Page 1
PrimeQHelp@bibby
-
scientific.com
| www.bibby
-
scientific.com
+44(01785) 810433
1
Technical Note T08-002A
Figure 1
:
The
Results Editor
home screen.
Relative Quantification Analysis
General Introduction to Data Analysis
The aim of this Technical Note is to explain the principle of relative quantification analysis and to guide you through
the experimental set up and analysis of data. It begins with a general introduction to data analysis.
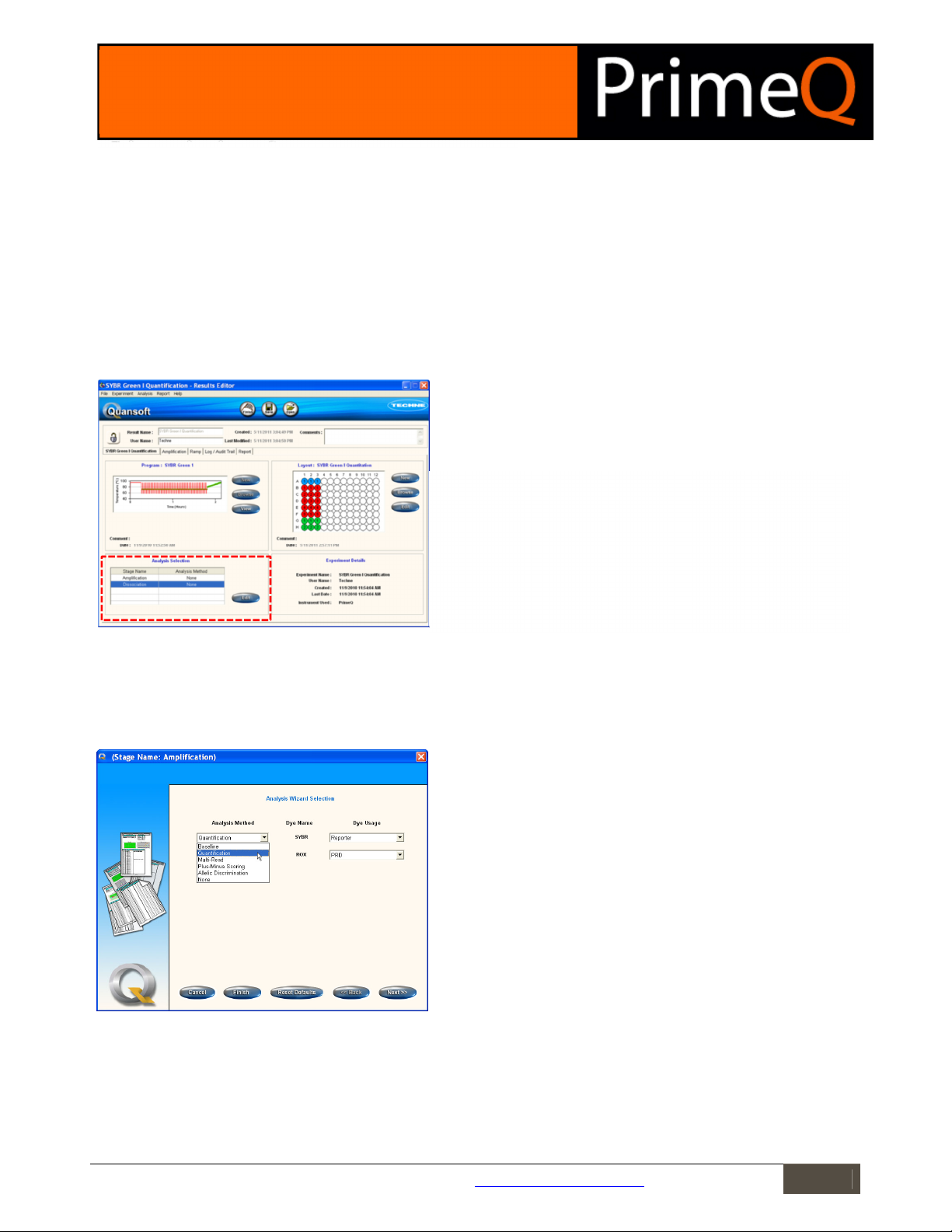
Results Editor
The starting point for data analysis is the Results Editor. Before an analysis method has been set, the Results Editor
home screen will display the plate layout and thermal cycling program. Each stage where readings have been made
will have its own tab showing a graph of raw data.
