucleic Acid Applications
Measure Oligo DNA or Oligo RNA
Setting Available Options Description
• Molecular Weight of oligo calculated from user-defined
base sequence.
• Number of Bases entered.
• Molar Ext. Coefficient (260 nm). Molar extinction
coefficient of oligo (in ng-cm/µL) at 260 nm calculated from
entered base sequence.
• %GC. Percentage of guanine and cytosine residues in
total number of bases entered.
Baseline
Correction
On or off
Enter baseline
correction
wavelength in nm or
use default value
(340 nm)
Corrects for any offset caused by light scattering particulates
by subtracting measured absorbance at specified baseline
correction wavelength from absorbance values at all
wavelengths in sample spectrum. As a result, absorbance of
sample spectrum is zero at specified baseline correction
wavelength.
Tip: If the sample has a modification that absorbs light at
340 nm, select a different correction wavelength or turn off
Baseline Correction.
Related Topics
• Instrument Settings
Detection Limits for Oligo DNA and Oligo RNA Measurements
The lower detection limits and reproducibility specifications for the oligonucleotide
sample types (ssDNA and RNA) are provided here. The upper detection limits are
dependent on the upper absorbance limit of the instrument and the extinction
coefficients for the user-defined base sequences.
To calculate upper detection limits for nucleic acid samples
To calculate upper detection limits in ng/µL, use the following equation:
(upper absorbance limit
For example, for a sample measurement using an extinction coefficient of 55, the
equation looks like this:
(550 AU * 55 ng-cm/µL) = 30,250 ng/µL
Note For measurements with 10 mm pathlength cuvettes, the upper
absorbance limit is 1.5 AU, which is approximately 75 ng/µL for dsDNA.
62 NanoDrop One User Guide Thermo Scientific
instrumen
* extinction coefficient
t
sample
)
Page 63

4
Nucleic Acid Applications
Measure Oligo DNA or Oligo RNA
Calculations for Oligo DNA and Oligo RNA Measurements
As with the other nucleic acid applications, the
Oligo applications use the Beer-Lambert
equation to correlate absorbance with
concentration based on the sample’s extinction
coefficient and pathlength. Because
oligonucleotides are short, single-stranded
molecules (or longer molecules of repeating
sequences), their spectrum and extinction
coefficient () are closely dependent on base
composition and sequence.
(The generally accepted extinction coefficients
and factors for single-stranded DNA and RNA
provide a reasonable estimate for natural,
essentially randomized, sequences but not for
short, synthetic oligo sequences.) To ensure the
most accurate results, we use the exact value of
to calculate oligonucleotide concentration.
260
The NanoDrop software allows you to specify
the base sequence of an oligonucleotide before
it is measured. For any entered base sequence,
the software uses the equation at the right to
calculate the extinction coefficient.
Tip: The extinction coefficient is wavelength
specific for each oligonucleotide and can be
affected by buffer type, ionic strength and pH.
Extinction Coefficients for Oligonucleotides
The software uses the nearest neighbor method and the
following formula to calculate molar extinction
coefficients for specific oligonucleotide base sequences:
N
2
+
3
1
where:
260
N1–
=
1
N1–
–
1
2
= molar extinction coefficient in L/mole-cm
=
1
nearest neighbor
=
2
individual bases
=
3
modifications, such as fluorescent dyes
Thermo Scientific NanoDrop One User Guide 63
Page 64

4 Nucleic Acid Applications
Measure Oligo DNA or Oligo RNA
Calculated nucleic acid concentrations are
based on the absorbance value at 260 nm, the
factor used and the sample pathlength. A
single-point baseline correction (or analysis
correction) may also be applied.
Concentration is reported in mass units.
Calculators are available on the Internet to
convert concentration from mass to molar units
based on sample sequence.
Absorbance values at 260 nm, 280 nm and
sometimes 230 nm are used to calculate purity
ratios for the measured nucleic acid samples.
Purity ratios are sensitive to the presence of
contaminants in the sample, such as residual
solvents and reagents typically used during
sample purification.
Measured Values
A260 absorbance
Note: For micro-volume absorbance measurements and
measurements taken with nonstandard (other than
10 mm) cuvettes, the spectra are normalized to a 10 mm
pathlength equivalent.
• Nucleic acid absorbance values are measured at
260 nm using the normalized spectrum. This is the
reported A260 value if Baseline Correction is not
selected.
• If Baseline Correction is selected, the absorbance
value at the correction wavelength is subtracted from
the sample absorbance at 260 nm. The corrected
absorbance at 260 nm is reported and used to
calculate nucleic acid concentration.
A230, A280 absorbance
• Normalized absorbance values at 230 nm, 260 nm
and 280 nm are used to calculate A260/A230 and
A260/A280 ratios.
Sample Pathlength
• For micro-volume measurements, the software
selects the optimal pathlength (between 1.0 mm and
0.03 mm) based on sample absorbance at the
analysis wavelength.
• For cuvette measurements, pathlength is determined
by the cuvette Pathlength setting in the software (see
General Settings).
• Displayed spectra and absorbance values are
normalized to a 10 mm pathlength equivalent.
64 NanoDrop One User Guide Thermo Scientific
Page 65

4
Nucleic Acid Applications
Measure Oligo DNA or Oligo RNA
The five nucleotides that comprise DNA and
RNA exhibit widely varying A260/A280 ratios.
Estimated A260/A280 ratios for each
independently measured nucleotide are
provided below:
Guanine: 1.15
Adenine: 4.50
Cytosine: 1.51
Uracil: 4.00
Thymine: 1.47
The A260/A280 ratio for a specific nucleic acid
sequence is approximately equal to the
weighted average of the A260/A280 ratios for
the four nucleotides present.
Note: RNA will typically have a higher 260/280
ratio due to the higher ratio of Uracil compared
to that of Thymine.
Reported Values
• Nucleic acid concentration. Reported in
selected unit (i.e., ng/µL, µg/uL or µg/mL).
Calculations are based on modified Beer’s Law
equation using corrected nucleic acid absorbance
value.
• A260/A280 purity ratio. Ratio of corrected
absorbance at 260 nm to corrected absorbance at
280 nm.
• A260/A230 purity ratio. Ratio of corrected
absorbance at 260 nm to corrected absorbance at
230 nm.
Note: The traditional purity ratios (A260/A280 and
A260/A230), which are used as indicators of the
presence of various contaminants in nucleic acid
samples, do not apply for oligonucleotides because the
shapes of their spectra are highly dependent on their
base compositions. See side bar for more information.
Related Topics
• Calculations for Nucleic Acid Measurements
Thermo Scientific NanoDrop One User Guide 65
Page 66

