
Ion AmpliSeq™ SARS‑CoV‑2 Research Panel
Instructions for use on an Ion GeneStudio™ S5 Series System
Pub. No. MAN0019277 Rev. B.0
80 samples per sequencing run), fast turnaround time, and
WARNING! Read the Safety Data Sheets (SDSs)
and follow the handling instructions. Wear appropriate
protective eyewear, clothing, and gloves. Safety Data
Sheets (SDSs) are available from thermofisher.com/
support.
minimal hands‑on time in SARS‑CoV‑2 research studies.
Ordering instructions
To order the Ion AmpliSeq™ SARS‑CoV‑2 Research Panel, follow
these steps.
QUICK REFERENCE
This quick reference provides guidelines and instructions for using
the Ion AmpliSeq™ SARS‑CoV‑2 Research Panel to prepare Ion
AmpliSeq™ libraries from SARS-CoV-2 samples, sequence the
libraries on an Ion GeneStudio™ S5 System, Ion GeneStudio™ S5
Plus System, or Ion GeneStudio™ S5 Prime System, then analyze
the sequencing results with Torrent Suite™ Software.
Product description .............................. 1
■
Ordering instructions ............................. 1
■
Isolate and quantify viral RNA ...................... 1
■
Prepare libraries with the Ion AmpliSeq™ SARS‑CoV‑2
■
Research Panel .................................. 3
Install plugins and import panel files ................. 5
■
Create a Planned Run for the Ion AmpliSeq™ SARS‑CoV‑2
■
Research Panel .................................. 6
Prepare template and load chips on the Ion Chef
■
Instrument ..................................... 7
Start a sequencing run ............................ 7
■
Analyze sequencing data from SARS‑CoV‑2 samples .... 7
■
(Optional) Re-run the SARS‑CoV‑2 plugins after the
■
sequencing run is complete ........................ 8
Guidelines for sample quality, viral copy number, and variant
■
calling ......................................... 9
Limited product warranty .......................... 9
■
™
1. Go to AmpliSeq.com and sign in, or register for a new
account.
2. In the navigation bar, go to the Fixed Panels dropdown
menu, then select Community Panels.
3. In the Research Area navigation pane on the left side of the
screen, select the Infectious Disease checkbox to filter the
list. Find the Ion AmpliSeq™ SARS‑CoV‑2 Research Panel in
the filtered list, then click Preview Order.
Note: Alternatively, enter SARS-CoV-2 in the search field at
the top of the screen to find the panel page.
4. In the Order options window, select GeneStudio in the
Choose instrument section, then click Next.
5. In the Format option window, select Manual or Ion Chef,
then click Next.
6. In the Order summary window, review the order, then
select Proceed to cart. As an option, select the List
recommended consumables checkbox, then click Preview
to see a list of additional products that you may need. Select
the items, then click Add all to cart.
7. Click Proceed to checkout to complete the order at
thermofisher.com.
Unless otherwise indicated, all other materials listed in this
quick reference are available at thermofisher.com.
Product description
The Ion AmpliSeq™ SARS‑CoV‑2 Research Panel consists of
two 5X primer pools that target 237 amplicons specific to the
SARS‑CoV‑2 (the virus that causes COVID-19) and 5 human
expression controls. With an amplicon length range of 125–
275 bp, the panel provides >99% coverage of the SARS‑CoV‑2
genome (~30 kb), and covers all potential serotypes. The panel
is a community Ion AmpliSeq™ panel available for order through
AmpliSeq.com.
When used in conjunction with the Ion Chef™ System and an Ion
GeneStudio™ S5 Series System, the Ion AmpliSeq™ SARS‑CoV‑2
Research Panel oers high sensitivity, high throughput (up to
Isolate and quantify viral RNA
Guidelines for RNA isolation and sample normalization
For Research Use Only. Not for use in diagnostic procedures.
• A sample containing as little as 20 copies of viral RNA
(10 copies per target amplification reaction) can be used
to prepare an Ion AmpliSeq™ SARS‑CoV‑2 Research Panel
library. For optimal results, we recommend a viral copy
number in the 200 to 200,000 range, or an amount of
total RNA between 1–10 ng. For more information, see
“Guidelines for sample quality, viral copy number, and variant
calling” on page 9.
• The amount of viral RNA among samples should be
approximately equivalent so that the target amplification
conditions you select are optimal for all samples.
• See “Recommended materials for isolation and
quantification” on page 2 for recommended Thermo Fisher
Scientific kits and master mix.
