
MycoSEQ™ Mycoplasma Real-Time PCR
Detection Kit
USER GUIDE
for use with the PrepSEQ™ Mycoplasma Sample Preparation Kit
Catalog Numbers 4460623, 4460626
Publication Number 4465874
Revision D.0
For Research Use Only. Not for use in diagnostic procedures.

Life Technologies Ltd | 7 Kingsland Grange | Woolston, Warrington WA1 4SR | United Kingdom
For descriptions of symbols on product labels or product documents, go to thermofisher.com/symbols-definition.
The information in this guide is subject to change without notice.
DISCLAIMER: TO THE EXTENT ALLOWED BY LAW, THERMO FISHER SCIENTIFIC INC. AND/OR ITS AFFILIATE(S) WILL NOT BE
LIABLE FOR SPECIAL, INCIDENTAL, INDIRECT, PUNITIVE, MULTIPLE, OR CONSEQUENTIAL DAMAGES IN CONNECTION WITH OR
ARISING FROM THIS DOCUMENT, INCLUDING YOUR USE OF IT.
Revision history: Pub. No. 4465874
Revision Date Description
D.0 28 September 2020 Update to include run and analysis information for AccuSEQ™ Real‑Time
C.0 24 May 2018 Updated template, legal, and content information. Reorganized
PCR Software v3.1.
content. Added information about using the AccuSEQ™ Software v2.0
Mycoplasma SEQ module.
Important Licensing Information: These products may be covered by one or more Limited Use Label Licenses. By use of these
products, you accept the terms and conditions of all applicable Limited Use Label Licenses.
TRADEMARKS: All trademarks are the property of Thermo Fisher Scientific and its subsidiaries unless otherwise specified.
©2020 Thermo Fisher Scientific Inc. All rights reserved.

Contents
■
CHAPTER 1 Product information .................................................. 6
Product description ............................................................. 6
Contents and storage ............................................................ 6
Required materials not supplied ................................................... 7
Workflow ...................................................................... 8
■
CHAPTER 2 Methods .............................................................. 9
Prepare the sample .............................................................. 9
Prepare the kit reagents and premix solution ........................................ 9
Prepare the PCR reactions ...................................................... 10
Setup, run, and anlayze samples with AccuSEQ™ Software v3.1 on the
QuantStudio™ 5 Instrument .................................................... 11
Sign in to the AccuSEQ™ Real‑Time PCR Software ............................. 11
Create a MycoSEQ™ experiment ............................................. 12
Start the run (QuantStudio™ 5 Real‑Time PCR Instrument) ...................... 16
Monitor the run ............................................................ 17
Analyze the results ......................................................... 18
Guidance for unknown samples ............................................. 25
Guidance for controls ...................................................... 26
Example results with AccuSEQ™ Software v3.1 or later ......................... 28
■
■
MycoSEQ
APPENDIX A Troubleshooting .................................................... 34
AccuSEQ™ software ............................................................ 34
MycoSEQ™ kit ................................................................. 36
APPENDIX B Acceptance criteria ................................................ 37
Cycle threshold value (Ct) ....................................................... 38
Melting temperature (Tm) ........................................................ 39
Derivative value (DV) ............................................................ 40
Results interpretation using the acceptance criteria ................................ 41
Other acceptance criteria considerations ......................................... 41
™
Mycoplasma Real-Time PCR Detection Kit User Guide
3

Contents
■
APPENDIX C Use the kit with 7500 Fast System AccuSEQ™ Real-Time
PCR Detection Software v2.x .................................................... 42
Required materials not supplied ................................................. 42
Prepare the sample ............................................................. 43
Prepare the kit reagents and premix solution ...................................... 43
Prepare the PCR reactions ...................................................... 44
Create a new experiment ........................................................ 44
Define the experiment properties ............................................ 45
Setup the samples and controls ............................................. 45
View the plate layout ....................................................... 46
Save the experiment ....................................................... 47
Start the run (7500 Fast Real‑Time PCR Instrument) ................................ 47
Analyze the results ............................................................. 47
Set the baseline and threshold values ........................................ 48
Review the Results Summary ............................................... 48
Guidance for unknown samples ............................................. 50
Guidance for controls ...................................................... 51
Guidance for inconclusive results with AccuSEQ™ software v2.0 ................ 52
Example results with AccuSEQ™ Software v. 2.1.1 ................................. 53
Positive control ............................................................ 53
Negative control ........................................................... 54
Blank extraction control .................................................... 55
Positive extraction control .................................................. 56
Inhibition control and positive control ........................................ 57
Unknown sample: Negative result ............................................ 58
Unknown sample: Positive result ............................................ 59
Unknown sample: Positive result with decreased detection of DPC .............. 60
PCR inhibition ............................................................. 61
Multicomponent plots ...................................................... 62
■
APPENDIX D Use the kit with 7500 System SDS Software v1.4 or later ..... 63
Prepare the kit reagents and premix solution ...................................... 63
Prepare the PCR reactions ...................................................... 63
Seal the plates ................................................................. 65
Prepare the plate document ..................................................... 66
Perform PCR .................................................................. 68
Analyze the results ............................................................. 68
Set the baseline and threshold values ........................................ 68
Guidance for unknown samples ............................................. 69
Guidance for controls ...................................................... 70
Example positive results with SDS v1.4 software .............................. 70
4
MycoSEQ™ Mycoplasma Real-Time PCR Detection Kit User Guide

Example positive control extraction results with SDS v1.4 software .............. 72
Example negative results with SDS v1.4 software .............................. 73
■
APPENDIX E Background information ........................................... 74
■
APPENDIX F Kit specificity ....................................................... 75
Sensitivity ..................................................................... 75
Kit specificity .................................................................. 75
Inclusivity – detectable species .................................................. 75
Exclusivity – undetectable organisms ............................................. 76
■
APPENDIX G Good PCR practices .............................................. 78
Good laboratory practices for PCR and RT-PCR ................................... 78
Plate layout suggestions ........................................................ 78
■
Documentation and support ....................................................... 79
Contents
Related documentation ......................................................... 79
Customer and technical support ................................................. 80
Limited product warranty ........................................................ 81
MycoSEQ™ Mycoplasma Real-Time PCR Detection Kit User Guide
5
Product information
1
IMPORTANT! Before using this product, read and understand the information in the
“Safety” appendix in this document.
Product description
The MycoSEQ™ Mycoplasma Real-Time PCR Detection Kit detects Mycoplasma
species simply, reliably, and rapidly. To detect the presence of these microorganisms,
the assay uses the polymerase chain reaction (PCR) to amplify a target unique to a
wide variety of Mycoplasma species.
Contents and storage
Table 1 MycoSEQ™ Mycoplasma Real-Time PCR Detection Kit (Cat. No. 4460623)
Contents
Box 1: MycoSEQ™ Mycoplasma Real‑Time PCR Detection Kit
10✕ Mycoplasma Real‑Time PCR
Primer Mix
Negative Control
2✕ Power SYBR™ Green PCR
Master Mix
Box 2: MycoSEQ™ Discriminatory Positive/Extraction Control
MycoSEQ™ Discriminatory Positive/
Extraction Control, 1,000 copies/µL
[1]
To purchase the MycoSEQ™ Mycoplasma Real-Time PCR Detection Kit that includes the PrepSEQ™ Mycoplasma Sample Preparation Kit, use
Catalog Number 4460626.
[2]
The kit contains reagents for 100 reactions.
[1]
Cap color Amount
blue
1,000 µL
white
2 × 1,000 µL –25°C to –15°C on receipt, protected
white
yellow
[2]
325 µL –25°C to –15°C on receipt
2–8°C after first use, protected from
700 µL –25°C to –15°C
Storage
2–8°C after first use
from light.
light
6
MycoSEQ™ Mycoplasma Real-Time PCR Detection Kit User Guide

Required materials not supplied
Unless otherwise indicated, all materials are available through thermofisher.com.
"MLS" indicates that the material is available from fisherscientific.com or another
major laboratory supplier.
Item Source
Instrument
Chapter 1 Product information
Required materials not supplied
1
QuantStudio™ 5 Real‑Time PCR System with AccuSEQ™ Software
v3.1 or later
Generic consumables
Disposable gloves MLS
Aerosol-resistant pipette tips MLS
Pipettors:
•
Positive-displacement
•
Air-displacement
•
Multichannel
•
Repeat (1µL–1mL)
Consumables for the QuantStudio™ 5 Real‑Time PCR System
MicroAmp™ Fast Optical 96‑Well Reaction Plate with Barcode, 0.1
mL
MicroAmp™ 96-Well Base N8010531
MicroAmp™ Optical Adhesive Film 4360954
MicroAmp™ Adhesive Film Applicator 4333183
Contact your local sales representative
MLS
4346906
MycoSEQ™ Mycoplasma Real-Time PCR Detection Kit User Guide
7

Chapter 1 Product information
1
Workflow
Workflow
Prepare the sample (page 9)
▼
Prepare the kit reagents and premix solution (page 9)
▼
Prepare the PCR reactions (page 63)
▼
Create a MycoSEQ™ experiment (page 12)
▼
Start the run (QuantStudio™ 5 Real‑Time PCR Instrument) (page 16)
▼
Monitor the run (page 17)
▼
Analyze the results (page 18)
8
MycoSEQ™ Mycoplasma Real-Time PCR Detection Kit User Guide

2
IMPORTANT! This chapter describes how to prepare and run PCR samples using
AccuSEQ™ Software v3.1 or later on the QuantStudio™ 5 Instrument (0.1 mL block).