4
This page is intentionally blank.
66 NanoDrop One User Guide Thermo Scientific
Page 67

Protein Applications
Measure Protein A280
Measures the concentration of purified
protein samples that absorb at 280 nm.
Measure A280 Proteins
Reported Results
Settings
Detection Limits
5
Calculations
Measure Protein Concentration at A280
Use the Protein A280 application to quantify purified protein samples that contain
amino acids such as tryptophan or tyrosine, or cys-cys disulfide bonds, which
exhibit absorbance at 280 nm. This application reports protein concentration
measured at 280 nm and one absorbance ratio (A260/A280). A single-point baseline
correction can also be used. This application does not require a standard curve.
Note If your samples contain mainly peptide bonds and little or no amino acids,
use the Protein A205 application instead of Protein A280.
To measure Protein A280 samples
NOTICE
• Do not use a squirt or spray bottle on or near the instrument as liquids will
flow into the instrument and may cause permanent damage.
• Do not use hydrofluoric acid (HF) on the pedestals. Fluoride ions will
permanently damage the quartz fiber optic cables.
Thermo Scientific NanoDrop One User Guide 67
Page 68

5 Protein Applications
Measure Protein A280
Before you begin...
Before taking pedestal measurements with the NanoDrop One instrument, lift the
instrument arm and clean the upper and lower pedestals. At a minimum, wipe the
pedestals with a new laboratory wipe. For more information, see Cleaning the
Pedestals.
To measure a Protein A280 sample
1. From the Home screen, select the Proteins tab and select Protein A280.
2. Specify a sample type and baseline correction if desired.
3. Pipette 1–2 µL blanking solution onto the lower pedestal and lower the arm, or
insert the blanking cuvette into the cuvette holder.
Tip: If using a cuvette, make sure to align the cuvette light path with the
instrument light path.
4. Tap Blank and wait for the measurement to complete.
Tip: If Auto-Blank is On, the blank measurement starts automatically after you
lower the arm. (This option is not available for cuvette measurements.)
5. Lift the arm and clean both pedestals with a new laboratory wipe, or remove the
blanking cuvette.
6. Pipette 2 µL sample solution onto the pedestal and lower the arm, or insert the
sample cuvette into the cuvette holder.
7. Start the sample measurement:
– Pedestal: If Auto-Measure is On, lower arm; if Auto-Measure is off, lower arm
and tap Measure.
– Cuvette: Tap Measure
When the sample measurement is completed, the spectrum and reported values
are displayed (see the next section).
8. When you are finished measuring samples, tap End Experiment.
9. Lift the arm and clean both pedestals with a new wipe, or remove the sample
cuvette.
68 NanoDrop One User Guide Thermo Scientific
Page 69

High concentration BSA sample
5
Protein Applications
Measure Protein A280
Low concentration BSA sample
Best practices for protein measurements
• Isolate and purify protein samples before measurement to remove impurities.
Depending on the sample, impurities could include DNA, RNA and some buffer
components. See Preparing Samples for more information.
Note Extraction reagents that contribute absorbance between 200 nm and
280 nm will affect measurement results if present in samples (even residual
amounts).
• Ensure the sample absorbance is within the instrument’s absorbance detection
limits.
Thermo Scientific NanoDrop One User Guide 69
Page 70

5 Protein Applications
Measure Protein A280
• Choosing a blank:
– For the Protein A280, Protein A205, and Proteins & Labels applications,
blank with the same buffer solution used to resuspend the analyte of interest.
The blanking solution should be a similar pH and ionic strength as the analyte
solution.
– For the Protein BCA, Protein Bradford, and Protein Lowry applications, blank
with deionized water (DI H
2
O).
– For the Protein Pierce 660 application, blank with the reference solution used
to make the standard curve (reference solution should contain none of the
standard protein stock). For more information, see Working with standard
curves.
• Run a blanking cycle to assess the absorbance contribution of your buffer
solution. If the buffer exhibits strong absorbance at or near the analysis
wavelength (typically 280 nm or 205 nm), you may need to choose a different
buffer or application, such as a colorimetric assay (for example, BCA or
Pierce 660). See Choosing and Measuring a Blank for more information.
Note Buffers such as Triton X, RIPA, and NDSB contribute significant
absorbance and are not compatible with direct A280 or A205
measurements.
• For micro-volume measurements:
– Ensure pedestal surfaces are properly cleaned and conditioned. (Proteins
tend to stick to pedestal surfaces.)
– Gently (but thoroughly) vortex samples before taking a measurement. Avoid
introducing bubbles when mixing and pipetting.
– Follow best practices for micro-volume measurements.
– Use a 2 µL sample volume. See Recommended Sample Volumes for more
information.
C
• For cuvette measurements (NanoDrop One
instruments only), use compatible
cuvettes and follow best practices for cuvette measurements.
Related Topics
• Best practices for protein measurements
• Measure a Micro-Volume Sample
• Measure a Sample Using a Cuvette
• Prepare Samples and Blanks
• Basic Instrument Operations
70 NanoDrop One User Guide Thermo Scientific
Page 71