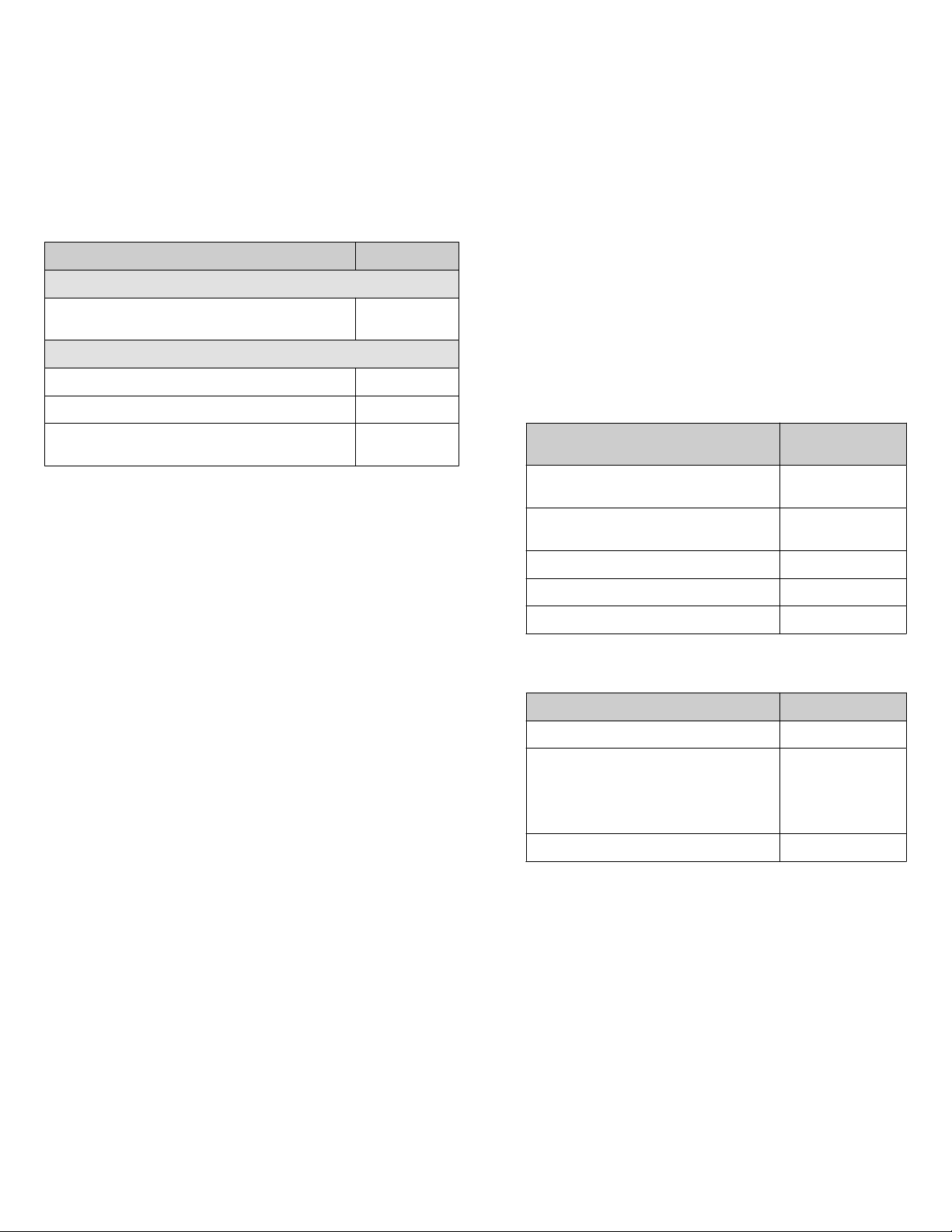
Recommended materials for isolation and quantification
We recommend the following Thermo Fisher Scientific kits and
master mix for the isolation and quantification of SARS‑CoV‑2
RNA.
Item Cat. No.
Isolation
MagMAX™ Viral/Pathogen Nucleic Acid Isolation Kit A42352 or
A48310
Quantify by real-time PCR
To determine the optimal number of target amplification cycles
to use in library preparation, quantify viral RNA copy number
in your SARS‑CoV‑2 samples using the TaqMan™ 2019-nCoV
Assay Kit v1, the TaqMan™ 2019-nCoV Control Kit v1, and the
TaqPath™ 1-Step RT-qPCR Master Mix, CG. For more information
on reaction set up, see the TaqMan™ 2019-nCoV Assay Kit v1
Product Information Sheet (Pub. No. MAN0019096).
If you are unable to quantify viral RNA copy number in your
samples, start with 16 target amplification cycles for manual
library preparation, or 17 target amplification cycles for library
preparation on the Ion Chef™ Instrument, then optimize if
needed. For more information, see “Prepare libraries manually” on
page 4, or “Prepare libraries on the Ion Chef™ Instrument” on
page 5.
Quantification
TaqMan™ 2019-nCoV Assay Kit v1 A47532
TaqMan™ 2019-nCoV Control Kit v1 A47533
TaqPath™ 1-Step RT-qPCR Master Mix, CG A15299 or
A15300
Additional positive controls are available at the BEI Resources
Repository at https://www.beiresources.org, or through other
commercial providers.
Isolate viral RNA
The MagMAX™ Viral/Pathogen Nucleic Acid Isolation Kit can be
used in either a manual or a high-throughput automated mode
using the MagMAX™ Express Magnetic Particle Processor or
KingFisher™ Purification System. Follow these basic steps to
isolate SARS-CoV-2 RNA using the MagMAX™ Viral/Pathogen
Nucleic Acid Isolation Kit (manual extraction). For detailed
information on how to use the kit, and required materials not
supplied, see the following user guides, which are available for
download at thermofisher.com.
• MagMAX™ Viral/Pathogen Nucleic Acid Isolation Kit (manual
extraction) User Guide (Pub. No. MAN0018072) or the
• MagMAX™ Viral/Pathogen Nucleic Acid Isolation Kit
(automated extraction) User Guide (Pub. No. MAN0018073)
1. For each 2019-nCoV assay (N Protein, S Protein, and
ORF1ab), combine the following components per reaction
to make a reaction mix for the total number of reactions, plus
10% overage.
Component
TaqPath™ 1-Step RT-qPCR Master Mix,
CG (4X)
2019-nCoV assay (20X; N Protein,
S Protein, or ORF1ab)
RNAse P assay (20X) 1.25 µL
RT-PCR Grade Water 11.25 µL
Total reaction mix volume 20.0 µL
Volume per
reaction
6.25 µL
1.25 µL
2. For each reaction, combine the following components in a
MicroAmp™ Optical 96-Well Reaction Plate (0.2‑mL) well.
Component
Reaction mix (from step 1) 20.0 µL
• Nucleic acid research sample or
• 1 µL 2019-nCoV Control v1 + 4 µL
RT-PCR Grade Water or
• NTC
Volume per well
5.0 µL
1. Digest 200–400 μL of each sample with Proteinase K in
a deep-well 96-well plate, then bind RNA to Nucleic Acid
Binding Beads.