If you are using AccuSEQ™ software v2.x on the 7500 Fast, see Appendix C, “Use
the kit with 7500 Fast System AccuSEQ™ Real-Time PCR Detection Software v2.x”.
If you are using SDS software v1.4 or later, see Appendix D, “Use the kit with 7500
System SDS Software v1.4 or later”.
IMPORTANT! For information on how to avoid PCR contamination, see Appendix G,
“Good PCR practices”.
Prepare the sample
Prepare the DNA template for the PCR reactions using the PrepSEQ™ Mycoplasma
Nucleic Acid Extraction Kit.
Methods
For more information, see:
•
The PrepSEQ™ Sample Preparation Kits for Mycoplasma, MMV, and Vesivirus
User Guide (Pub. No. 4465957)
•
The PrepSEQ™ Express Nucleic Acid Extraction Kit for Mycoplasma, MMV, and
Vesivirus Detection User Guide (Pub. No. MAN0016799)
Prepare the kit reagents and premix solution
1.
Thaw all kit reagents completely.
2.
Vortex briefly, then spin down the reagents.
MycoSEQ™ Mycoplasma Real-Time PCR Detection Kit User Guide
9

Chapter 2
2
Prepare the PCR reactions
Methods
3.
Prepare the Premix Solution according to the following table.
Component for premix
solution
Power SYBR™ Green PCR
Master Mix, 2✕
Mycoplasma Real‑Time
PCR Primer Mix, 10✕
Total premix solution
volume
[1]
Includes 10% excess to compensate for pipetting errors.
4.
Mix the Premix Solution by gently pipetting up and down, then cap the tube.
Prepare the PCR reactions
1.
Dispense the following into each well to be used, gently pipetting at the bottom
of the well.
Negative control reaction
To prepare…
Volume for one 30‑µL
reaction
15.0 µL 66.0 µL
3.0 µL 13.2 µL
18.0 µL 79.2 µL
•
Add 18 µL of Premix Solution
•
Add 12 µL of Negative Control
(water)
Volume for four 30‑µL
reactions
In each tube or well…
[1]
Unknown or spiked sample reaction
Inhibition-control reaction
Positive control reaction
•
Add 18 µL of Premix Solution
•
Add 10 µL of unknown sample
•
Add 2 µL of Negative Control
(water)
•
Add 18 µL of Premix Solution
•
Add 10 µL of unknown sample
•
Add 2 µL of the Discriminatory
Positive Control (DPC)
•
Add 18 µL of Premix Solution
•
Add 2 µL of the DPC
•
Add 10 µL of Negative Control
(water)
Note: The MycoSEQ™ Mycoplasma Discriminatory Positive/Extraction Control
can be used as a spike control that is added to the unknown sample or lysate
before sample preparation.
2.
Mix each sample by gently pipetting up and down.
10
MycoSEQ™ Mycoplasma Real-Time PCR Detection Kit User Guide

Chapter 2
Setup, run, and anlayze samples with AccuSEQ™ Software v3.1 on the QuantStudio™ 5 Instrument
3.
Seal the plate with MicroAmp™ Optical Adhesive Film. See “Seal the plates” on
page 65.
4.
Briefly centrifuge the reaction plate.
Methods
Setup, run, and anlayze samples with AccuSEQ™ Software v3.1 on
the QuantStudio™ 5 Instrument
Sign in to the AccuSEQ™ Real‑Time PCR Software
Thermo Fisher Scientific recommends configuring the Windows™ 10 screen save
feature to require sign in when the screensaver is activated. This prevents other users
from accessing the AccuSEQ™ Real‑Time PCR Software and making changes.
1.
Launch the AccuSEQ™ Real‑Time PCR Software by double-clicking the
AccuSEQ icon .
2.
Enter the Username, then Password.
(First login only) The default username is Administrator and the default
password is Administrator.
2
3.
Click Sign in.
The following restrictions may be seen in the software:
•
Access to functions in the software is based on the permissions associated with
the individual user account.
•
If a user account does not have permission to perform a function, the function is
grayed out in the software.
•
If the system is configured for password expiration, you will be periodically
prompted to change your password. If the system is configured to monitor failed
log in attempts, you will be locked out of the software if you incorrectly enter
your user name or password more than a specified number of times.
MycoSEQ™ Mycoplasma Real-Time PCR Detection Kit User Guide
11
2
1
Chapter 2 Methods
2
Setup, run, and anlayze samples with AccuSEQ™ Software v3.1 on the QuantStudio™ 5 Instrument
Create a MycoSEQ™ experiment
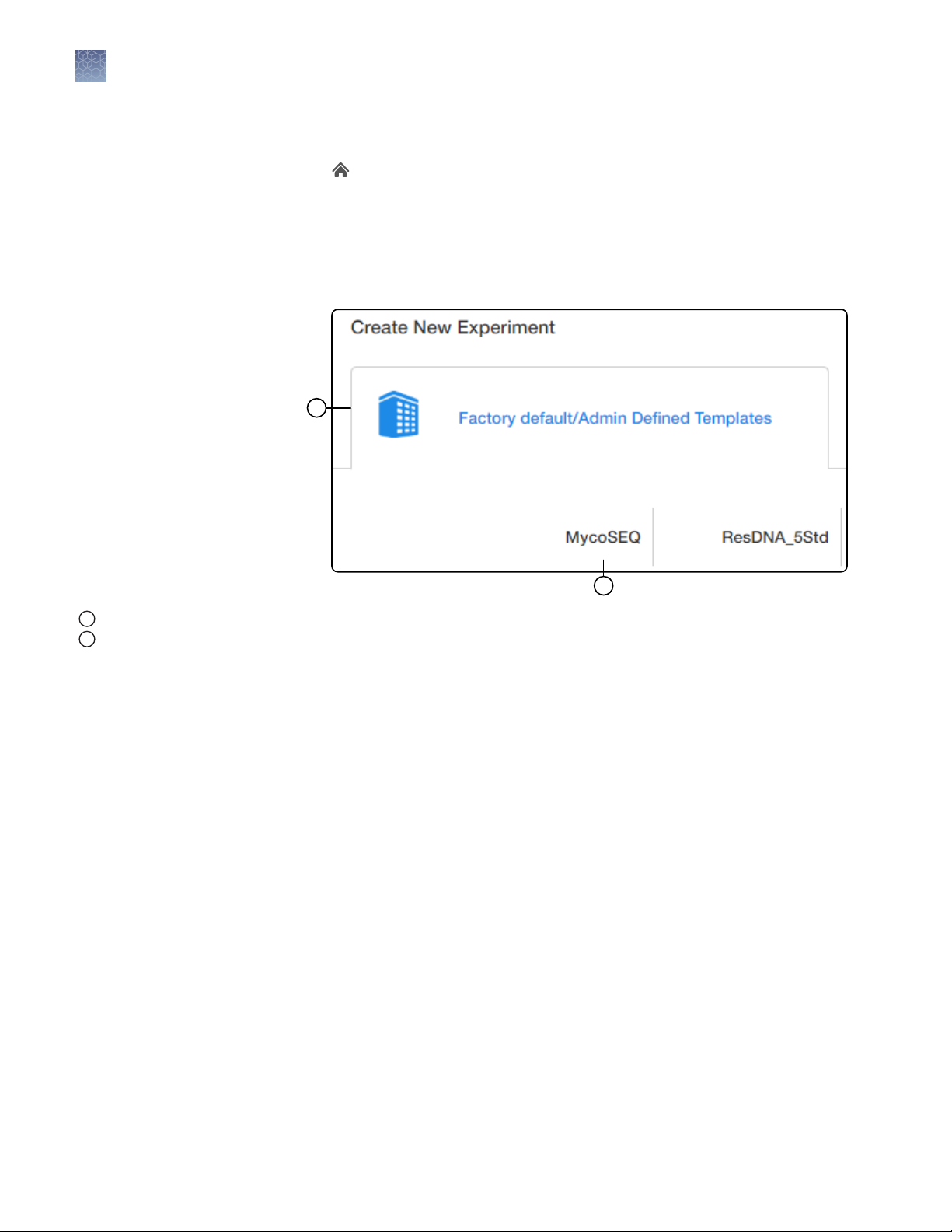
1.
In the Home screen, click the Factory default/Admin Defined Template
tab, then select MycoSEQ.
Note: To create a copy of an existing MycoSEQ™ experiment, see chapter 6 of
the AccuSEQ™ Real‑Time PCR Software v3.1 User Guide (Pub. No. 100094287).
Custom MycoSEQ™ experiments must be set up as Quantitation-Standard
Curve type experiments with SYBR™ chemistry.
Factory default/Admin Defined Template tab
1
MycoSEQ template
2
2.
In the Experiment Properties pane of the Setup tab:
a.
(Optional) Change the system‑generated name of the experiment.
b.
(Optional) Enter the plate Barcode, then add Comments.
Note: Comments are not editable post analysis.
Default MycoSEQ™ settings (These settings cannot be changed in the
default template. See the AccuSEQ™ Real‑Time PCR Software v3.1 User
Guide (Pub. No. 100094287) for creating custom templates).
•
Experiment Type—Quantitation-Standard Curve
•
Chemistry—SYBR™ Green Reagents
•
Ramp Speed—Standard - 2hrs
c.
Click Next.