Protein A280 Reported Results
Protein A280 measurement screen (Local Control)
For each measured sample, this application shows the absorbance spectrum and a
summary of the results. Here is an example of the Local control screen:
5
Protein Applications
Measure Protein A280
tap to open
Sample name;
tap to edit
UV spectrum
Protein
concentration
Tap to select unitMenu of options;
Absorbance at
280 nm
Purity ratio
Tap r ow to select
sample and
update spectrum;
tap more rows to
overlay up to five
spectra. Press
and hold sample
row to view
measurement
details.
Pinch and zoom to
adjust axes; double-tap
to reset
Swipe screen left to view
table with more
measurement results
Tap to end
experiment and
export data
Drag tab down/up
to see more/less
sample data
Note Micro-volume absorbance measurements and measurements taken with
nonstandard cuvettes are normalized to a 10.0 mm pathlength equivalent.
Thermo Scientific NanoDrop One User Guide 71
Page 72

5 Protein Applications
Measure Protein A280
Protein A280 reported values
The initial screen that appears after each measurement (see previous image) shows
a summary of the reported values. To view all reported values, press and hold the
sample row. Here is an example:
Application
Baseline correction
wavelength
Sampling
method
Baseline correction
absorbance
Sample name;
tap to edit
Date/time
measured
Protein conc.
Absorbance at
280 nm
Purity ratio
Sample type
Related Topics
• Basic Instrument Operations
• Protein A280 Calculations
Settings for Protein A280 Measurements
To show the Protein A280 settings, in local instrument control, from the Protein A280
measurement screen, tap > Protein A280 Setup.
From the PC control software, from the Protein A280 measurement screen, select
the settings icon to view Protein A280 Setup.
72 NanoDrop One User Guide Thermo Scientific
Page 73

Protein A280 settings
The Protein A280 application provides a variety of sample type options for purified
protein analysis.
Each sample type applies a unique extinction coefficient to the protein calculations. If
the extinction coefficient of the sample is known, choose the
(mass) option and enter the value. Otherwise, calculate the extinction coefficient or
choose the option that best matches the sample solution. If you only need a rough
estimate of protein concentration and the sample extinction coefficient is unknown,
select the 1 Abs=1 mg/mL sample type option.
5
Protein Applications
Measure Protein A280
+ MW (molar) or 1%
Tip Ideally, the extinction coefficient should be determined empirically using a
solution of the study protein at a known concentration using the same buffer.
Setting Available Options
Sample
a
type
1 Abs = 1 mg/mL General reference Recommended when extinction coefficient is
BSA 6.7 Calculates BSA (Bovine Serum Albumin)
Mass Ext.
Coefficient
(L/gm-cm)
Description
unknown and rough estimate of protein
concentration is acceptable for a solution with
no other interfering substances. Assumes
0.1% (1 mg/mL) protein solution produces
1.0A at 280 nm (where pathlength is 10 mm),
1% = 10.
i.e.,
protein concentration using mass extinction
coefficient (
) of 6.7 L/gm-cm at 280 nm for
1% (i.e., 10 mg/mL) BSA solution. Assuming
MW is 66,400 daltons (Da), molar extinction
coefficient at 280 nm for BSA is approximately
43,824 M
-1cm-1
.
Thermo Scientific NanoDrop One User Guide 73
Page 74

5 Protein Applications
Measure Protein A280
Setting Available Options
IgG 13.7 Suitable for most mammalian antibodies (i.e.,
Lysozyme 26.4 Calculates lysozyme protein concentration
Mass Ext.
Coefficient
(L/gm-cm)
Description
immunoglobulin G or IgG). Calculates protein
concentration using mass extinction
coefficient (
1% (i.e., 10 mg/mL) IgG solution. Assuming
MW is 150,000 Da, molar extinction coefficient
at 280 nm for IgG is approximately
210,000 M
using mass extinction coefficient (
26.4 L/gm-cm at 280 nm for
1% (i.e., 10 mg/mL) lysozyme solution.
Assumes molar extinction coefficient for egg
white lysozyme ranges between
36,000 M
) of 13.7 L/gm-cm at 280 nm for
-1cm-1
.
) of
-1cm-1
and 39,000 M-1cm-1.
Other protein
+ MW)
(
Other protein
1%)
(
a
To add or edit a custom protein, use Protein Editor.
User entered molar
extinction coefficient
and molecular
weight
User entered mass
extinction coefficient
Assumes protein has known molar extinction
coefficient (
where:
(
molar
Enter MW in kiloDaltons (kDa) and molar
extinction coefficient (
1000 (i.e.,
with molar extinction coefficient of 210,000
-1cm-1
M
Assumes protein has known mass extinction
coefficient (). Enter mass extinction
coefficient in L/gm-cm for 10 mg/mL (
protein solution.
) and molecular weight (MW),
)*10=(
percent
)*(MW
) in M
protein
-1cm-1
)
/1000). For example, for protein
, enter 210.
divided by
1%)
74 NanoDrop One User Guide Thermo Scientific
Page 75

Setting Available Options
Mass Ext.
Coefficient
(L/gm-cm)
Description
5
Protein Applications
Measure Protein A280
Baseline
On or off
Correction
Enter baseline
correction
wavelength in nm or
use default value
(340 nm)
Protein editor
N/A Corrects for any offset caused by light
scattering particulates by subtracting
measured absorbance at specified baseline
correction wavelength from absorbance
values at all wavelengths in sample spectrum.
As a result, absorbance of sample spectrum is
zero at specified baseline correction
wavelength.
Tip: If the sample has a modification that
absorbs light at 340 nm, select a different
correction wavelength or turn off Baseline
Correction.
Use the Protein Editor to add a custom protein to the list of available protein sample
types in Protein A280 Setup and Proteins & Labels Setup.
To access the Protein Editor:
• From the PC control software Home screen, select Settings >
Protein Editor.
PC Control software Protein Editor
Click to add
custom protein
Click to edit
selected custom
protein
Click to delete
selected custom
Custom proteins (will appear in Sample Type list in
Protein A280 Setup and Proteins & Labels Setup)
Thermo Scientific NanoDrop One User Guide 75
protein
Page 76