2. Wash the Nucleic Acid Binding Beads.
3. Elute the RNA from the Nucleic Acid Binding Beads.
Use 1–10 ng total RNA in library target amplification reactions.
We recommend quantifying viral copy number by real-time PCR,
described in “Quantify by real-time PCR”.
2 Ion AmpliSeq
™
SARS‑CoV‑2 Research Panel on an Ion GeneStudio™ S5 Series System Quick Reference
Total reaction volume 25.0 µL
3. Set up and run the reactions on a real-time PCR instrument
using the following settings:
• Analysis method: Comparative C
t
Note: You must use Comparative Ct to analyze 2019-
nCoV assay data using QuantStudio™ Design and
Analysis Software v2 and ExpressionSuite™ Software.
• Cycling mode: Standard
• Thermal cycling protocol:
Stage Step Temperature Time
Hold
Hold
Hold Activation
UNG
incubation
Reverse
transcription
[2]
[1]
25°C 2 minutes
50°C 15 minutes
95°C 2 minutes
Denaturation 95°C 3 seconds
Cycling
(40 cycles)
[1]
Heat-labile UNG in TaqPath™ 1-Step RT-qPCR Master Mix, CG is
completely inactivated during the first ramp to 95°C.
[2]
Required for RT inactivation, first denaturation, and activation of the DNA
polymerase.
Anneal/
Extension
60°C 30 seconds
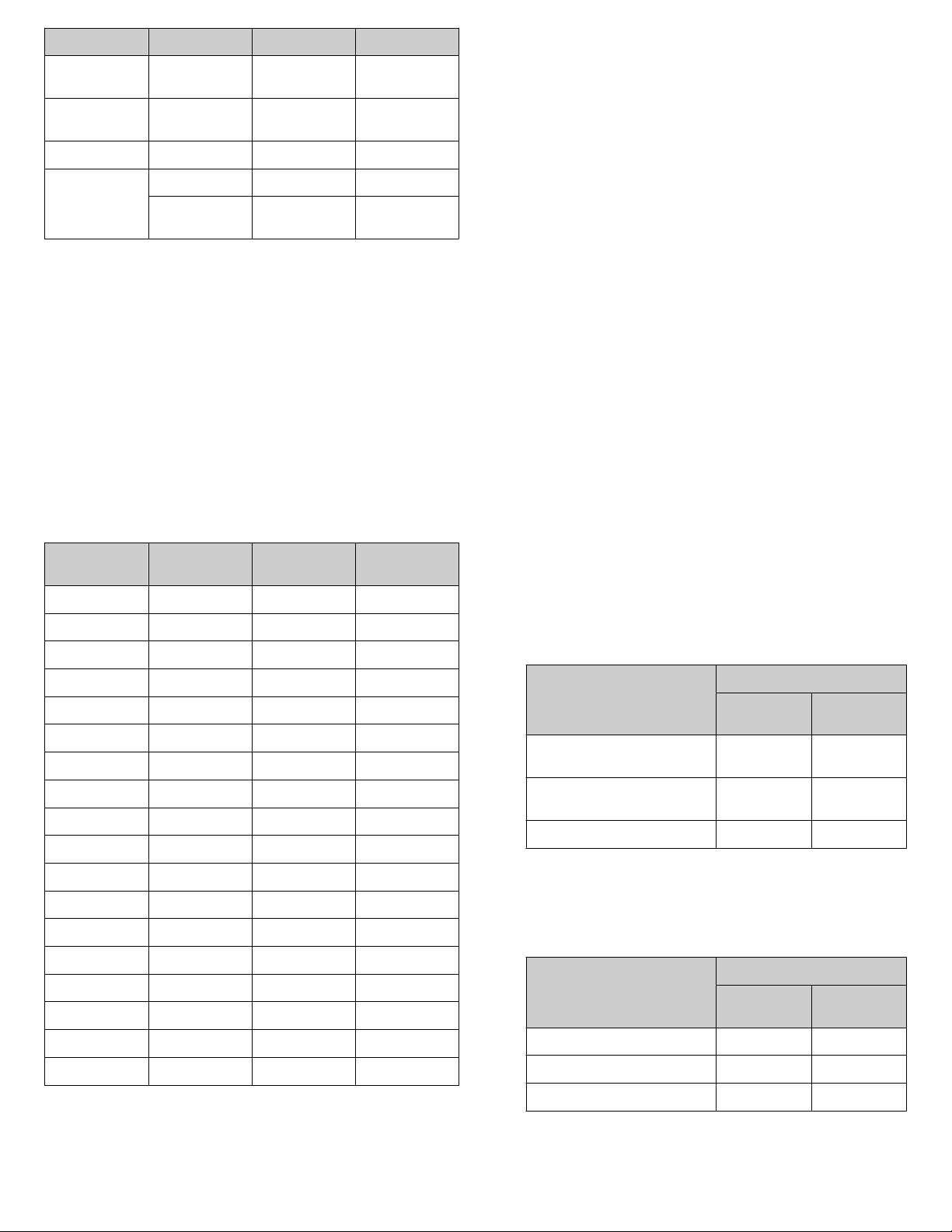
Use the Ct result for each 2019-nCoV assay to estimate copy
number in your sample. See example data in the table on
page 3.
Copy number determination by qPCR
Note: If your qPCR data give a dierent relationship between
Ct and copy number, this is likely a result of dierences in the
baseline or threshold selected. Determine the copy number of a
sample according to the known copy number in control reactions.