12
MycoSEQ™ Mycoplasma Real-Time PCR Detection Kit User Guide
1
2
Chapter 2 Methods
Setup, run, and anlayze samples with AccuSEQ™ Software v3.1 on the QuantStudio™ 5 Instrument
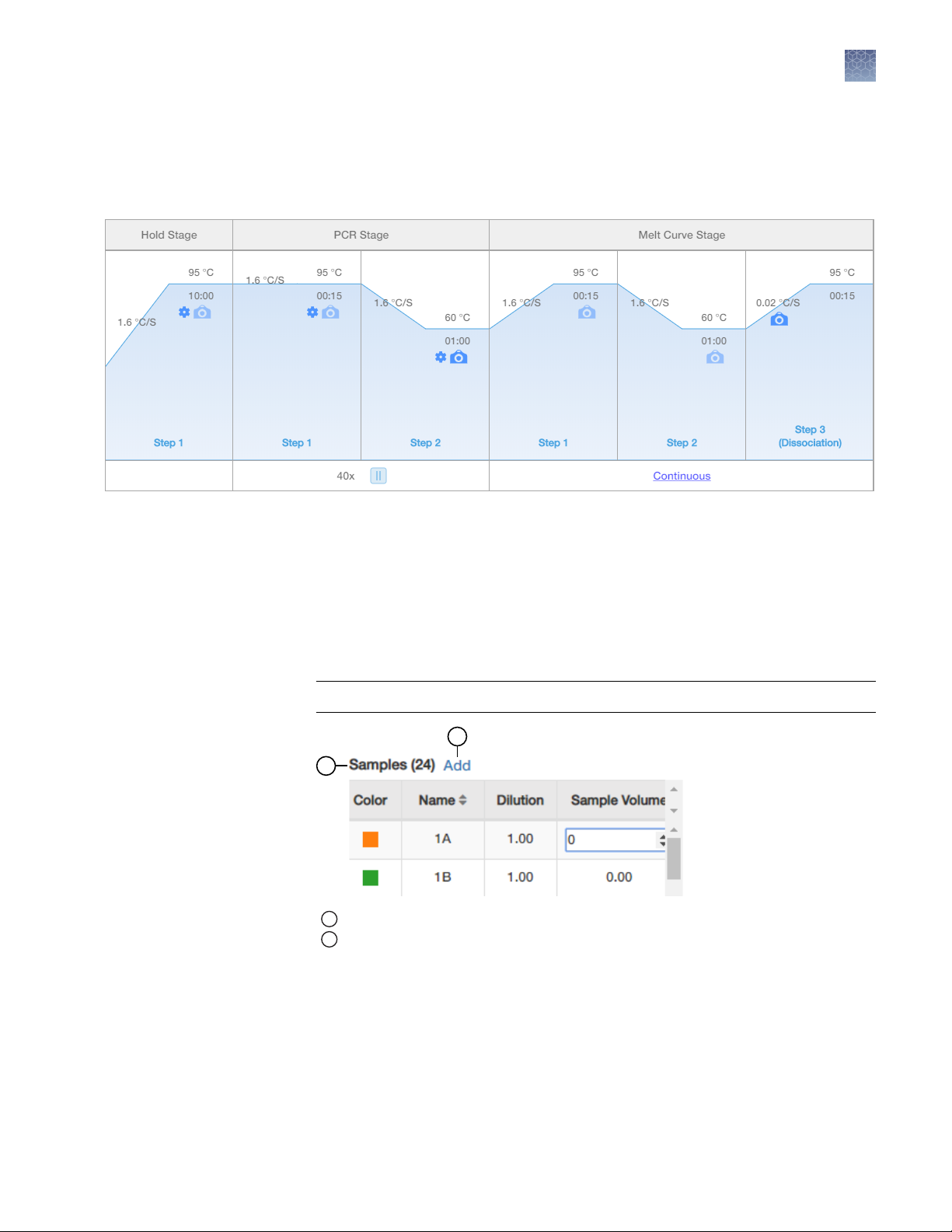
3.
In the qPCR Method pane of the Setup tab, view the default volume and
cycling conditions (These settings cannot be changed in the default template.
See the AccuSEQ™ Real‑Time PCR Software v3.1 User Guide (Pub. No.
100094287) for creating custom templates).
2
Figure 1 MycoSEQ™ template default cycling conditions
4.
Click Next.
5.
In the Samples pane of the Setup tab, enter the sample Name. Add additional
Samples if needed.
Note: Only the sample Name is necessary for experiments run from the factory
default MycoSEQ template.
IMPORTANT! Do not change the Targets.
Samples pane
1
Add—adds additional samples
2
MycoSEQ™ Mycoplasma Real-Time PCR Detection Kit User Guide
13
108
7
1
9
64 5
2
3
Chapter 2 Methods
2
Setup, run, and anlayze samples with AccuSEQ™ Software v3.1 on the QuantStudio™ 5 Instrument
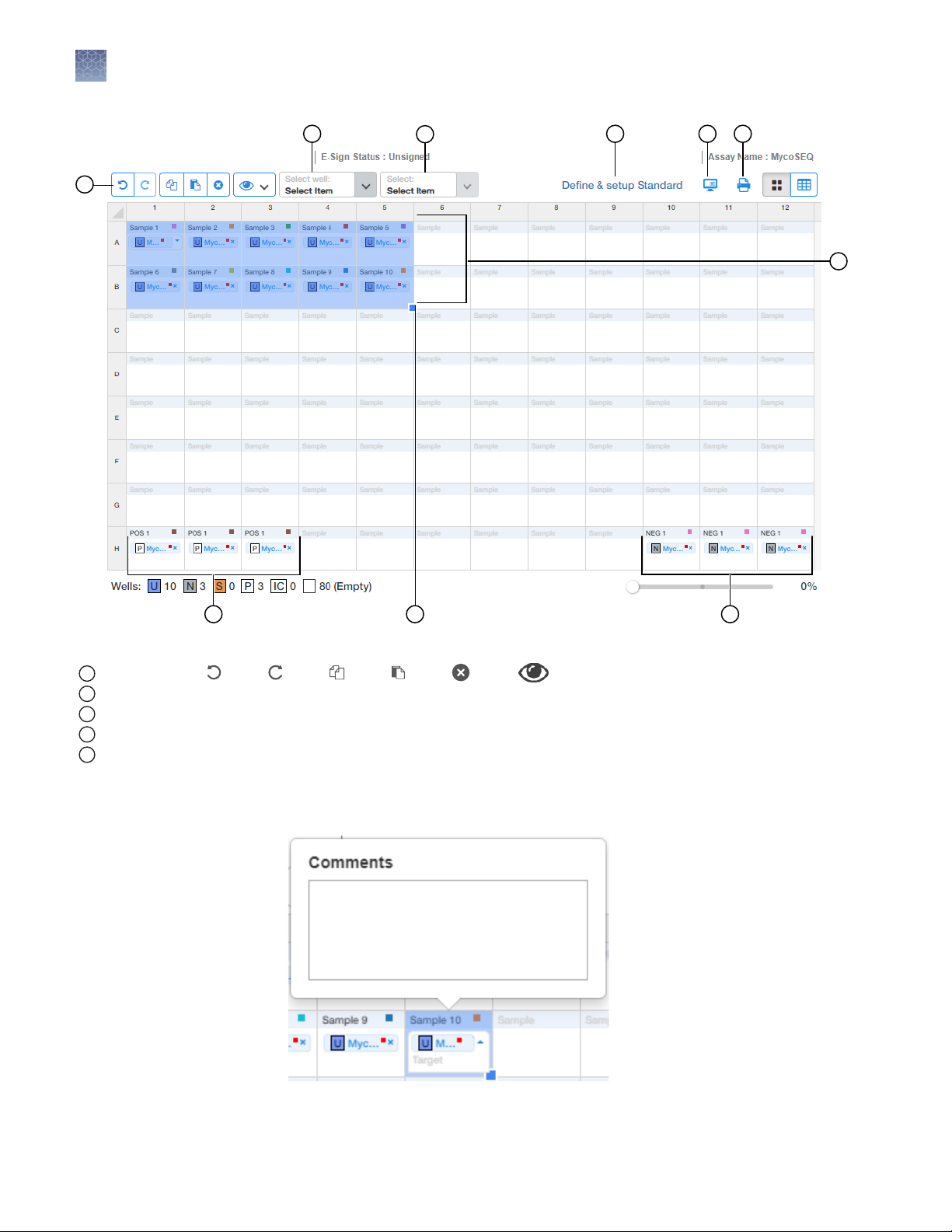
Figure 2 MycoSEQ™ template default sample plate layout
Toolbar (in order: Undo, Redo, Copy, Paste, Delete, View)
1
5 default Samples with and without an inhibition control (IC)
2
Positive control (P)
3
Selected samples. Click-drag to add additional samples.
4
No template control (N)
5
6.
(Optional) Double-click wells to add Comments. Comments can also be added
post-analysis.
14
MycoSEQ™ Mycoplasma Real-Time PCR Detection Kit User Guide

Chapter 2 Methods
Setup, run, and anlayze samples with AccuSEQ™ Software v3.1 on the QuantStudio™ 5 Instrument
7.
Click Next.
The Run tab is displayed.
8.
Experiments are auto‑saved in the software. To save additional changes made to
the experiment, exit the experiment. The software prompts you to save changes.
Click Yes.
Note: Clicking Save As will create a copy of the experiment.