5 Protein Applications
Measure Protein A280
• From the local control Home screen, tap > Protein Editor. Alternatively,
from the Protein A280 or Proteins & Labels measurement screen, tap >
Settings > Protein Editor.
Local Control Protein Editor
Tap to add custom protein
Custom proteins (will appear in Sample
Type list in Protein A280 Setup and
Proteins & Labels Setup)
Tap to edit
selected custom
protein
Tap to delete
selected custom
protein
Tap to close Protein Editor
These operations are available from the Protein Editor:
Add custom protein
1. In Protein Editor, select or to show the New Protein Type
box.
2. Enter a unique Name for the new protein (from local control, tap field to display
keyboard, tap Done key to close keyboard).
3. Enter a Description for the new protein.
4. Specify whether to enter Molar Extinction coefficient or Mass Extinction
coefficient for custom protein.
– If Mass Extinction coefficient is selected, enter mass extinction
coefficient in L/gm-cm for 10 mg/mL (
76 NanoDrop One User Guide Thermo Scientific
1%) protein solution.
Page 77

5
Protein Applications
Measure Protein A280
Tap a field to show keyboard; to
close, tap Done key
– if Molar Extinction is selected,
– enter molar extinction coefficient (
/1000). For example, for protein with molar extinction coefficient
is,
of 210,000 M
-1cm-1
, enter 210.
) in M
-1cm-1
divided by 1000 (that
– Enter molecular weight (MW) in kiloDaltons (kDa)
5. From local control, tap OK to close the New Protein Type box. From PC
control software, select Save.
6. Enter your password to sign the changes if prompted.
Enter your
password to
sign the
changes
The new custom protein appears in the Type list in Protein A280 Setup and
Proteins & Labels Setup.
Thermo Scientific NanoDrop One User Guide 77
Page 78

5 Protein Applications
Measure Protein A280
Edit custom protein
1. In Protein Editor, tap to select custom protein
2. Tap to show the Edit Protein Type box
3. Edit any entries or settings
4. Tap OK
Delete custom protein
1. In Protein Editor, tap to select a custom protein to delete
2. Tap
Note Deleting a custom protein permanently removes the protein and all
associated information from the software.
Detection Limits for Protein A280 Measurements
Detection limits and reproducibility specifications for purified BSA proteins are
provided here. The BSA lower detection limit and reproducibility values apply to any
protein sample type. The upper detection limits are dependent on the upper
absorbance limit of the instrument and the sample’s extinction coefficient.
To calculate upper detection limits for other (non-BSA) protein sample types
To calculate upper detection limits in ng/µL for proteins, use the following equation:
(upper absorbance limit
For example, if the sample’s mass extinction coefficient at 280 nm is 6.7 for a 1%
(10 mg/mL) solution, the equation looks like this:
(550 / 6.7) * 10 = 824.6 (or ~825)
instrumen
/mass extinction coefficient
t
sample
) * 10
78 NanoDrop One User Guide Thermo Scientific
Page 79

Calculations for Protein A280 Measurements
5
Protein Applications
Measure Protein A280
The Protein A280 application uses the
Beer-Lambert equation to correlate
absorbance with concentration. Solving
Beer’s law for concentration yields the
equation at the right.
The extinction coefficient of a peptide or
protein is related to its tryptophan (W),
tyrosin (Y) and cysteine (C) amino acid
composition.
Tip: The extinction coefficient is
wavelength specific for each protein and
can be affected by buffer type, ionic
strength and pH.
Beer-Lambert Equation (solved for concentration)
c = A / ( * b)
where:
A = UV absorbance in absorbance units (AU)
= wavelength-dependent molar absorptivity coefficient (or
extinction coefficient) in liter/mol-cm
b = pathlength in cm
c = analyte concentration in moles/liter or molarity (M)
Note: Dividing the measured absorbance of a sample
solution by its molar extinction coefficient yields the molar
concentration of the sample. See Published Extinction
Coefficients for more information regarding molar vs. mass
concentration values.
Extinction Coefficients for Proteins
At 280 nm, the extinction coefficient is approximated by the
weighted sum of the 280 nm molar extinction coefficients of
the three constituent amino acids, as described in this
equation:
= (nW * 5500) + (nY * 1490) + (nC * 125)
where:
= molar extinction coefficient
n = number of each amino acid residue
5500, 1490 and 125 = amino acid molar absorptivities at
280 nm
Thermo Scientific NanoDrop One User Guide 79
Page 80

5 Protein Applications
Measure Protein A280
This application offers six options (shown at
right) for selecting an appropriate extinction
coefficient for each measured sample, to
be used in conjunction with Beer’s Law to
calculate sample concentration.
If the extinction coefficient of the sample is
known, choose the
+ MW (molar) or
1% (mass) option and enter the value.
Otherwise, calculate the extinction
coefficient or choose the option that best
matches the sample solution.
Tip: Ideally, the extinction coefficient
should be determined empirically using a
solution of the study protein at a known
concentration using the same buffer.
Most sources report extinction coefficients
for proteins measured at or near 280 nm in
phosphate or other physiologic buffer.
These values provide sufficient accuracy for
routine assessments of protein
concentration.
Available Options for Extinction Coefficient
• 1 Abs = 1 mg/mL, where sample type and/or ext.
coefficient is unknown (produces rough estimate of
protein concentration)
• BSA (Bovine Serum Albumin, 6.7 L/gm-cm)
• IgG (any mammalian antibody, 13.7 L/gm-cm)
• Lysozyme (egg white lysozyme, 26.4 L/gm-cm)
• Other protein (
+ MW), user-specified molar ext.
coefficient
• Other protein (
1%), user-specified mass ext.
coefficient
•
Note: See Sample Type for details.
Published Extinction Coefficients
Published extinction coefficients for proteins may be reported
as:
• wavelength-dependent molar absorptivity (or extinction)
coefficient (
) with units of M
-1cm-1
• percent solution extinction coefficient (1%) with units of
(g/100 mL)
-1cm-1
(i.e., 1% or 1 g/100 mL solution
measured in a 1 cm cuvette)
• protein absorbance values for 0.1% (i.e., 1 mg/mL)
solutions
Tip: Assess published values carefully to ensure unit of
measure is applied correctly.
80 NanoDrop One User Guide Thermo Scientific
Page 81