Copy number determination of SARS-CoV-2 with TaqMan
2019-nCoV Assay Kit v1 and TaqMan™ 2019-nCoV Control Kit
v1
Ct of N Protein
Ct of S Protein Ct of ORF1ab
qPCR reaction
34 36 37 5
33 35 36 10
32 34 35 20
31 33 34 39
30 32 33 78
29 31 32 156
28 30 31 312
27 29 30 625
26 28 29 1,250
25 27 28 2,500
24 26 27 5,000
23 25 26 10,000
22 24 25 20,000
21 23 24 40,000
20 22 23 80,000
19 21 22 160,000
18 20 21 320,000
17 19 20 640,000
™
Copies in
Prepare libraries with the Ion AmpliSeq
™
SARS‑CoV‑2 Research Panel
After reverse transcription, two options are available for preparing
libraries using the Ion AmpliSeq™ SARS‑CoV‑2 Research Panel:
• manual preparation using the Ion AmpliSeq™ Library Kit Plus
(Cat. Nos. 4488990, A35907, or A38875)—follow steps listed
in “Prepare libraries manually” on page 4
• automated preparation on the Ion Chef™ Instrument using the
Ion AmpliSeq™ Kit for Chef DL8 (Cat. No. A29024)—follow
steps listed in “Prepare libraries on the Ion Chef™ Instrument”
on page 5
For detailed information on using these kits, see the Ion
AmpliSeq™ Library Kit Plus User Guide (Pub. No. MAN0017003),
or the Ion AmpliSeq™ Library Preparation on the Ion Chef™ System
User Guide (Pub. No. MAN0013432), available for download at
thermofisher.com.
Reverse transcribe RNA with the SuperScript™ VILO
cDNA Synthesis Kit
Use the SuperScript™ VILO™ cDNA Synthesis Kit (Cat. No.
11754050) to reverse transcribe SARS‑CoV‑2 RNA for both
manual library preparation and automated library preparation on
the Ion Chef™ Instrument.
1. Warm the 5X VILO™ Reaction Mix to room temperature for at
least 20 minutes, then vortex to mix.
Note: If there is visible precipitate in the 5X VILO™ Reaction
Mix, vortex further until the mix is completely dissolved.
2. Combine the following components per reaction to make
a master mix for the total number of reactions, plus
10% overage. Up to 8 sample libraries can be prepared in
one Ion Chef™ Instrument run.
Volume
Component
5X VILO™ Reaction Mix (blue
cap)
10X SuperScript III™ Enzyme
Blend (black cap)
Total volume per well 3 µL 4.5 µL
3. For each reaction, combine the following components in a
MicroAmp™ Optical 96-Well Reaction Plate (0.2‑mL) well for
manual library preparation, or wells A1 to H1 of an IonCode
96 Well PCR Plate from the Ion AmpliSeq™ Kit for Chef DL8
for library preparation on the Ion Chef™ Instrument.
Component
Master mix (from step 2) 3 µL 4.5 µL
RNA (1–10 ng) 7 µL 10.5 µL
Total volume per well 10 µL 15 µL
Manual
2 µL 3 µL
1 µL 1.5 µL
Volume
Manual
™
Ion Chef
Instrument
Ion Chef
Instrument
™
™
™
Ion AmpliSeq™ SARS‑CoV‑2 Research Panel on an Ion GeneStudio™ S5 Series System Quick Reference 3

4. Seal the plate with MicroAmp™ Clear Adhesive Film,
vortex thoroughly, then briefly centrifuge to collect droplets.
Load the plate in a thermal cycler, place a MicroAmp
™
Compression Pad on the plate, then run the following
program to synthesize cDNA.
Temperature Time
42°C 30 minutes
85°C 5 minutes
10°C Hold
STOPPING POINT Samples can be stored at 10°C for up
to 16 hours in the thermal cycler. For longer term, store at
−30°C to −10°C.
Prepare libraries manually
See the Ion AmpliSeq™ Library Kit Plus User Guide (Pub. No.
MAN0017003) for detailed instructions for amplifying targets and
completing the manual library preparation workflow for a 2-pool
RNA panel using the Ion AmpliSeq™ Library Kit Plus. Abbreviated
steps and panel-specific guidelines are provided here.
1. Set up an amplification master mix for each sample using
the entire volume of the reverse transcription reaction.
Component
5X Ion AmpliSeq™ HiFi Mix (red cap) 4.5 µL
cDNA (from previous step) 10 µL
Nuclease-free Water 3.5 µL
Total volume 18 µL
2. Mix by pipetting up and down 5 times, then transfer 8 µL
of each sample-specific master mix into 2 wells of a 96-well
PCR plate.
3. Add 2 µL of Ion AmpliSeq™ SARS‑CoV‑2 Research Panel
primer pool 1 into the first well, and 2 µL of primer pool 2 to
the second well for a total of 10 µL in each well.
Volume
Viral copy number
Number of amplification cycles
Manual Ion Chef™ Instrument
10 26 27
20 25 26
39 24 25
78 23 24
156 22 23
312 21 22
625 20 21
1,250 19 20
2,500 18 19
5,000 17 18
10,000 16 17
20,000 15 16
40,000 14 15
80,000 13 14
160,000 12 13
320,000 11 12
640,000 10 11
Cycle number recommendations in the preceding table
are based on qPCR quantification of viral copy number.
Without qPCR quantification, use the following guidelines to
determine optimal cycle number empirically.
• Low viral load suspected: 24-27 cycles
• High viral load suspected: 14-21 cycles
• Isolates or enriched viral particles: ~12 cycles for 2 ng
input
If you are working with samples with unknown viral load,
and cannot quantify using qPCR, use 16 target amplification
cycles as a starting point for manual library preparation.
4. Seal the plate with a MicroAmp™ Clear Adhesive Film, then
place a MicroAmp™ Compression Pad on the plate.
5. To amplify targets, run the following program.
Stage
Hold Activate the
Cycle; set
number
according to the
following table
Hold — 10°C Hold
4 Ion AmpliSeq
Step Temperature Time
enzyme
Denature 98°C 15 seconds
Anneal and
extend
98°C 2 minutes
60°C 4 minutes
™
SARS‑CoV‑2 Research Panel on an Ion GeneStudio™ S5 Series System Quick Reference
STOPPING POINT
Target amplification reactions can be
stored at 10°C overnight on the thermal cycler. For longer
term, store at −30°C to −10°C.