2
MycoSEQ™ Mycoplasma Real-Time PCR Detection Kit User Guide
15
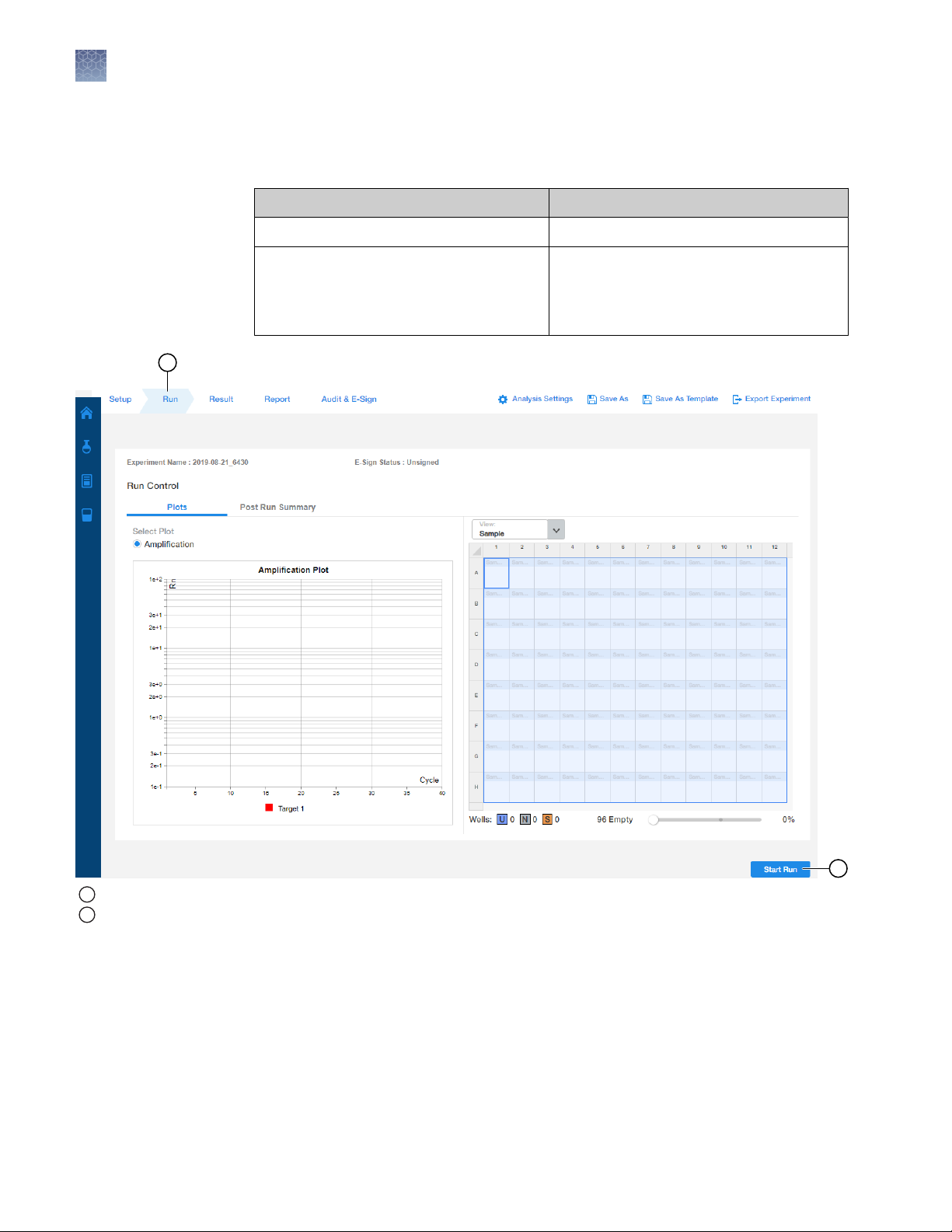
2
1
Chapter 2 Methods
2
Setup, run, and anlayze samples with AccuSEQ™ Software v3.1 on the QuantStudio™ 5 Instrument
Start the run (QuantStudio™ 5 Real‑Time PCR Instrument)
Start the run in the AccuSEQ™ Software v3.1 or later.
Option Description
If the experiment is open Click Start Run.
If the experiment is closed
1.
Open the experiment.
2.
Click the Run tab.
3.
Click Start Run.
1
Run tab
2
Start Run button
A message stating Run has been started successfully is displayed when the run
has started.
16
MycoSEQ™ Mycoplasma Real-Time PCR Detection Kit User Guide
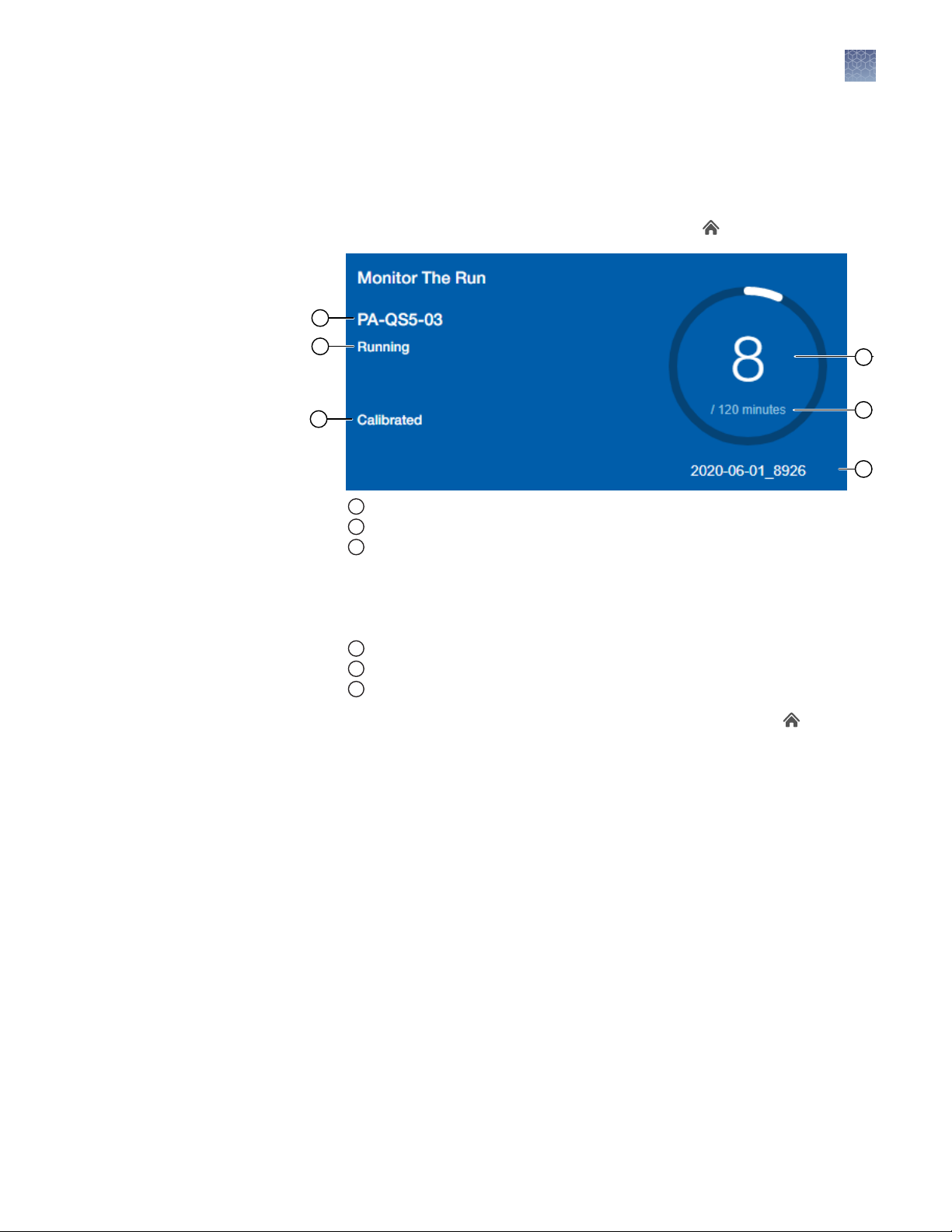
Monitor the run
6
3
1
2
5
4
Chapter 2 Methods
Setup, run, and anlayze samples with AccuSEQ™ Software v3.1 on the QuantStudio™ 5 Instrument
During the instrument run, you can monitor the run from the following places:
• On the instrument touchscreen.
• In the Monitor the Run pane of the AccuSEQ™ Software (Home) screen.
1
Instrument name
2
Instrument status (Ready, Running, Oine)
3
Calibration status
– Calibrated—All required calibrations are "Current". Required calibrations include:
Background, ROI/Uniformity, and Dyes.
– Not calibrated—None of the required calibrations are complete.
– Requires calibration—One or more dyes are not calibrated.
4
Time lapsed (if a run is in progress)
5
Total run time
6
Experiment name
• In the Open Existing Experiments pane of the AccuSEQ™ Software (Home)
screen. The experiment being run is the first experiment listed. Status is Run.
2
Note: You cannot start another run while the instrument status is Running.