5
Protein Applications
Measure Protein A280
The equation at the right shows the
relationship between molar extinction
coefficient (
coefficient (1%).
To determine concentration (c) of a sample
in mg/mL, use the equation at the right and
a conversion factor of 10.
Tip: The NanoDrop One software includes
the conversion factor when reporting
protein concentrations.
) and percent extinction
molar
Conversions Between
(
Example: To determine percent solution extinction coefficient
1%) for a protein that has a molar extinction coefficient of
(
43,824 M
daltons (Da), rearrange and solve the above equation as
follows:
1% = (
) * 10 = (1%) * (MW
molar
-1cm-1
and a molecular weight (MW) of 66,400
* 10) / (MW
molar
molar
and 1%
)
protein
)
protein
1% = (43,824 * 10) / 66,400 Da)
1% = 6.6 g/100 mL
Conversions Between g/100 mL and mg/mL
C
Example: If measured absorbance for a protein sample at
280 nm relative to the reference is 5.8 A, protein
concentration can be calculated as:
in mg/mL = (A / 1%) * 10
protein
C
protein
C
protein
C
protein
= (A / 1%) * 10
= (5.8/6.6 g/100 mL) * 10
= 8.79 mg/mL
Thermo Scientific NanoDrop One User Guide 81
Page 82

5 Protein Applications
Measure Protein A280
Calculated protein concentrations are
based on the absorbance value at 280 nm,
the selected (or entered) extinction
coefficient and the sample pathlength. A
single-point baseline correction (or analysis
correction) may be applied.
Concentration is reported in mass units.
Calculators are available on the Internet to
convert concentration from mass to molar
units based on sample sequence.
Absorbance values at 260 nm and 280 nm
are used to calculate purity ratios for the
measured protein samples.
Purity ratios are sensitive to the presence of
contaminants in the sample, such as
residual solvents and reagents typically
used during sample purification.
Measured Values
A280 absorbance
Note: For micro-volume absorbance measurements and
measurements taken with nonstandard (other than 10 mm)
cuvettes, the spectra are normalized to a 10 mm pathlength
equivalent.
• Protein absorbance values are measured at 280 nm
using the normalized spectrum. If Baseline Correction is
not selected, this is the reported A280 value and the
value used to calculate protein concentration.
• If Baseline Correction is selected, the normalized and
baseline-corrected absorbance value at 280 nm is
reported and used to calculate protein concentration.
Sample Pathlength
• For micro-volume measurements, the software selects
the optimal pathlength (between 1.0 mm and 0.03 mm)
based on sample absorbance at the analysis wavelength.
• For cuvette measurements, pathlength is determined by
the cuvette Pathlength setting in the software (see
General Settings).
• Displayed spectra and absorbance values are normalized
to a 10 mm pathlength equivalent.
82 NanoDrop One User Guide Thermo Scientific
Page 83

5
Protein Applications
Measure Protein A280
Reported Values
• Protein concentration. Reported in selected unit
(mg/mL or µg/mL). Calculations are based on
Beer-Lambert equation using corrected protein
absorbance value.
• A260/A280 purity ratio. Ratio of corrected
absorbance at 260 nm to corrected absorbance at
280 nm. An A260/A280 purity ratio of ~0.57 is generally
accepted as “pure” for proteins.
Note: Although purity ratios are important indicators of
sample quality, the best indicator of protein quality is
functionality in the downstream application of interest (e.g.,
real-time PCR).
Thermo Scientific NanoDrop One User Guide 83
Page 84

5 Protein Applications
Measure Protein A205
Measure Protein A205
Measures the concentration of purified
protein populations that absorb at
205 nm.
Measure A205 Proteins
Reported Results
Settings
Detection Limits
Calculations
Measure Protein Concentration at A205
Use the Protein A205 application to quantify purified peptides and other proteins that
contain peptide bonds, which exhibit absorbance at 205 nm. This application
reports protein concentration and two absorbance values (A205 and A280). A
single-point baseline correction can also be used. This application does not require a
standard curve.
Note If your samples contain mainly amino acids such as tryptophan or
tyrosine, or cys-cys disulfide bonds, use the Protein A280 application instead of
Protein A205.
To measure Protein A205 samples
NOTICE
• Do not use a squirt or spray bottle on or near the instrument as liquids will
flow into the instrument and may cause permanent damage.
• Do not use hydrofluoric acid (HF) on the pedestals. Fluoride ions will
permanently damage the quartz fiber optic cables.
Before you begin...
Before taking pedestal measurements with the NanoDrop One instrument, lift the
instrument arm and clean the upper and lower pedestals. At a minimum, wipe the
pedestals with a new laboratory wipe. For more information, see Cleaning the
Pedestals.
84 NanoDrop One User Guide Thermo Scientific
Page 85

5
Protein Applications
Measure Protein A205
To measure a Protein A205 sample
1. From the Home screen, from the Proteins tab, select Protein A205.
2. Specify a sample type and baseline correction if desired.
3. Pipette 1–2 µL of the blanking solution onto the lower pedestal and lower the
arm, or insert the blanking cuvette into the cuvette holder.
Tip: If using a cuvette, make sure to align the cuvette light path with the
instrument light path.
4. Tap Blank and wait for the measurement to complete.
Tip: If Auto-Blank is On, the blank measurement starts automatically after you
lower the arm. (This option is not available for cuvette measurements.)
5. Lift the arm and clean both pedestals with a new laboratory wipe, or remove
the blanking cuvette.
6. Pipette 2 µL sample solution onto the pedestal and lower the arm, or insert the
sample cuvette into the cuvette holder.
7. Start the sample measurement:
– Pedestal: If Auto-Measure is On, lower arm; if Auto-Measure is off, lower
arm and tap Measure.
– Cuvette: Tap Measure
When the sample measurement is completed, the spectrum and reported
values are displayed (see the next section).
8. When you are finished measuring samples, tap End Experiment.
9. Lift the arm and clean both pedestals with a new wipe, or remove the sample
cuvette.
Thermo Scientific NanoDrop One User Guide 85
Page 86