Each stage where readings have been made will have a separate results
tab enabling analysis of the data for that particular stage.
The Analysis Selection box displays the stage name as assigned in the
program setup. Only those stages that have been assigned with reads are
displayed since stages without reads have no data to analyse. Double-click
on a stage name, or highlight a stage name then click on Edit to launch the
Analysis Wizard Selection screen.
Selecting the Analysis Method
The Analysis Selection box on the Results Editor home screen allows the user to define the method of analysis to
be applied to the readings gathered during the PCR run. Highlighting a stage name and pressing the Edit button will
launch the Analysis Wizard Selection screen and allow an Analysis Method to be assigned for that stage.
Figure 2: The Analysis Wizard Selection screen.
Analysis Method: The drop-down menu lists analysis types appropriate to
the selected stage. These will be dependent on the number of reads and
the number of cycles programmed into the run.
Dye name: The name of the dyes selected in the program setup will be
displayed.
Dye Usage: Assign a Dye Usage from the list in the drop-down menu. There
will be one dye usage box for each read present in the stage.
Cancel: Aborts the procedure and takes the user back to the Results Editor.
Finish: Accepts all the default analysis settings for the analysis method
chosen and closes the Wizard.
Reset defaults: Returns all analysis parameter settings to the defaults.
Back/Next: Allows the user to move between screens in the Analysis
Wizards.
The Analysis Wizards
Once an analysis method has been defined, a series of default settings will automatically analyse the data. The
defaults can be viewed in the Analysis Wizards and edited if required. The intuitive Analysis Wizards explain in
detail the mathematics behind the analysis settings and will lead you through each stage of the analysis setup.
With experience, analysis can be quickly performed using the Analysis Properties displayed in the Parameters (PAR)
box feature found on each data graph.
Page 2

PrimeQHelp@bibby
-
scientific.com
|www.bibby
-
scientific.com
+44(01785) 810433
2
T08-002A: Relative Quantification Analysis
Analysis Method Options
The following analysis options are available in Quansoft:
Baseline
This simple analysis method allows for correction of differences in background fluorescence. It is also incorporated
into many of the other analysis methods.
Quantification
Quantification analysis is used to determine the absolute or relative quantity of a target DNA template in a given
test sample by measuring the cycle-to-cycle change in the fluorescent signal. The fluorescent signal increases
proportionately to the amount of amplified DNA and quantification is performed either by comparison of the
fluorescence of a PCR product of unknown concentration with that of several dilutions of an external standard, or
by comparing the fluorescence of one product relative to another. To be able to make this comparison, the
fluorophore is measured at a point in the amplification where the reaction efficiency can be considered optimal.
This is generally around the cycle at which an increase in fluorescence is first detected.
Dissociation curve
Dissociation curve analysis can add to the information obtained from the PCR. Also known as melting curve
analysis, it measures the temperature at which the DNA strands separate into single strands. This provides a
measurement of the melting temperature or Tm, taken as the point at which 50% of the double stranded DNA
(dsDNA) molecules are dissociated. Using the easy-to-program ‘ramp’ function, PrimeQ will perform a thermal
ramping program that can be used to determine the Tm of the PCR product. This analysis provides the user with
extra confidence in experiments using intercalating dye chemistry for identifying amplification of non-specific
products or contamination.
Plus/minus scoring
This analysis exploits PrimeQ’s fluorescence technology to determine with ease and accuracy the presence or
absence of a PCR product in any given sample. Input data can either be kinetic (where readings are taken
throughout the amplification stage) or end-point (readings taken at the end of the run). The software scores the
samples as positive or negative according to user-defined thresholds.
Allelic discrimination
Users of PrimeQ have the option of this powerful technique capable of detecting single nucleotide differences
(SNPs). It can be used to discriminate between genotypes, mutations and polymorphisms within or between
samples simply by comparing the fluorescence signal obtained using allele-specific, dye-labeled probes.
Multi-read
This is a simple end point analysis method which will report the average fluorescence of a selected number of
readings. It is useful for assays other than PCR, for example fluorescence-based DNA assays, where just the
fluorescence of a sample needs to be measured; in this way PrimeQ can be used as a simple fluorescence plate
reader or fluorimeter.