MycoSEQ™ Mycoplasma Real-Time PCR Detection Kit User Guide
17
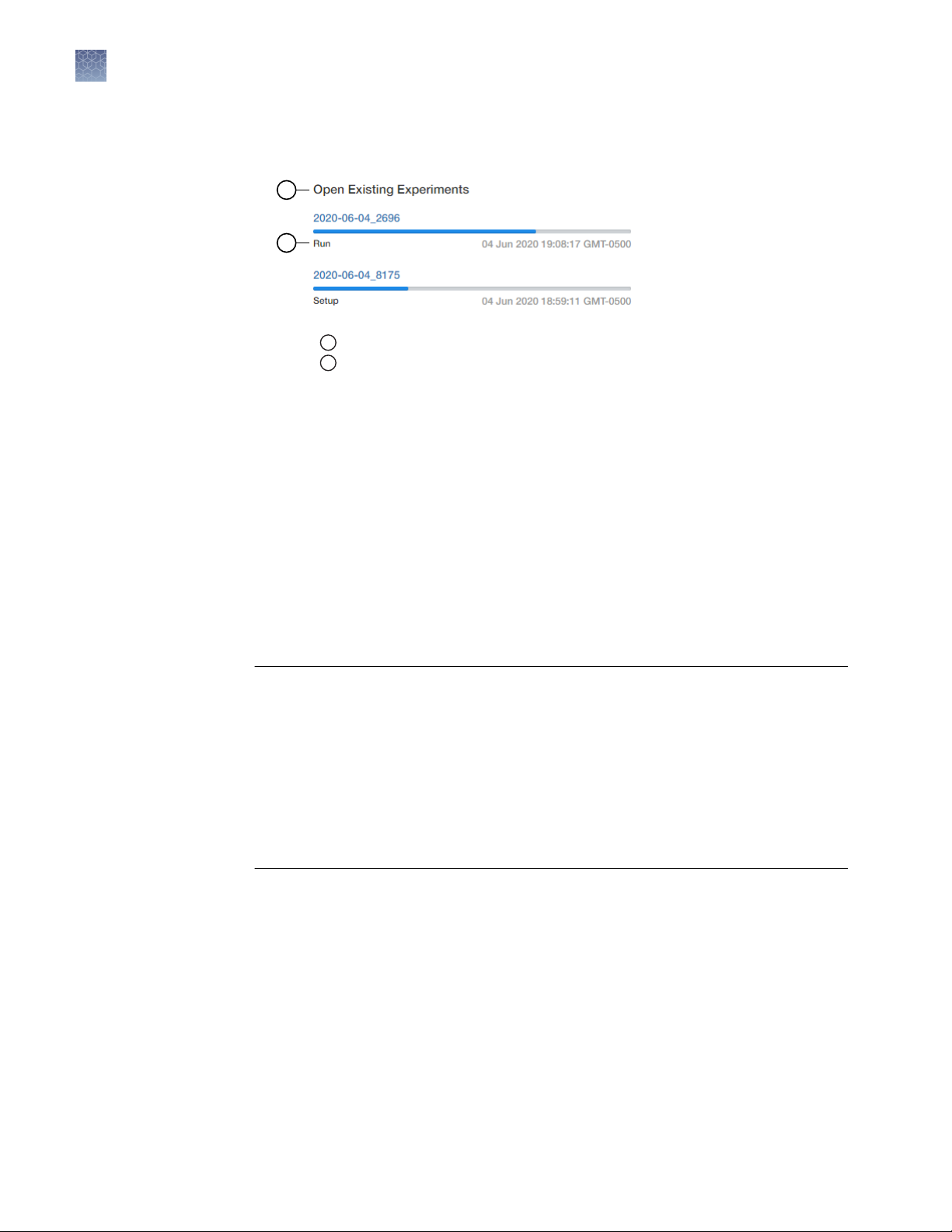
2
1
Chapter 2 Methods
2
Setup, run, and anlayze samples with AccuSEQ™ Software v3.1 on the QuantStudio™ 5 Instrument
When the run is complete, the status changes to Analysis and the status bar
displays as complete.
1
Open Existing Experiments pane
2
Experiment status
• In the Run tab of the AccuSEQ™ Real‑Time PCR Software.
You can perform the following actions.
– Select wells in the plate layout to highlight respective curves in the plot.
– Hover over curves in the plot for well information.
– (Optional) Change what is displayed in the table wells, by selecting Sample
Name, Sample Color, or Target in the View dropdown list.
Analyze the results
On run completion, the Post Run Summary displays the run length, user and
instrument information, and a list of any errors that occurred.
When the run is complete, unload the plate from the instrument.
IMPORTANT! The acceptance criteria that are provided in this section are based on
our current knowledge of assay performance in detection of Mycoplasma recovered
from a wide variety of unknown sample matrices. We recommend that you qualify
and validate the assay internally using samples that are specific to your process and
manufacturing environment (raw materials, bioreactor, or cell line samples) to ensure
that these criteria are appropriate.
For specific sample types, it may be necessary to make slight changes to the
acceptance criteria based on specific results. We can provide you with one-on-one
support during this process.
Workflow:
Review MycoSEQ™ experiments
“View the Result Summary” on page 19
▼
“Evaluate the overall shape of the Amplification Plot curves” on page 20
▼
“View and evaluate the Melt Curve Plot” on page 22
18
MycoSEQ™ Mycoplasma Real-Time PCR Detection Kit User Guide
1
4
3
2
Chapter 2
Setup, run, and anlayze samples with AccuSEQ™ Software v3.1 on the QuantStudio™ 5 Instrument
Methods
See the AccuSEQ™ Real‑Time PCR Software v3.0 User Guide (Pub. No. 100084348)
for more information on analyzing results.
View the Result Summary
The Result tab is populated when the run is complete.
Calls are made based on the customizable Rule Settings within the Analysis
Settings for MycoSEQ™ experiments. See the AccuSEQ™ Real‑Time PCR Software
v3.1 User Guide (Pub. No. 100094287) for more information.
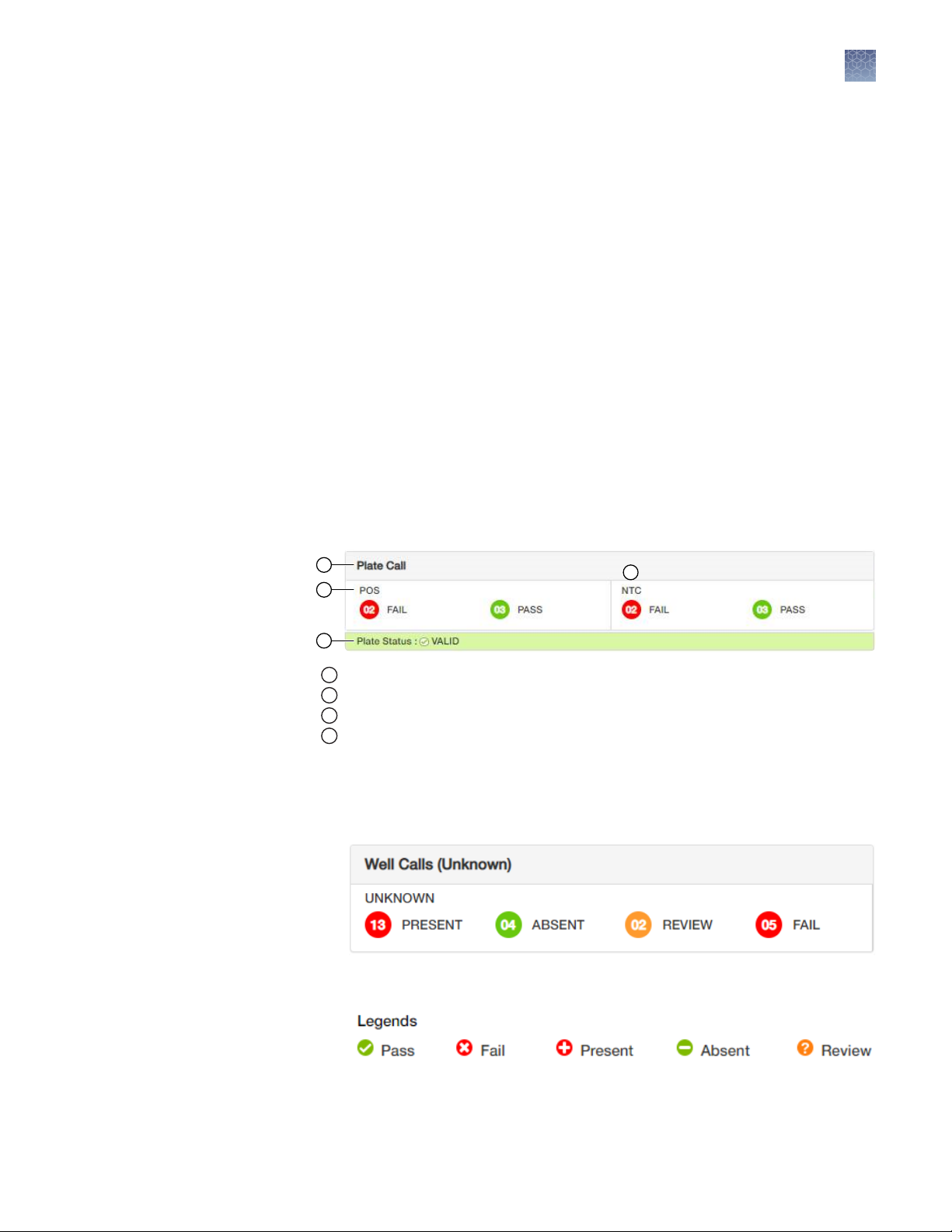
1.
View the Plate Call.
a.
View the calls for the positive controls (POS).
b.
View the calls for the no template controls (NTC).
The number inside the circle indicates the number of samples that passed or
failed. The overall Plate Status (VALID or INVALID) is determined by the POS
and NTC calls.