5 Protein Applications
Measure Protein A205
Protein A205 Reported Results
Protein A205 measurement screen (Local Control)
For each measured sample, this application shows the absorbance spectrum and a
summary of the results. Here is an example:
tap to open
Sample name;
tap to edit
UV spectrum
Protein
concentration
Tap to select unitMenu of options;
Absorbance at
205 nm
Absorbance at
280 nm
Tap r ow to select
sample and update
spectrum; tap
more rows to
overlay up to five
spectra. Press and
hold sample row to
view measurement
details.
Drag tab down/up
Pinch and zoom to
adjust axes; double-tap
to reset
Swipe screen left to view
table with more
measurement results
Tap to end
experiment and
export data
to see more/less
sample data
Note Micro-volume absorbance measurements and measurements taken with
nonstandard cuvettes are normalized to a 10.0 mm pathlength equivalent.
Protein A205 reported values
The initial screen that appears after each measurement (see previous image) shows
a summary of the reported values. To view all reported values, press and hold the
sample row. Here is an example:
86 NanoDrop One User Guide Thermo Scientific
Page 87

5
Protein Applications
Measure Protein A205
Application
Baseline Correction
wavelength
Sampling
method
Baseline Correction
absorbance
Sample name;
tap to edit
Date/time
measured
Protein conc.
Absorbance at
205 nm
Absorbance at
280 nm
Sample type
Thermo Scientific NanoDrop One User Guide 87
Page 88

5 Protein Applications
Measure Protein A205
Protein A205 measurement screen (PC Control)
For each measured sample, this application shows the absorbance spectrum and
reported results. Here is an example:
Click to select cuvette or
pedestal measurement
Sample name;
Click to edit
Portein
A205
Setup
Run
Blank
Measure
sample
Click to end
experiment and
export data
Menu of options;
click to open
Protein
concentration
UV spectrum
Absorbance
values
Click and Drag
to adjust axes;
double-click to
reset
Click to
select unit
Click row to
select sample
and update
spectrum; click
more rows to
overlay up to
five spectra.
Related Topics
• Basic Instrument Operations
• Protein A205 Calculations
88 NanoDrop One User Guide Thermo Scientific
Page 89

Settings for Protein A205 Measurements
To show the Protein A205 settings, in local instrument control, from the Protein A205
measurement screen, tap > Protein A205 Setup.
From the PC control software, from the Protein A205 measurement screen, select
the settings icon to view Protein A205 Setup.
Protein A205 settings
The Protein A205 application provides a variety of method options for protein
analysis.
5
Protein Applications
Measure Protein A205
Mass Ext.
Setting Available Options
Sample type 31 31 Assumes 0.1% (1 mg/mL) at 205 nm = 31
Scopes 27 + 120 *
Coefficient
(L/gm-cm)
(A280/A205)
Description
Assumes
120 * (A280/A205)
0.1% (1 mg/mL) at 205 nm = 27 +
Thermo Scientific NanoDrop One User Guide 89
Page 90

5 Protein Applications
Measure Protein A205
Setting Available Options
Mass Ext.
Coefficient
(L/gm-cm)
Description
Baseline
Correction
Other protein
1%)
(
On or off
Enter baseline
correction
wavelength in nm or
use default value
(340 nm)
User entered mass
extinction coefficient
N/A Corrects for any offset caused by light
Assumes protein has known mass extinction
coefficient (). Enter mass extinction
coefficient in L/gm-cm for 1 mg/mL (
protein solution.
scattering particulates by subtracting
measured absorbance at specified baseline
correction wavelength from absorbance
values at all wavelengths in sample spectrum.
As a result, absorbance of sample spectrum is
zero at specified baseline correction
wavelength.
Tip: If the sample has a modification that
absorbs light at 340 nm, select a different
correction wavelength or turn off Baseline
Correction.
0.1%)
90 NanoDrop One User Guide Thermo Scientific
Page 91

Calculations for Protein A205 Measurements
5
Protein Applications
Measure Protein A205
As with the other protein applications,
Proteins A205 uses the Beer-Lambert
equation to correlate absorbance with
concentration based on the sample’s
extinction coefficient and pathlength.
This application offers three options (shown
at right) for selecting an appropriate
extinction coefficient for each measured
sample, to be used in conjunction with
Beer’s Law to calculate sample
concentration.
If the extinction coefficient of the sample is
known, choose the 1% (mass) option and
enter the value. Otherwise, calculate the
extinction coefficient or choose the option
that best matches the sample solution.
Tip: Ideally, the extinction coefficient
should be determined empirically using a
solution of the study protein at a known
concentration using the same buffer.
Calculated protein concentrations are
based on the absorbance value at 205 nm,
the selected (or entered) extinction
coefficient and the sample pathlength. A
single-point baseline correction may also
be applied.
Concentration is reported in mass units.
Calculators are available on the Internet to
convert concentration from mass to molar
units based on the sample sequence.
Available Options for Extinction Coefficient
• 31, assumes
• Scopes, assumes
0.1% (1 mg/mL) at 205 nm = 31
0.1% (1 mg/mL) at 205 nm = 27 +
120 * (A280/A205)
• Other protein, enter mass extinction coefficient in
L/gm-cm for 1 mg/mL (
0.1%) protein solution
Note: See Sample Type for details.
Measured Values
A205 absorbance
Note: For micro-volume absorbance measurements and
measurements taken with nonstandard (other than 10 mm)
cuvettes, the spectra are normalized to a 10 mm pathlength
equivalent.
• Protein absorbance values are measured at 205 nm
using the normalized spectrum. If Baseline Correction is
not selected, this is the reported A205 value and the
value used to calculate protein concentration.
• If Baseline Correction is selected, the normalized and
baseline-corrected absorbance value at 205 nm is
reported and used to calculate protein concentration.
A280 absorbance
• Normalized and baseline-corrected (if selected)
absorbance value at 280 nm is also reported.
Thermo Scientific NanoDrop One User Guide 91
Page 92