Page 3

PrimeQHelp@bibby
-
scientific.com
|www.bibby
-
scientific.com
+44(01785) 810433
3
T08-002A: Relative Quantification Analysis
Absolute
quantification
Uses known standards to generate a standard curve and calculates the “absolute”
Relative quantification
Uses known standards to generate a standard curve for each reporter and compares
Relative Cq
Subtracts the Cq value
of a reference from
an unknown amplified from the same
Analysis method: Relative Quantification
This analysis method determines the concentration of one target relative to another and is often used to compare
the expression of a gene of interest with that of a reference or housekeeping gene amplified from the same
sample. Automatic relative quantification analysis in Quansoft can only be applied to a multiplex reaction where
the two targets are amplified in the same well. This ensures that the amount of sample added to the well is
consistent for both the gene of interest and reference.
There are two methods available in Quansoft to compare the relative amounts of two samples. Firstly, Relative
Quantification is similar to absolute quantification analysis but uses two reporter dyes and two standard curves to
compare the concentration of one DNA template relative to a second template. This method is useful for
optimizing reactions for multiplex assays. Secondly, Relative Cq is used in the comparison of the relative amount of
samples in different wells. This approach is particularly useful for screening assays where it is necessary to compare
a fold difference of sample B to a calibrator sample A, for example. In such an assay, information about absolute
amounts is not required as the value relative to the calibrator provides the necessary information. No standards
are required. This is commonly known as the 2
Both approaches use quantification cycle (Cq) calculation as the basis of the quantification. The Cq is the cycle
number at which the concentration of the amplicon (measured by fluorescence) reaches a set threshold. The Cq is
inversely proportional to the initial template concentration. Quansoft can use one of two methods to calculate Cq
values: fit points or first derivative maximum; further details are given below.
-ΔΔCq
method.
value of the unknowns e.g. copies/well, µg/ml etc.
the absolute concentrations of the two reporters in the same well (ratio of one to the
other).
sample and compares to a control sample (2
Table 1: Quantification analysis options. Relative quantification methods are described in this Technical Note.
-ΔΔCp
method).
Setting up and running the experiment
Quantification analysis is a kinetic analysis method and requires fluorescence readings to be taken at each cycle of
the amplification stage. Therefore a reading for each reporter and a passive reference dye (PRD), if used must be
included at the end of the extension step of each cycle.
For relative quantification using a standard curve, standards of know concentration or copy number amplified by
the same primers as the target must also be prepared. This is to ensure that the reaction efficiency is the same
between the standards and unknowns. We recommend that you use at least three standards diluted serially at 10fold dilutions. For increased precision, all reactions should be performed in duplicate or triplicate. The standards
need to be defined in the plate layout and the concentrations added to the Well Information table in the Plate
Layout Editor. To enable the relative quantification analysis, standards of the same concentration for each reaction
of the multiplex must be amplified in the same wells. For example, if standards of 1000, 10,000 and 100,000 copies
for reaction A are placed in wells B1, C1 and D1, then standards of 1000, 10,000 and 100,000 copies for reaction B
must also be amplified in wells B1, C1 and D1. The same applies to unknowns.
Page 4

PrimeQHelp@bibby
-
scientific.com
|www.bibby
-
scientific.com
+44(01785) 810433
4
T08-002A: Relative Quantification Analysis
For Relative Cq analysis, no standards are required but the gene of interest and reference (or housekeeping) gene
must be amplified in the same well for Quansoft to be able to analyse the results automatically.
During the run, the real-time collection of data can be monitored on the Run Screen. The plate layout shows the
fluorescence curve on a per-well basis and the temperature profile plot indicates how far the run has progressed.
Fluorescence from the different reporter dyes can be viewed by clicking on the Dye to View drop down menu.
When the run has completed, results can be viewed in the Results Editor with data from each stage of the run
located under its own tab.
Quantification Analysis Wizard setup
Once the PCR has completed, open the results file and from the Results Editor home screen set up the
Quantification analysis as described below.
• In the Analysis Selection box, highlight the stage name for analysis to be applied and click Edit. The Analysis
Wizard will launch.
• Select Quantification from the drop-down menu in the Analysis Method selection box.
• Assign a dye usage for each of the reads (e.g. Reporter, REF etc.) and click Next. The Quantification Analysis
Wizard will launch.
• If a PRD was assigned in the dye usage menu, the next screen will offer the option of PRD correction. Click Next.
Figure 3: Analysis Wizard Selection.