Note: One passing POS and NTC call are required for a Plate Status to be
VALID. This requirement is not editable.
2
Plate Call
1
Positive controls (POS)
2
Plate Status
3
No Template Controls (NTC)
4
2.
View the Well Calls (Unknown).
a.
View the total number of wells for each call—Present, Absent, Review, or
Fail.
b.
Use the Legends to view the calls in the plate
MycoSEQ™ Mycoplasma Real-Time PCR Detection Kit User Guide
19
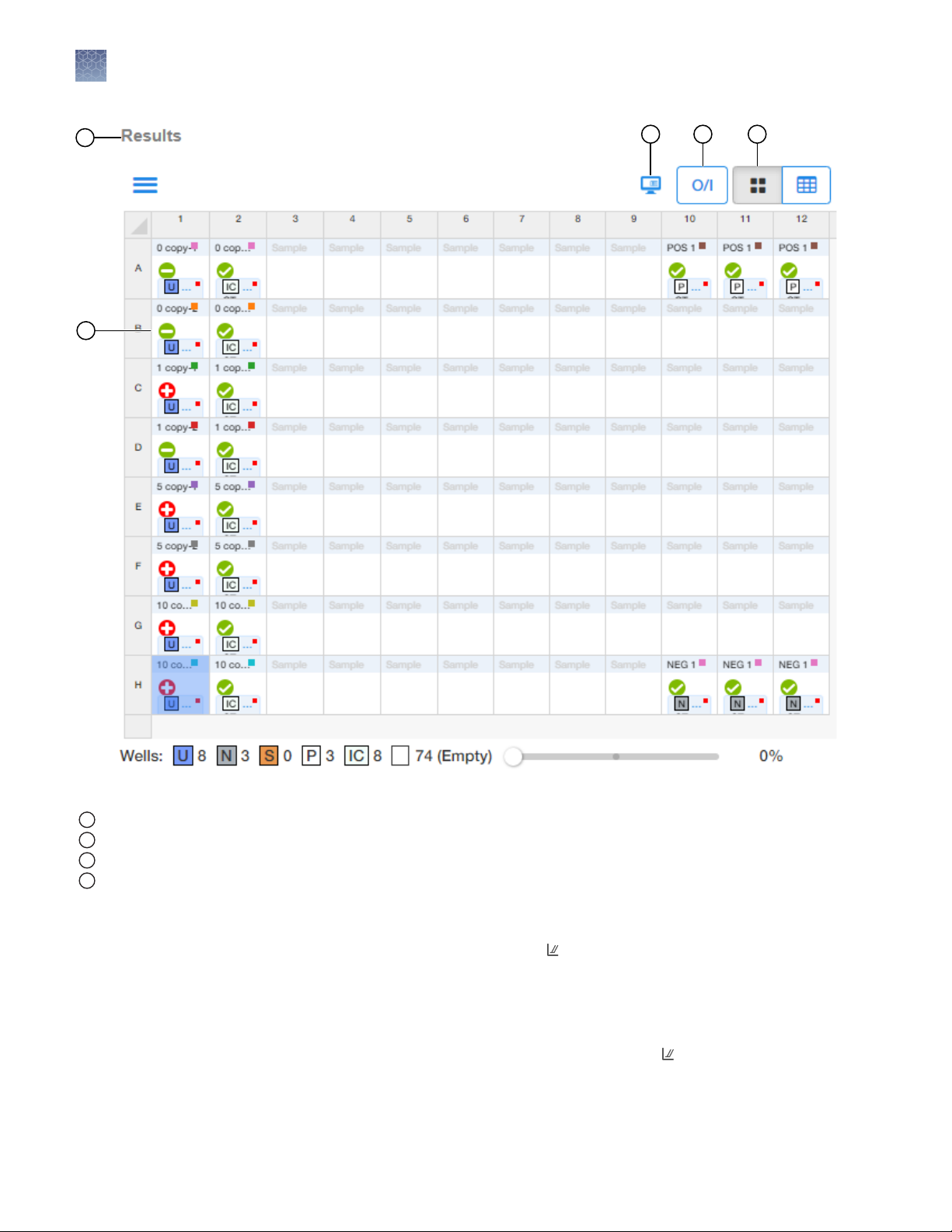
1
4
2
3
5
Chapter 2
2
Setup, run, and anlayze samples with AccuSEQ™ Software v3.1 on the QuantStudio™ 5 Instrument
Methods
Figure 3 Example MycoSEQ™ Results (Grid View)
Results pane
1
Well call
2
Flags—the number within the triangle indicates the number of QC flag calls in the well; review in QC Summary.
3
Grid View
4
Evaluate the overall shape of the Amplification Plot curves
You can evaluate the overall shape of the Amplification curves in the Result tab.
Evaluating the amplification curve in wells with a Review call can help determine if
the sample should be rerun, or whether to edit the call to Present or Absent.
Ensure that the experiment is open in the AccuSEQ™ Software.
20
1.
In the Results Analysis pane of the Result tab, click Amplification in the
horizontal scroll bar.
Note: If no data are displayed in the Result Analysis pane, then click Analyze.
MycoSEQ™ Mycoplasma Real-Time PCR Detection Kit User Guide
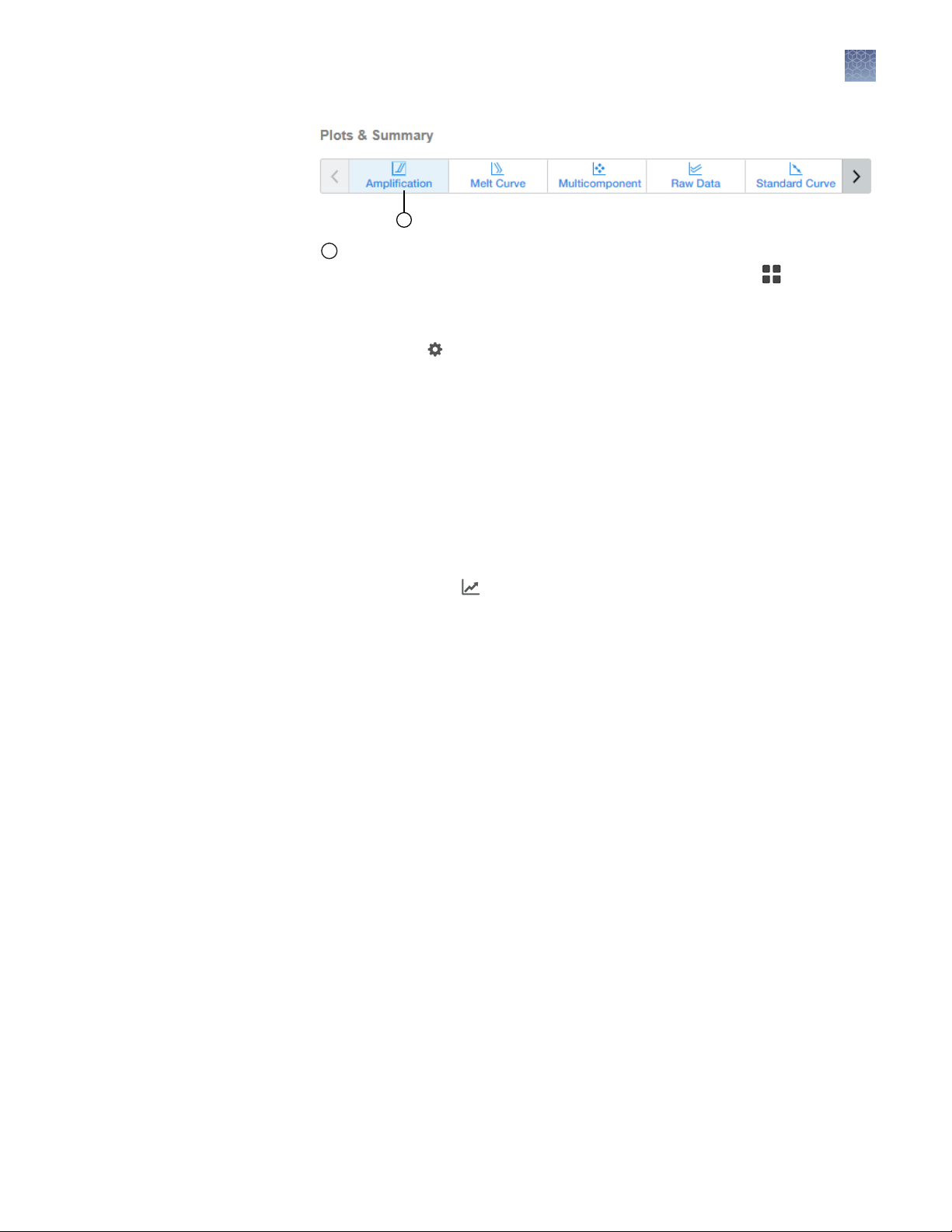
1
Chapter 2 Methods
Setup, run, and anlayze samples with AccuSEQ™ Software v3.1 on the QuantStudio™ 5 Instrument
Amplification Plot
1
The Amplification Plot is displayed for the selected wells in the (Grid View) .
2.
Ensure that the Target selected is Mycoplasma.
3.
(Optional) Click (Settings) , then make the following selections:
•
Plot Type: ΔRn, Rn, or C
•
Graph Type: Log or Linear
•
Show: Legend, Cq Mark (the cycle at which the curvature of the
T
amplification curve is maximal), or Unselected
•
Plot Color: Target, Sample, Well, or Flag_Status
•
Threshold: Select Auto or specify a threshold, then decide whether to Show
Threshold.