5 Protein Applications
Measure Protein A205
Sample Pathlength
• For micro-volume measurements, the software selects
the optimal pathlength (between 1.0 mm and 0.03 mm)
based on sample absorbance at the analysis wavelength.
• For cuvette measurements, pathlength is determined by
the cuvette Pathlength setting in the software (see
General Settings).
• Displayed spectra and absorbance values are normalized
to a 10 mm pathlength equivalent.
Reported Values
• Protein concentration. Reported in selected unit
(mg/mL or µg/mL). Calculations are based on
Beer-Lambert equation using corrected protein
absorbance value.
92 NanoDrop One User Guide Thermo Scientific
Page 93

Measure Proteins and Labels
Measures the concentration of purified
proteins that have been labeled with up
to two fluorescent dyes.
Measure Labeled Proteins
Reported Results
Settings
Detection Limits
Calculations
Measure Labeled Protein Samples
5
Protein Applications
Measure Proteins and Labels
Use the Proteins and Labels application to quantify proteins and fluorescent dyes for
protein array conjugates, as well as metalloproteins such as hemoglobin, using
wavelength ratios. This application reports protein concentration measured at
280 nm, an A269/A280 absorbance ratio, and the concentrations and measured
absorbance values of the dyes, allowing detection of dye concentrations as low as
0.2 picomole per microliter. This information is useful for evaluating protein/dye
conjugation (degree of labeling) for use in downstream applications.
To measure labeled protein samples
NOTICE
• Do not use a squirt or spray bottle on or near the instrument as liquids will
flow into the instrument and may cause permanent damage.
• Do not use hydrofluoric acid (HF) on the pedestals. Fluoride ions will
permanently damage the quartz fiber optic cables.
Before you begin...
Before taking pedestal measurements with the NanoDrop One instrument, lift the
instrument arm and clean the upper and lower pedestals. At a minimum, wipe the
pedestals with a new laboratory wipe. For more information, see Cleaning the
Pedestals.
To measure a labeled protein sample
1. From the Home screen, select the Proteins tab and then select Protein &
Labels.
Thermo Scientific NanoDrop One User Guide 93
Page 94

5 Protein Applications
Measure Proteins and Labels
2. Specify the sample type and the type of dye(s) used.
3. Pipette 1–2 µL of the blanking solution onto the lower pedestal and lower the
4. Tap Blank and wait for the measurement to complete.
5. Lift the arm and clean both pedestals with a new laboratory wipe, or remove the
6. Pipette 2 µL sample solution onto the pedestal and lower the arm, or insert the
Tip: Select a dye from the pre-defined list or add a custom dye using the
Dye/Chromophore Editor.
arm, or insert the blanking cuvette into the cuvette holder.
Tip: If using a cuvette, make sure to align the cuvette light path with the
instrument light path.
Tip: If Auto-Blank is On, the blank measurement starts automatically after you
lower the arm. (This option is not available for cuvette measurements.)
blanking cuvette.
sample cuvette into the cuvette holder.
7. Start the sample measurement:
– Pedestal: If Auto-Measure is On, lower arm; if Auto-Measure is off, lower arm
and select Measure.
– Cuvette: select Measure
When the sample measurement is completed, the spectrum and reported values
are displayed (see the next section).
8. When you are finished measuring samples, select End Experiment.
9. Lift the arm and clean both pedestals with a new wipe, or remove the sample
cuvette.
94 NanoDrop One User Guide Thermo Scientific
Page 95

5
Protein Applications
Measure Proteins and Labels
Peptide backbone
Dye absorbance peak used to
calculate dye concentration
A280 absorbance peak used to
calculate protein concentration
Typical sample spectrum measured with Proteins & Labels
Related Topics
• Best practices for protein measurements
• Measure a Micro-Volume Sample
• Measure a Sample Using a Cuvette
• Prepare Samples and Blanks
• Basic Instrument Operations
Thermo Scientific NanoDrop One User Guide 95
Page 96

5 Protein Applications
Measure Proteins and Labels
Proteins & Labels Reported Results
Proteins & Labels measurement screen
For each measured sample, this application shows the absorbance spectrum and a
summary of the results. Below is an example of the measurement screen of the PC
control software:
Application
Sampling
method
Sample name of
next measurement;
select to edit
UV spectrum
Run Blank
Proteins &
Labels setup
Measure sample
End experiment
Menu of options;
click to open
Right click graph
area to view display
options
Menu of table options;
click to choose which
columns to report
.
Sample name;
select to edit
Protein
concentration
Dye concentration(s)
Click row to select sample
and update spectrum.
Measurement screen of PC Control software
Note
• A baseline correction is performed at 850 nm (absorbance value at 850 nm is
subtracted from absorbance values at all wavelengths in sample spectrum).
• Micro-volume absorbance measurements and measurements taken with
nonstandard cuvettes are normalized to a 10.0 mm pathlength equivalent.
96 NanoDrop One User Guide Thermo Scientific
Page 97