Select the Quantification analysis method and assign dye usages.
Baseline Correction
Follow the directions in the Operator’s Manual in section 4.5 to select an appropriate baseline correction method.
Note that the default parameters may not always be suited to the fluorescent data, especially if an increase in
fluorescence is first seen to occur prior to 10 cycles of amplification. It is recommended to check the baseline
correction parameters whilst viewing the fluorescent data graphs and adjusting as necessary using the Analysis
Properties displayed in the PAR box (see Viewing and Changing the Parameters below). Click Next to progress to
the next screen.
Cq Calculation Method
Quansoft offers two methods to calculate Cq values: fit points or first derivative maximum. The latter is an
automatic method dependent entirely on mathematical calculations and requires no user input. The fit points
method is more flexible and involves the setting of thresholds. See the Technical Note T08-001A: Absolute
Quantification Analysis for a further explanation of Cq calculation.
Once you have set the Cq calculation method, click Next to progress to the Relative Quantification screen.
Page 5

PrimeQHelp@bibby
-
scientific.com
|www.bibby
-
scientific.com
+44(01785) 810433
5
T08-002A: Relative Quantification Analysis
calibrator sample.
Relative Quantification
When there are two or more reporters (i.e. a multiplex assay), the Quantification Analysis Wizard will offer the
choice of Relative quantification which compares the concentration of two reporters and requires standards, or
Relative Cq which compares the Cq values of two reporters and does not require standards.
For Relative Cq (2
the Analysis Wizard. In addition, for a full analysis to be calculated, the calibrator or control sample, to which all
other samples are to be compared, must be defined as CAL (Calibrator) in the plate layout.
-ΔΔCp
method), one of the reporters is usually assigned a REF (reference) dye usage at the start of
Figure 4: Relative Quantification analysis. Relative Quantification compares the concentration of two reporters and calculates the ratio of
one to the other. Relative Cq compares the Cq of two reporters by normalising to a reference and calculating the fold difference relative to a
Once the analysis has been selected, click Next to progress to the Report Options.
Report Options and Summary
The Report Options screen allows you to select data to appear in a printable report. The selection can be changed
from the Report tab in the Results Editor if necessary. The Summary screen provides a quick checklist of the
parameters set up in the Analysis Wizard.
• Click Back to change any settings or Cancel to abort the procedure.
• Click Finish to complete the set up.
Viewing the analysed results
Once the analysis has been set up, click on the results tab for the stage which has been set up for quantification
analysis. The analysed data will be displayed as a series of graphs which will vary according to view selection.
Individual graphs can be closed or enlarged for easier viewing. Clicking an individual well or a selection of wells in
the plate layout will highlight just the selected well(s) on the graphs. Clicking the Show All Wells button will reselect all wells.
Relative Quantification
The Results Editor provides three display options for the relative quantification data which can be selected using
the Dye to View drop down menu. The Relative Quantification option displays the individual standard curve
graphs for each reporter.
Page 6

PrimeQHelp@bibby
-
scientific.com
|www.bibby
-
scientific.com
+44(01785) 810433
6
T08-002A: Relative Quantification Analysis
Figure
7: Combined standard curves displays the standard curves of each
Figure
5: Relative Quantification in the Dye to View options displays
the individual standard curves for each reporter.
To view the full results data for an individual reporter, select the individual reported dye name. The results will be
displayed in the same format as absolute quantification with a baseline correction graph (if chosen in the analysis
setup), a quantification cycles graph and the standard curve. This display can be used to find the best fit for the
crossing line. Note that the crossing line must be in the same position for both standard curves, so each reporter
will need to be reviewed in turn to obtain the best fit for each.
Figure 6: Selecting an individual reporter displays the full results data as
in absolute quantification analysis.
The Combined standard curves view is used to display the standard curves for each reporter on the same plot. The
details of each standard curve can be useful when optimizing reactions in preparation for multiplexing.
reporter on the same plot for comparison.
• y = the slope; ideally this should be approximately -3.32.
• c = the value at which the line intercepts the y axis.
• R2 = Correlation coefficient (mean squared error of the determination). A perfect correlation has a value of 1.
• E = efficiency of the reaction whereby E = 10
target every cycle).
Parallel standard curves indicate reactions of similar efficiency.