•
Baseline: Decide whether to Show Baseline.
2
4.
(Optional) Adjust the
a.
(Optional) In the General tab, add a Plot Title, adjust the Font and Color,
then click Apply.
b.
In the X Axis or Y Axis tabs, you can:
•
Add a Label
•
Select whether you want Tick Marks
•
Select Auto-adjust range or enter minimum and maximum values
Note: The minimum value must be greater than 0.
(Plot Properties).
MycoSEQ™ Mycoplasma Real-Time PCR Detection Kit User Guide
21
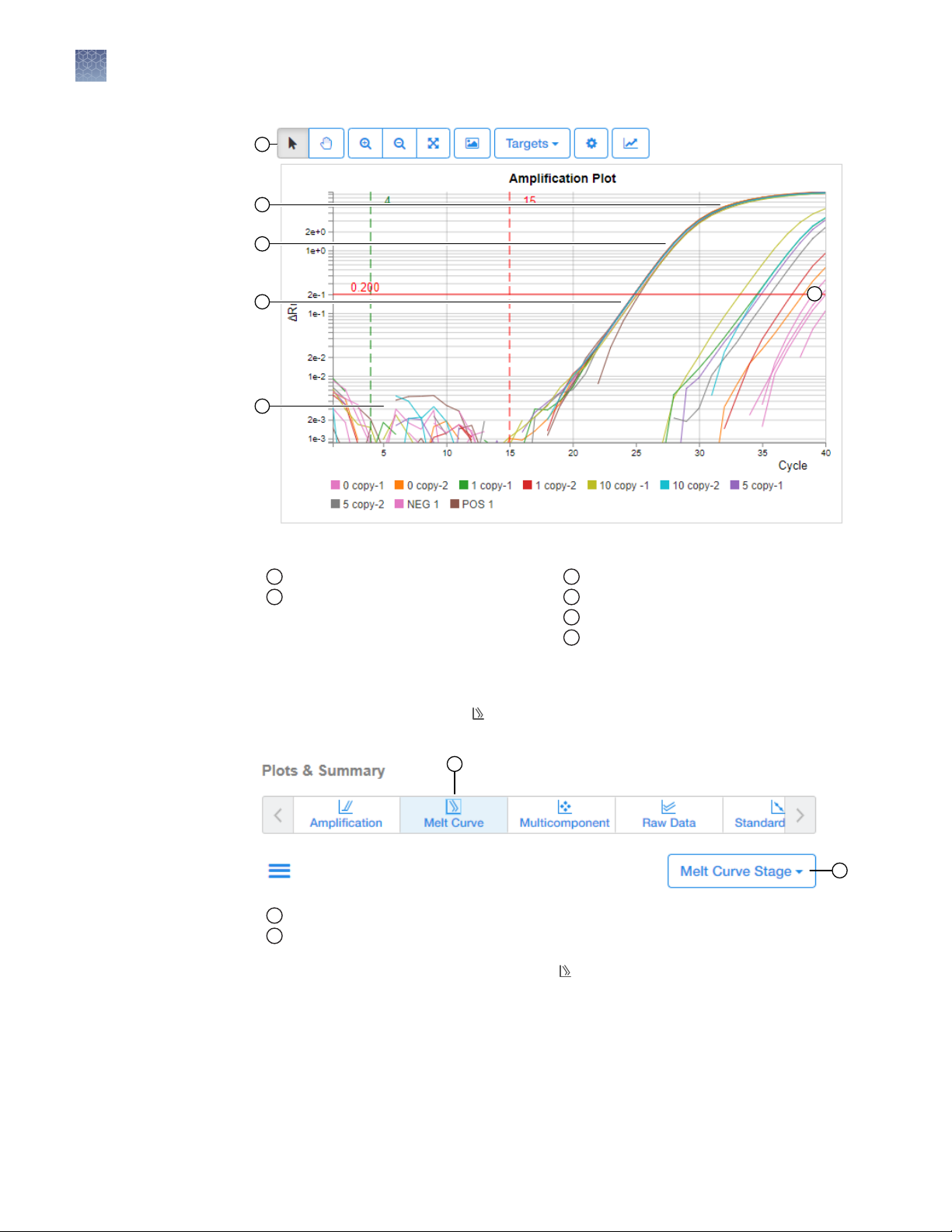
1
2
3
4
5
6
2
1
Chapter 2
2
Setup, run, and anlayze samples with AccuSEQ™ Software v3.1 on the QuantStudio™ 5 Instrument
Methods
Figure 4 Typical Amplification Plot (4 phases)
Amplification Plot tools
1
Plateau phase
2
Linear phase
3
Exponential (geometric) phase
4
Baseline
5
Threshold
6
View and evaluate the Melt Curve Plot
You can view and evaluate the
the Result tab.
Melt Curve plot
1
Melt Curve Stage dropdown
2
1.
In the Result Analysis window, select Melt Curve Plot from the horizontal
scroll bar.
2.
Keep the default Melt Curve Stage.
Custom experiments with multiple data collections in the Melt Curve Stage, can
select the stage that they want displayed.
Melt Curve Plot in the Result Analysis window in
22
MycoSEQ™ Mycoplasma Real-Time PCR Detection Kit User Guide
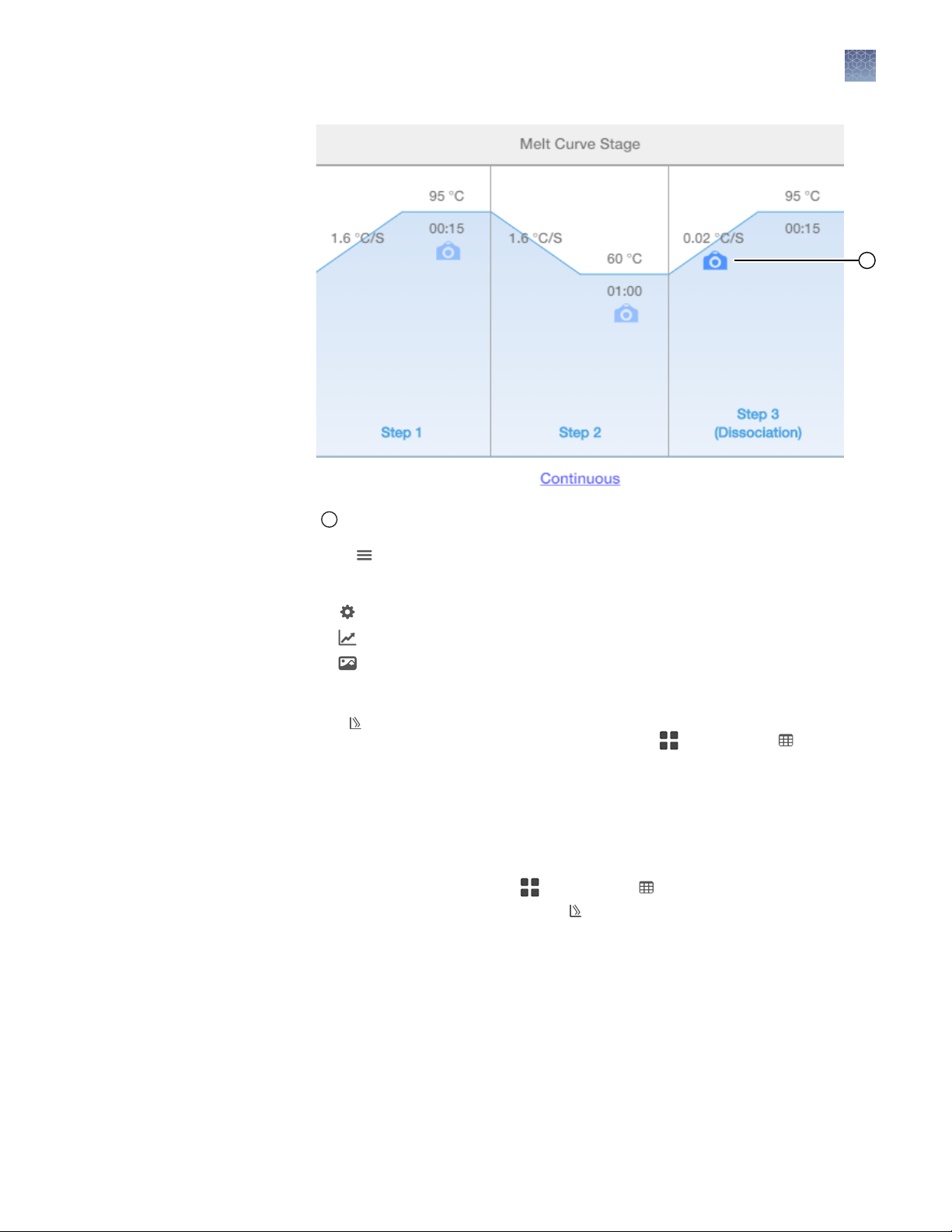
1
Chapter 2 Methods
Setup, run, and anlayze samples with AccuSEQ™ Software v3.1 on the QuantStudio™ 5 Instrument
2
Stage 3 data collection point
1
3.
Click to configure the plot, then make the following selections:
•
Targets: Select Mycoplasma.
•
Plot Settings: Select the Plot Type and Plot Color.
•
Plot Properties: Edit Plot Title, change fonts, colors, and labels.
•
Save Image: Save the image (PNG or SVG).
•
Use the select, pan, and zoom options to interact with the plot.