5
Protein Applications
Measure Proteins and Labels
Menu of options;
tap to open
UV-visible spectrum
Sample name;
tap to edit
Protein
concentration
Tap to select unit
Dye
concentration(s)
Tap r ow to select
sample and
update spectrum;
tap more rows to
overlay up to five
spectra. Press
and hold sample
row to view
measurement
details.
Pinch and zoom to
adjust axes; double-tap
to reset
Swipe screen left to view
table with more
measurement results
Measurement screen of NanoDrop One local control
Proteins & Labels reported values
The initial screen that appears after each measurement (see previous image) shows
a summary of the reported values. To view all reported values, press and hold the
sample row. Here is an example:
Reported values for Proteins & Labels application
• Sample details (application and sampling method used, i.e., pedestal or cuvette)
• Sample Name
• Creation date
• Protein
Drag tab down/up
to see more/less
sample data
Tap to end
experiment and
export data
• A280
Thermo Scientific NanoDrop One User Guide 97
Page 98

5 Protein Applications
Measure Proteins and Labels
• Sample Type
• Dye 1/Dye 2
• Sloping Dye Correction
• Analysis Correction
Related Topics
• Basic Instrument Operations
• Proteins & Labels calculations
Settings for Proteins and Labels Measurements
To show the Proteins & Labels settings, from the Proteins & Labels measurement
screen, tap > Proteins & Labels Setup.
Setting Available Options
Sample
a
type
1 Abs = 1 mg/mL
BSA
IgG
Lysozyme
Other protein
+ MW)
(
Other protein
1%)
(
Mass Ext.
Coefficient
(L/gm-cm)
General reference
6.7
13.7
26.4
user-entered molar
extinction
coefficient/
molecular weight
User entered mass
extinction
coefficient
Description
Tap here for detailed description of each
available setting.
Each sample type applies a unique extinction
coefficient to the protein calculations. If the
extinction coefficient of the sample is known,
choose the
option and enter the value. Otherwise,
calculate the extinction coefficient or choose
the option that best matches the sample
solution. If you only need a rough estimate of
protein concentration and the sample
extinction coefficient is unknown, select the
1 Abs=1 mg/mL sample type option.
Tip: Ideally, the extinction coefficient should
be determined empirically using a solution of
the study protein at a known concentration
using the same buffer.
+ MW (molar) or 1% (mass)
98 NanoDrop One User Guide Thermo Scientific
Page 99

Setting Available Options
Analysis
Correction
On or off
b
Enter analysis
correction wavelength
in nm or use default
value (340 nm)
5
Protein Applications
Measure Proteins and Labels
Mass Ext.
Coefficient
(L/gm-cm)
Description
N/A Corrects sample absorbance measurement
for any offset caused by light scattering
particulates by subtracting absorbance value
at specified analysis correction wavelength
from absorbance value at analysis
wavelength. Corrected value is used to
calculate sample concentration.
Tip: If the sample has a modification that
absorbs light at 340 nm, select a different
correction wavelength or turn off Analysis
Correction.
Dye 1/Dye 2
c
Type
Cy3, 5, 3.5, or 5.5,
Alexa Fluor 488, 546,
555, 594, 647, or 660
See
Dye/Chromophore
Editor for specific
values for each
dye
Dye 1/Dye 2
Unit
picomoles/microliter
(pmol/uL), micromoles
not applicable Select unit for reporting dye concentrations.
(uM), or millimoles
(mM)
Sloping Dye
Correction
a
To add or edit a custom protein, use Protein Editor.
b
Analysis Correction affects calculation for protein concentration only.
c
To add custom dye or edit list of available dyes, use Dye/Chromophore Editor.
d
Sloping Dye Correction affects calculations for dye concentration only.
On or off Corrects dye absorbance measurements for
d
Select pre-defined dye used to label sample
material, or one that has been added using
Dye/Chrom. Editor.
any offset caused by light scattering
particulates by subtracting absorbance value
of a sloping baseline from 400 nm to 850 nm
from absorbance value at dye’s analysis
wavelength.
Related Topics
• Instrument Settings
• Protein Editor
• Dye/Chromophore Editor
Thermo Scientific NanoDrop One User Guide 99
Page 100

5 Protein Applications
Measure Proteins and Labels
Detection Limits for Proteins and Labels Measurements
Detection limits and reproducibility specifications for purified BSA proteins and dyes
that are pre-defined in the software are provided here. The BSA lower detection limit
and reproducibility values apply to any protein sample type. The upper detection
limits are dependent on the upper absorbance limit of the instrument and the
sample’s extinction coefficient.
To calculate upper detection limits for other (non-BSA) protein sample types
To calculate upper detection limits in mg/mL for proteins, use the following equation:
(upper absorbance limit
instrument
/mass extinction coefficient
For example, if the sample’s mass extinction coefficient at 280 nm is 6.7 for a 1%
(10 mg/mL) solution, the equation looks like this:
(550 / 6.7) * 10 = 824.6 (or ~825)
Related Topics
• Detection Limits for All Applications
Calculations for Proteins and Labels Measurements
As with the other protein applications,
Proteins & Labels uses the Beer-Lambert
equation to correlate absorbance with
concentration based on the sample’s
extinction coefficient and pathlength.
This application offers six options (shown at
right) for selecting an appropriate extinction
coefficient for each measured sample, to
be used in conjunction with Beer’s Law to
calculate sample concentration.
Available Options for Extinction Coefficient
• 1 Abs = 1 mg/mL, where sample type and/or ext.
coefficient is unknown (produces rough estimate of
protein concentration)
• BSA (Bovine Serum Albumin, 6.7 L/gm-cm)
• IgG (any mammalian antibody, 13.7 L/gm-cm)
• Lysozyme (egg white lysozyme, 26.4 L/gm-cm)
sample
) * 10
If the extinction coefficient of the sample is
known, choose the + MW (molar) or
1% (mass) option and enter the value.
Otherwise, calculate the extinction
coefficient or choose the option that best
matches the sample solution.
• Other protein (
coefficient
• Other protein (
coefficient
+ MW), user-specified molar ext.
1%), user-specified mass ext.
Note: See Sample Type for details.
Tip: Ideally, the extinction coefficient
should be determined empirically using a
solution of the study protein at a known
concentration using the same buffer.
100 NanoDrop One User Guide Thermo Scientific
 Loading...
Loading...