6. See the Ion AmpliSeq™ Library Kit Plus User Guide to
complete the remaining steps in library preparation:
a. Combine primer pool 1 and 2 target amplification
reactions.
b. Partially digest amplicons.
c. Ligate barcode adapters and purify.
Note: We recommend the Ion Xpress™ Barcode
Adapters 1–96 Kit (Cat. No. 4474517) or the IonCode
Barcode Adapters 1–384 Kit (Cat. No. A29751).
d. Quantify libraries (using the Ion Library TaqMan
™
Quantitation Kit (Cat. No. 4468802), or by
Qubit™fluorometry).
You can store libraries at 4–8°C for up to 1 month. For longer
lengths of time, store at –30°C to –10°C.
™

Prepare libraries on the Ion Chef™ Instrument
B
A
C
D
1
2
4
3
See the Ion AmpliSeq™ Library Preparation on the Ion Chef
System User Guide (Pub. No. MAN0013432) for detailed
instructions for setting up the Ion Chef™ Instrument for library
preparation for a 2-pool 5X RNA panel using the Ion AmpliSeq
Kit for Chef DL8. Abbreviated steps and panel-specific guidelines
are provided here.
1. Uncap all 4 tubes in positions A, B, C, and D in the Ion
AmpliSeq™ Chef Reagents DL8 cartridge. Save the caps.
Position A (150 µL Primer Pool 1 at 2X concentration)
1
Position B (150 µL Primer Pool 2 at 2X concentration)
2
Position C (Empty tube)
3
Position D (Output tube)
4
2. Pipet 60 µL of the 5X Ion AmpliSeq™ SARS‑CoV‑2 Research
Panel primer pool 1 into the Position A tube, and 60 µL of
the 5X primer pool 2 into the Position B tube, then add 90 µL
of Nuclease-free Water to each tube to dilute primers to 2X.
Using a new tip for each tube, pipet up and down 5 times to
mix.
™
™
cycles as a starting point for library preparation on the
Ion Chef™ Instrument.
5. After run setup, tap Start Run on the instrument screen.
At completion of the run, the tube in Position D of the Ion
AmpliSeq™ Chef Reagents DL8 cartridge contains 700 µL
of combined barcoded libraries. We recommend that you
quantify the concentration by qPCR using the Ion Library
TaqMan™ Quantitation Kit (Cat. No. 4468802).
You can store libraries at 4–8°C for up to 1 month. For longer
lengths of time, store at –30°C to –10°C.
Install plugins and import panel files
You can analyze the sequencing results in Torrent Suite™ Software
with SARS‑CoV‑2 plugins.
Install the SARS-CoV-2 Research Plug-in Package
On Connect, you can download the SARS-CoV-2 Research Plugin Package. Then, in Torrent Suite™ Software, an administrator can
install the plugin package.
1. Sign in to apps.thermofisher.com.
2. Click
3. Under Resource Libraries, click Plugins.
4. Click SARS-CoV-2 Research Plug-in Package.
5. Click Download Plugin.
A compressed file that contains the plugin package is
downloaded to your local machine.
(AppConnect).
3. After adding the primer pools, load the Ion AmpliSeq
™
Chef Reagents DL8 cartridge, with uncapped tubes, in the
Ion Chef™ Instrument, then load the following components:
• Ion AmpliSeq™ Chef Solutions DL8 cartridge
• Ion AmpliSeq™ Tip Cartridge L8
• Enrichment Cartridge
• IonCode™ 96 Well PCR Plate in which you prepared
cDNA (“Reverse transcribe RNA with the SuperScript
VILO™ cDNA Synthesis Kit” on page 3)—remove the
seal from the plate.
• PCR Frame Seal
4. Set up the run on the Ion Chef™ Instrument following the
instructions in Ion AmpliSeq™ Library Preparation on the
Ion Chef™ System User Guide. On the AmpliSeq Workflow
Options screen, make the following selections:
• # of primer pools: 2
• Target amplification cycles: select the number of
cycles recommended for library preparation on the
Ion Chef™ Instrument in step 5 of “Prepare libraries
manually”
• Anneal & extension time: 4 minutes
If you are working with samples with unknown viral load,
and cannot quantify using qPCR, use 17 target amplification
6. Extract the downloaded package file.
The SARS-CoV-2 Research Plug-in Package contains
four plugins: COVID19AnnotateSnpE, IRMAreport,
AssemblerTrinity, and SARS_CoV_2_coverageAnalysis.
7. In Torrent Suite™ Software, click
(Settings)4Plugins4
Install or Upgrade Plugin.
™
8. For each of the four plugins, perform the following steps.
Repeat these installation steps for each of the four plugins.
a. Click Select File, browse to the file location, select a
plugin file, then click Open.
b. In the Install or Upgrade Plugin dialog box, click
Upload and Install.
Ion AmpliSeq™ SARS‑CoV‑2 Research Panel on an Ion GeneStudio™ S5 Series System Quick Reference 5

Configure the AssemblerTrinity Plugin
1.
In Torrent Suite™ Software, in the Plan tab, in the Templates
screen, click Plan New Run.
You must complete these steps before you run the
AssemblerTrinity plugin.
1. In a command prompt window on the Ion Torrent™ Server,
enter this command:
$ cd /results/plugins/AssemblerTrinity
2. In a command prompt window, enter one of these
commands depending on the operating system and Torrent
Suite™ Software version.