The Melt Curve Plot is displayed for data points that are selected in the plot
settings. The data points for selected wells in the Grid View or Table
View are highlighted in the plot.
4.
(Optional) View the default Melt Curve Stage.
5.
Confirm that amplification in the no template control and IC control wells is as
expected. Use one of the following options:
•
Select control wells in the
Grid View or Table View, then confirm the
location of the data points in the Melt Curve Plot.
•
View the amplification plots for the no template controls.
MycoSEQ™ Mycoplasma Real-Time PCR Detection Kit User Guide
23
1
2
Chapter 2 Methods
2
Setup, run, and anlayze samples with AccuSEQ™ Software v3.1 on the QuantStudio™ 5 Instrument
6.
In the Melt Curve Plot, view the signal intensity and calls for the unknown
samples.
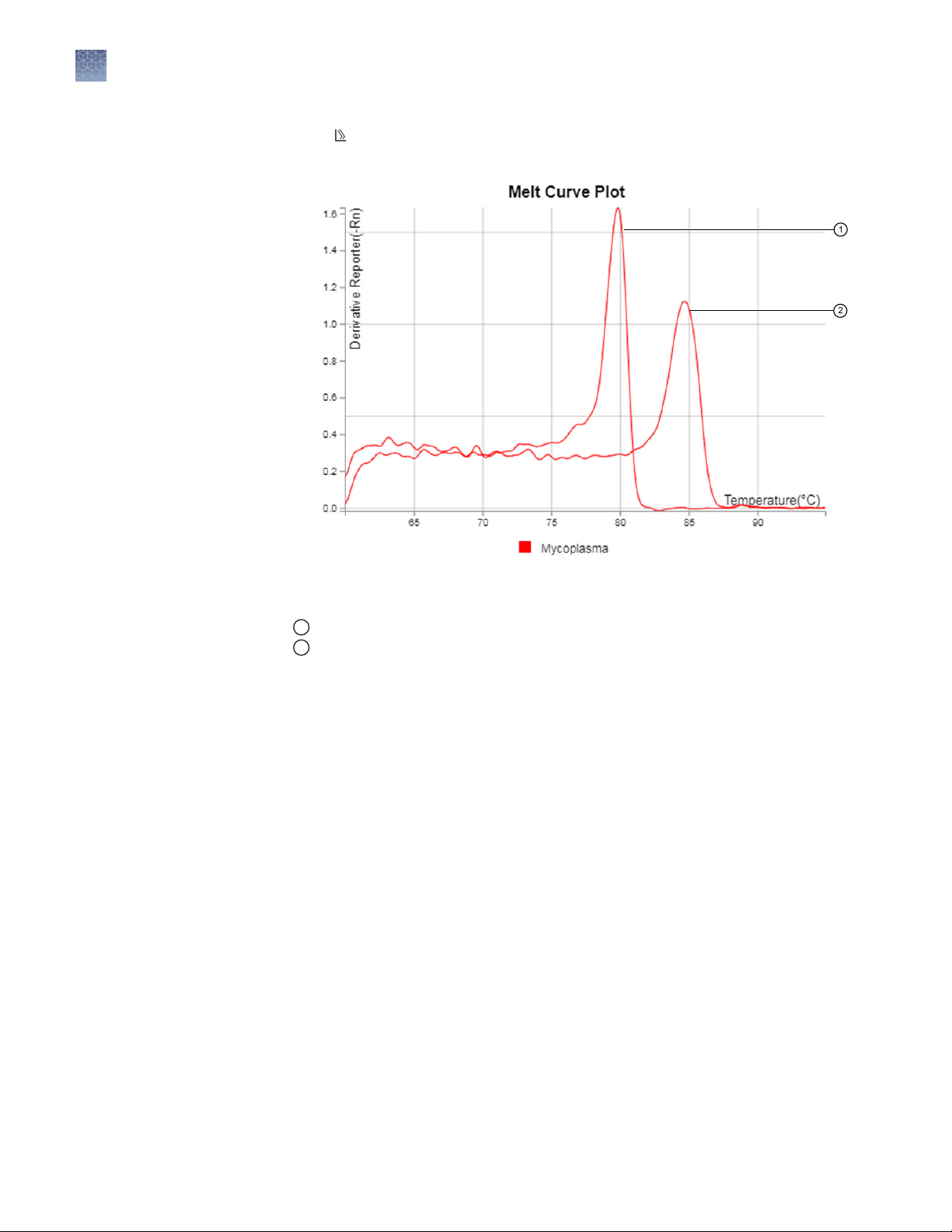
Figure 5 Example Melt Curve Plot
Melting temperature (Mycoplasma)
1
Melting temperature (positive control)
2
24
MycoSEQ™ Mycoplasma Real-Time PCR Detection Kit User Guide
Setup, run, and anlayze samples with AccuSEQ™ Software v3.1 on the QuantStudio™ 5 Instrument
Guidance for unknown samples
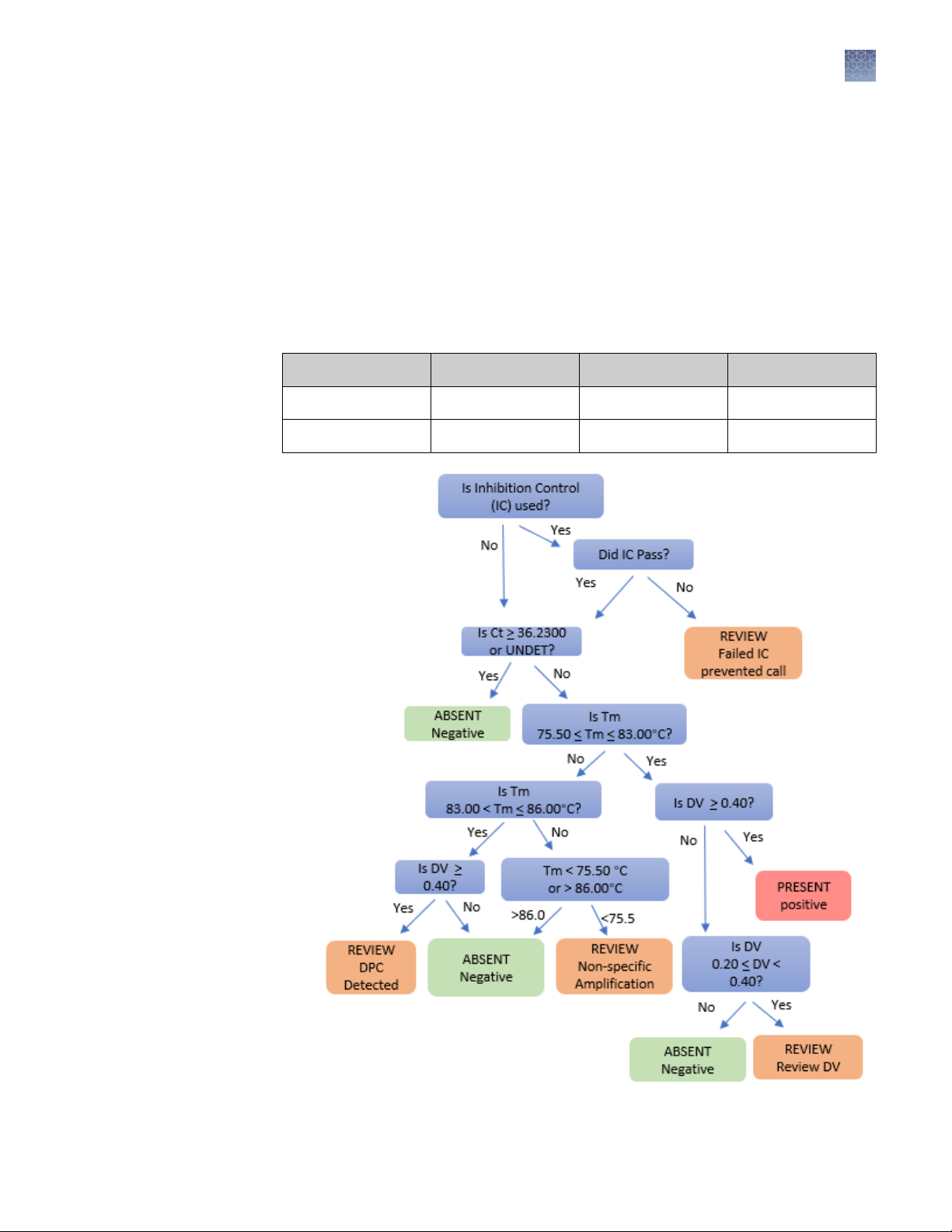
The table shows criteria for positive and negative calls. A positive call indicates that
at least one genome copy of Mycoplasma DNA was present in the unknown reaction
and the sample is positive for the presence of Mycoplasma.
Note: Tm and DV assay acceptance criteria are only relevant if Ct value for
present acceptance criteria are met. The AccuSEQ™ Software v3.1 flags these
as "Review".
Table 2 Example acceptance criteria for unknown samples: AccuSEQ™ Software
v3.1 or later
Chapter 2
Methods
2
Result C
Present < 36.2300 C
Absent ≥ 36.2300 C
t
Tm (°C) DV
t
t
75.50 < Tm < 83.00 ≥0.40
< 75.50 <0.20
Figure 6 Decision tree for unknown sample calls (with or without an inhibition control [IC])
MycoSEQ™ Mycoplasma Real-Time PCR Detection Kit User Guide
25
 Loading...
Loading...