-1/slope
. The ideal value is 2 for 100% efficiency (i.e. a doubling of
Page 7

PrimeQHelp@bibby
-
scientific.com
|www.bibby
-
scientific.com
+44(01785) 810433
7
T08-002A: Relative Quantification Analysis
Figure
8: Results table for Relative Quantification.
Figure
9: Relative Cq displays the Cq graphs for each reporter.
The results table displays the calculated results for each reporter and compares the ratio of one to the other.
No: Well number.
Well ID: Location of well.
Sample ID: User-supplied or default name of well.
Cq (Dye 1): Quantification cycle for Dye 1.
Conc (Dye 1): Calculated concentration of sample for Dye 1.
Cq (Dye 2): Quantification cycle for Dye 2.
Conc (Dye 2): Calculated concentration of sample for Dye 2.
Ratio: Dye 1 divided by Dye 2.
Comments: User inputted text.
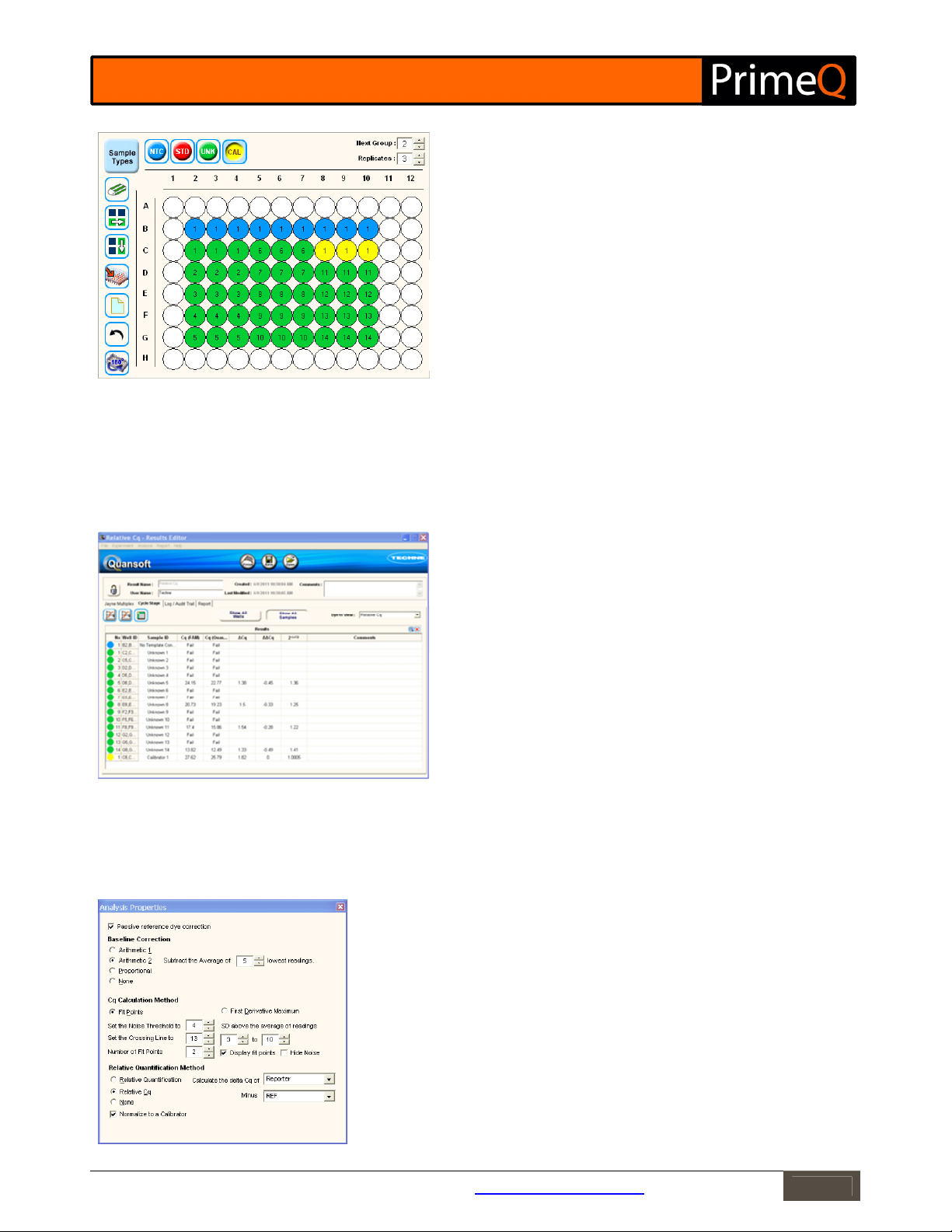
Relative Cq
Clicking on Relative Cq in the Dye to view drop down menu will display the quantification cycle graphs for both
reporters. To view the full results data for an individual reporter, including a baseline correction graph, select the
individual reported dye name.