• For servers with Ubuntu™ operating system 18.04 and
Torrent Suite™ Software version 5.12, run this command
in a command prompt window:
$ sudo dpkg -i Singularity/Ubuntu18.04/
ion- singularity_3.5.3-2_amd64.deb
• For servers with Ubuntu™ operating system 14.04 and
all Torrent Suite™ Software versions from 5.4 and later,
including 5.12, run this command in a command prompt
window:
$ sudo dpkg -i Singularity/ Ubuntu14.04/
ion- singularity_3.5.3-2_amd64.deb
Import the SARS‑CoV‑2 reference sequence and target
region files
Your Ion Torrent™ Server account must be linked to
AmpliSeq.com before you download the Ion AmpliSeq
SARS‑CoV‑2 Research Panel reference sequence and target
region files from AmpliSeq.com. For more information, see the
Torrent Suite™ Software 5.14 User Guide (Pub. No. MAN0019144).
Note: You can also download panel files from the panel page on
AmpliSeq.com, then upload to the Ion Torrent™ Server from your
drive.
1. In Torrent Suite™ Software, click
(Settings)4Reference
Sequences.
2. Click Import Preloaded Reference Sequences.
3. Click Import in the Ion AmpliSeq™ SARS‑CoV‑2 Reference
row.
4. When the Status is Successfully Completed, in the Short
Name column, click ion_ampliseq_sars-cov2.
5. Scroll to the Available Target Regions and Hotspot
Files section and ensure that the Ion_AmpliSeq_SARSCoV-2.2020323.Designed.bed file is available.
Create a Planned Run for the Ion AmpliSeq
™
™
SARS‑CoV‑2 Research Panel
These instructions include the specific settings and selections
required for a Planned Run that includes the Ion AmpliSeq
SARS‑CoV‑2 Research Panel. For more information, see the
Torrent Suite™ Software 5.14 User Guide (Pub. No. MAN0019144).
Alternatively, you can use one of the preconfigured Planned Run
templates in the panel file set imported from AmpliSeq.com. A
template is provided for each chip type. If you use a Planned Run
template, you need only confirm that correct settings are selected.
™
2. In the Ion Reporter step, click Next.
3. In the Research Application step:
a. In Research Application, select DNA.
b. In Target Technique, select AmpliSeq DNA.
c. Click Next.
4. In the Kits step, make the following selections.
a. In Chip Type, select an appropriate chip.
Compatible chips (ordered separately) include the Ion
510™ Chip, Ion 520™ Chip, Ion 530™ Chip, and the Ion
540™ Chip.
b. In Barcode Set (optional), select the appropriate
barcode.
Compatible barcode sets are IonXpress and IonCode.
c. In Template Kit, select the IonChef radio button, then
select either Ion 510 & Ion 520 & Ion 530 Kit-Chef, or
Ion 540 Kit-Chef.
d. In Flows, set the number to 550.
e. Click Next.
5. In the Plugins step, select the following plugins.
• coverageAnalysis
• variantCaller. Next to variantCaller, click Configure.
In Parameter settings ensure that the default setting,
Germ Line - Low Stringency, is selected, then click
Save Changes.
• COVID19AnnotateSnpE
• IRMAreport
• SARS_CoV_2_coverageAnalysis
• (Optional) AssemblerTrinity
6. In the Projects step, click Next.
7. Complete the Plan step.
a. Enter a name for the plan.
b. In Reference Library, select ion_ampliseq_sars-
cov2(AmpliSeq SARS-CoV-2 Reference).
c. In Target Regions, select Ion_Ampliseq_SARS-
CoV-2.2020323.Designed.bed.
We recommend that you allow 1 M reads per sample for
the Ion AmpliSeq™ SARS‑CoV‑2 Research Panel. Use the
following chip capacity table to set the maximum number of
sample libraries loaded per chip.
Chip type
Ion 510™ Chip 2–3 M
Ion 520™ Chip 4–6 M
Ion 530™ Chip 15–20 M
Ion 540™ Chip 60–80 M
Reads per chip
6 Ion AmpliSeq
™
SARS‑CoV‑2 Research Panel on an Ion GeneStudio™ S5 Series System Quick Reference

8. Review the Summary pane, then click Plan Run.
Note: You need to create a Planned Run for each chip that
you load in an Ion Chef™ templating run.
Prepare template and load chips on the Ion Chef™ Instrument
After you have created a Planned Run, see the one of the
following user guides for detailed information on setting up and
running the Ion Chef™ Instrument to prepare template and load
chips, depending on the Ion Torrent™ chip you are using. The user
guides are available for download at thermofisher.com.
• Ion 510™ & Ion 520™ & Ion 530™ Kit – Chef User Guide (Pub.
No. MAN0016854)
• Ion 540™ Kit – Chef User Guide (Pub. No. MAN0010851)
Use the Ion 510™ & Ion 520™ & Ion 530™ Kit – Chef (Cat. No.
A34461), or the Ion 540™ Kit – Chef (Cat. No. A30011), and
the Ion Chef™ Instrument to template Ion Sphere™ Particles with
Ion AmpliSeq™ SARS‑CoV‑2 Research Panel libraries that you
prepared, and load one or two Ion 510™, Ion 520™, Ion 530™,
or Ion 540™ Chips. Follow the guidelines in the user guides for
combining and diluting libraries. The kits also contain the reagents
and components required for sequencing loaded chips on an Ion
GeneStudio™ S5 Series Sequencer.
Recommended library input concentration (library quantification
using the Ion Library TaqMan™ Quantitation Kit (Cat. No.
4468802)
Chip
Ion 510™ Chip
Ion 520™ Chip
Ion 530™ Chip
Ion 540™ Chip 70 pM 1050 × 10
Note:
If the library concentration is lower than the concentration
·
recommended in the table, the volume of input library can be
increased to 50 μL to reach the number of total molecules.
If your lab has been using other library quantification methods
·
such the Qubit™ dsDNA HS Assay Kit, and has an established
protocol for templating, follow that protocol.