During the analysis set up, you need to select which reporters to compare and allocate a dye usage. The first is
usually the gene of interest and the second the REF gene amplified from the same sample. The first step of the
relative Cq calculation is normalization of the gene of interest to the reference gene. This is calculated by
subtracting the Cq of the reference from that of the gene of interest (ΔCq). This value can then be compared to
that of a calibrator which is a sample selected to compare all the other samples to i.e. the expression of the
calibrator will be 1.0. To define a calibrator you will need to go to the plate layout and assign a CAL sample type to
the chosen calibrator sample (see Figure 10). The calculation, which is performed by the software, can be
summarized as follows:
•
Calculate Cq for each sample (using either fit points or first derivative maximum).
•
Calculate the ΔCq for each sample e.g. ΔCq = Cq Reporter – Cq REF.
•
If a calibrator sample is present, then assign this sample as a CAL sample type in the plate layout.
•
Calculate ΔΔCq e.g. ΔΔCq = ΔCq sample – ΔCq calibrator.
•
Calculate 2
-ΔΔCq
Page 8
PrimeQHelp@bibby
-
scientific.com
|www.bibby
-
scientific.com
+44(01785) 810433
8
T08-002A: Relative Quantification Analysis
Figure 10: Designate a Calibrator sample in the plate layout.
Click on Sample Types to open the Sample Type screen. Select Calibrator -
CAL and transfer to the Active list.
Next, select the CAL sample type and click on the well(s) containing the
chosen calibrator.
This value represents the amount of target normalized to a control gene (REF) and relative to a calibrator (CAL)
sample. If this number is more than 1, it represents a fold increase in expression over the calibrator which in theory
has no upper limit. However, be aware that a decrease can only be represented by a value between 0 and 1 such
that a seemingly small decrease may in fact represent a significant decrease in expression relative to the calibrator.
The results of the calculation are displayed in the Results table.
Figure 11: Results table for Relative Cq.
No: Well number.
Well ID: Location of well.
Sample ID: User-supplied or default name of well.
Cq (Dye 1): Quantification cycle for Dye 1.
Cq (Dye 2): Quantification cycle for Dye 2.
ΔCq: Calculated ΔCq.
ΔΔCq: Calculated ΔΔCq.
-ΔΔCq
2
: Calculated 2
Comments: User inputted text.
-ΔΔCq
Viewing and Changing the Parameters
The analysis parameters as set up in the Quantification Wizard can be changed at any time and can be easily
accessed via the PAR box on any of the graphs. You can change any of the parameters, and in doing so, the changes
will be reflected immediately in the adjacent graphs.
Figure 12: Analysis Parameters.
Click the PAR button next to any of the graphs to bring up the analysis parameters for
Relative Quantification. If any settings are changed, the data will be recalculated and
the graphs and results table updated accordingly.
PRD and baseline correction parameters can be adjusted as required.
Changing the Cq Calculation Method will automatically update your graphs and in turn
this will result in different Cq determinations.
The relative quantification method and reporters to compare can be changed.
Page 9

PrimeQHelp@bibby
-
scientific.com
|www.bibby
-
scientific.com
+44(01785) 810433
9
T08-002A: Relative Quantification Analysis
PrimeQ Report
If a printed report of the analysed data is required, open the Report tab of the Results Editor to view the
Quantification analysis report. To change any of the report contents, click on the Report Options icon to open up
the Report Options box. Tabs will display the report options relevant for each stage. Change as appropriate and
click Done to finish. The report can be printed or saved as a PDF file for future reference.
Saving
Saving will overwrite any previous analysis, therefore to preserve a particular analysis set up, click on File followed
by Save As… to save as a different file name.
Trademarks
FAM™ and ROX™ are trademarks of Applera Corporation or its subsidiaries in the U.S. and certain other countries.
Quasar® is a registered trademark of Biosearch Technologies, Inc.
SYBR® is a registered trademark of Life Technologies Corporation.
 Loading...
Loading...