If needed, use polyclonality and low-quality filter results from a
·
sequencing run performed with ISPs templated at the starting
concentration, then titrate up or down to achieve optimal
concentrations.
Library conc. Volume
30 pM
25 μL
Total
molecules
450 × 10
6
6
Analyze sequencing data from SARS‑CoV‑2
samples
You can annotate variants, create consensus sequences, and
generate a de novo full-length transcriptome reconstruction with
SARS‑CoV‑2 plugins in Torrent Suite™ Software. Results from
plugins that are added to a planned run are displayed in the report
summary. For more information, see “Create a Planned Run for
the Ion AmpliSeq™ SARS‑CoV‑2 Research Panel” on page 6.
Review COVID19AnnotateSnpE plugin results
The COVID19AnnotateSnpE plugin generates an annotated list
of variants. You can use this plugin to identify and annotate
variants with public or private databases and perform multisample comparisons.
1. In the Data tab, click Completed Runs & Reports, then in
the Report Name column, click the report name link for the
completed sequencing run.
2. In the left navigation menu, click COVID19AnnotateSnpE.
3. (Optional) Click Download PDF4Plugins PDF to download
a plugin report summary when the status for the plugin is
Completed.
The PDF contains the information that is shown on the
screen. Hyperlinks to the data are not active in the PDF.
4. Click the COVID19AnnotateSnpE.zip link to download a
ZIP file.
The ZIP file contains annotated variants in VCF output files
for each barcode.
Review IRMAreport plugin results
The IRMAreport plugin generates FASTA files that contain the
consensus sequence for each barcode. The Iterative Refinement
Meta-Assembler (IRMA) plugin identifies low-frequency variants
for highly variable RNA viruses.
1. In the Data tab, click Completed Runs & Reports, then in
the Report Name column, click the report name link for the
completed sequencing run.
2. In the left navigation menu, click IRMAreport.
3. (Optional) Click Download PDF4Plugins PDF to download
a plugin report summary when the status for the plugin is
Completed.
4. Click the IRMAreport.html link to open the IRMA
Start a sequencing run
Use the sequencing components in the Ion 510™ & Ion 520
& Ion 530™ Kit – Chef, or the Ion 540™ Kit – Chef to set up
and start a sequencing run with a loaded Ion 510™, Ion 520™,
Ion 530™, or Ion 540™ Chip on an Ion GeneStudio™ S5 Series
Sequencer. Continue using the appropriate user guide listed
above for detailed information.
Ion AmpliSeq™ SARS‑CoV‑2 Research Panel on an Ion GeneStudio™ S5 Series System Quick Reference 7
™
Assembled Report.
5. Click the FASTA link to view the consensus sequence for
each sample.

Review AssemblerTrinity plugin results
The AssemblerTrinity plugin generates a genome-guided or de
novo viral sequence. The AssemblerTrinity plugin uses inchworm,
chrysalis, and butterfly software modules sequentially, then builds
de Bruijn graphs to resolve alternatively spliced isoforms and
transcripts derived from paralogous genes to generate contig
sequences.
1. In the Data tab, click Completed Runs & Reports, then in
the Report Name column, click the report name link for the
completed sequencing run.
(Optional) Re-run the SARS‑CoV‑2 plugins after
the sequencing run is complete
After the sequencing run is complete, you may
want to reconfigure the plugin settings and run the
COVID19AnnotateSnpE, IRMAreport, AssemblerTrinity, and the
SARS_CoV_2_coverageAnalysis plugins again to perform a new
analysis.
Run the COVID19AnnotateSnpE plugin
Follow these steps if you want to run the COVID19AnnotateSnpE
plugin after the sequencing run is complete.
2. In the left navigation menu, click AssemblerTrinity.
3. (Optional) Click Download PDF4Plugins PDF to download
a plugin report summary when the status for the plugin is
Completed.
4. Click the Trinity_report.html link to open the Trinity De
novo Assembly Report.
5. Click the FASTA link to view the individual contigs and the
longest contig for each sample.
6. (Optional) Change the configuration settings and run
the plugin again. For more information, see “Run the
AssemblerTrinity plugin” on page 9.
Review SARS_CoV_2_coverageAnalysis plugin results
The SARS_CoV_2_coverageAnalysis plugin generates statistics,
downloadable data files, and interactive visualization of coverage
over targeted regions of the SARS-CoV-2 reference genome.
Reads mapped to human endogenous controls (specified by the
target regions) are filtered out and ignored. The report summary
lists:
• the barcodes
• the samples
• the number of mapped
reads
• the percentage of filtered
reads
Additional details regarding read coverage are also provided on a
per-barcode basis.
1. In the Data tab, click Completed Runs & Reports, then in
the Report Name column, click the report name link for the
completed sequencing run.
2. In the left navigation menu, click
SARS_CoV_2_coverageAnalysis.
3. In the SARS_CoV_2_coverageAnalysis barcode summary, in
the Barcode Name column, click a barcoded sample link
to open a detailed SARS-CoV-2 Analysis Report for that
sample.
4. In the SARS-CoV-2 Analysis Report, review the plugin
results. Click the links at the bottom of the SARS-CoV-2
Analysis Report to download associated statistics and
summary files for each barcoded sample in the run.
• the percentage of on target
reads
• mean base coverage depth
• base coverage uniformity
The variantCaller plugin provides input for the
COVID19AnnotateSnpE plugin. The variantCaller plugin analysis
must be complete before you run the COVID19AnnotateSnpE
plugin.
1. In the report summary for the completed sequencing run, in
the left navigation menu, click variantCaller.
2. Copy the variantCaller ID, which is the number in
parentheses next to the variantCaller version.
For example, the variantCaller ID in this image is 1525.
3. Click Plugins4Select Plugins to Run, then click
COVID19AnnotateSnpE.
4. In the Torrent COVID19AnnotateSnpE Plugin dialog
box, in ID of VariantCaller results, paste or enter the
variantCaller ID.
5. Click Submit.
Run the IRMAreport plugin
Follow these steps if you want to run the IRMAreport plugin after
the sequencing run is complete. You can select some barcodes
for a new analysis and change the configuration settings, if
desired.
1. In the report summary for the completed sequencing
run, click Plugins4Select Plugins to Run, then click
IRMAreport.
2. (Optional) In the Configure Plugin dialog box, make custom
configuration changes.
a. Click Barcode Sample Settings ₊, then deselect the
checkbox in the row of a Barcode to remove it from
analysis.
All selected barcodes will be included in the analysis.
b. Click Show advanced options, then customize
advanced options.
You can adjust plugin parameters. For example, one
of the default parameters that can be adjusted is Skip
barcodes with fewer than 1000 reads.
3. Click Submit.
8 Ion AmpliSeq
™
SARS‑CoV‑2 Research Panel on an Ion GeneStudio™ S5 Series System Quick Reference

Run the AssemblerTrinity plugin
Follow these steps if you want to run the AssemblerTrinity
plugin after the sequencing run is complete. You can change
configuration settings and perform an analysis that is genomeguided or an analysis that is based on de novo viral sequence.
1. In the report summary for the completed sequencing
run, click Plugins4Select Plugins to Run, click
AssemblerTrinity.
2. In the Genome-Guided dropdown list:
a. Keep the Yes selection to run the plugin with reads
aligned to the reference genome or select No to run the
plugin genome-free.
b. (Optional) Click Show Advanced Options to view
default settings.
For optimal performance, the Min count of K-mers to
be assembled is set to 2.
3. Click Submit.
Run the SARS_CoV_2_coverageAnalysis plugin
Follow these steps if you want to run the
SARS_CoV_2_coverageAnalysis plugin after the sequencing run
is complete.
1. In the report summary for the completed sequencing
run, click Plugins4Select Plugins to Run, click
SARS_CoV_2_coverageAnalysis.
2. Click Show Advanced Options to view default settings.
3. Confirm or edit the Tier 1 Coverage Depth, Tier 2
Coverage Depth, and the Tier 3 Coverage Depth.
4. Click Submit.
Guidelines for sample quality, viral copy number, and variant calling
Sample quality and viral copy number
Viral copy number Recommendations and guidelines
200 to 200,000 copies Recommended range for optimal results
120 to 199 copies Only for high-quality samples without
degradation. We recommend sequencing
and variant detection with a minimum allele
frequency of 20%. For more information
about the minimum allele frequency, see
the Torrent Suite™ Software 5.14 User
Guide (Pub. No. MAN0019144).
20 to 119 copies Only for high-quality samples without
degradation that contain a very small
quantity of human RNA. Low-frequency,
random, false-positives are likely due to
errors occurring in reverse transcription. We
recommend ignoring heterozygous variant
calls from samples in this range.
• To reliably sequence low quality samples, the samples must
have a viral copy number ≥200 copies per reaction. For
partially degraded samples, which likely includes low titer
samples, the eective copy number that can be amplified
by the Ion AmpliSeq™ SARS‑CoV‑2 Research Panel is lower
than the viral copy number detected by qPCR because
the qPCR products are shorter than the 250 bp fragments
generated by the panel.
• Even for samples with viral titer >200 copies per reaction,
you may observe reverse transcription-derived false positives
if you decrease the minimum allele frequency cuto
below 0.2 (20%). Reverse transcription-related errors occur
randomly across the genome. To minimize calling falsepositives, be certain to amplify a sucient number of RNA
molecules and set the minimum allele frequency to at least
20%.
Limited product warranty
Life Technologies Corporation and/or its aliate(s) warrant their
products as set forth in the Life Technologies' General Terms
and Conditions of Sale at www.thermofisher.com/us/en/home/
global/terms-and-conditions.html. If you have any questions,
please contact Life Technologies at www.thermofisher.com/
support.
Life Technologies Corporation | 5781 Van Allen Way | Carlsbad, CA 92008
For descriptions of symbols on product labels or product documents, go to thermofisher.com/symbols-definition.
The information in this guide is subject to change without notice.
DISCLAIMER: TO THE EXTENT ALLOWED BY LAW, THERMO FISHER SCIENTIFIC INC. AND/OR ITS AFFILIATE(S) WILL NOT BE LIABLE FOR SPECIAL, INCIDENTAL, INDIRECT,
PUNITIVE, MULTIPLE, OR CONSEQUENTIAL DAMAGES IN CONNECTION WITH OR ARISING FROM THIS DOCUMENT, INCLUDING YOUR USE OF IT.
Revision history: Pub. No. MAN0019277
Revision Date Description
B.0 8 October 2020 • Updated ordering instructions for AmpliSeq.com.
• Added new guidance for sample quality, viral copy number, and variant calling.
• Added instructions to run and view results related to the SARS_CoV_2_coverageAnalysis plugin.
A.0 24 April 2020 New quick reference for using the Ion AmpliSeq™ SARS‑CoV‑2 Research Panel on an Ion GeneStudio™ S5 Series System.
Important Licensing Information: This product may be covered by one or more Limited Use Label Licenses. By use of this product, you accept the terms and conditions of all
applicable Limited Use Label Licenses.
©2020 Thermo Fisher Scientific Inc. All rights reserved. All trademarks are the property of Thermo Fisher Scientific and its subsidiaries unless otherwise specified. Ubuntu is a
registered trademark of Canonical Limited.
Ion AmpliSeq™ SARS‑CoV‑2 Research Panel on an Ion GeneStudio™ S5 Series System Quick Reference 9

thermofisher.com/support | thermofisher.com/askaquestion
thermofisher.com
8 October 2020
 Loading...
Loading...