Page 1
Data Explorer™ Software
Version 4 Series Software
User Guide
Page 2

© Copyright 2001, Applied Biosyst ems . All right s reserved.
For Research Use Only. Not for use in diagnostic procedures.
Information in this document is subject to change without notice. Applied Biosystems assumes no responsibility for any errors that
may appear in this docume nt . T hi s do cument is believed to be com pl et e a nd accurate at the time of publica ti on. In no event shall
Applied Biosystems be liable for incidental, special, multiple, or consequential damages in connection with or arising from the use
of this document.
Applied Biosystems is a registered trade m ark of Applera Corporation or its subsidiaries in the U.S. and certain other countries.
AB (Design), Applera, Biospectrometry, CombiSolv, Data Explorer, Mariner, and Voyager are trademarks of Applera Corporation
or its subsidiaries in the U.S. and cer tain other co untries.
HP and Laserjet ar e registered tradem arks of Hewlett-Packard Co.
Microsoft, PowerPoint, Visual Basic, and Windows NT are registered trade ma rks of Mic rosoft Corporation.
All other tradem arks are the sole property of their res p ective owners.
Printed in the USA, 7/2001
Part Number 4317717 Rev. C
Page 3

Table of Contents
Table of Contents
How to Use This Guide.............................................................. xi
Chapter 1 Data Explorer™ Basics
1.1 Overview .............................................................................. 1-2
1.2 File Formats and Types
1.2.1 Software Applications Compatibility ....................................... 1-5
1.2.2 Data (.DAT) File Format ......................................................... 1-5
1.3 Parts of the Data Explorer Window
1.4 Customizing the Data Explorer Window
1.4.1 Setting Default Values ...........................................................1-17
1.4.2 Customizing Processing and Graphic Settings (.SET) ...........1-17
1.4.3 Customizing Toolbars ............................................................1-21
1.5 Setting Graphic Options
1.5.1 Changing Background Color .................................................1-23
............................................................ 1-5
............................................. 1-11
....................................... 1-17
.......................................................... 1-23
1.5.2 Customizing Graphic Options ................................................1-24
1.5.3 Reverting to Previous Graphic Options .................................1-29
1.6 Managing Files
1.6.1 Converting .SPC File Format to .DAT File Format
(Mariner Data Only) ..............................................................1-30
1.6.2 Converting Data from Profile to Centroid
(Mariner Data Only) ..............................................................1-33
1.6.3 Converting to and Exporting ASCII Data ..............................1-34
1.6.4 Importing a Trace in ASCII Format ........................................ 1-35
1.6.5 Extracting and Saving Information
from .DAT, .RSD, and .RCD Files ..........................................1-36
1.6.6 Copying from Data Files ........................................................1-38
...................................................................... 1-30
™
Data Explorer
Software User’s Guide iii
Page 4

Table of Contents
Chapter 2 Using Chromatog ram and S pectrum Windows
2.1 Opening and Closing Data Files ................................................. 2-2
2.1.1 Opening Data Files ................................................................ 2-2
2.1.2 Displaying Mariner DAD Traces ............................................. 2-6
2.1.3 Displaying Voyager Chromatograms ...................................... 2-7
2.1.4 Viewing Read-Only Files ........................................................ 2-7
2.1.5 Moving Between Open Files .................................................. 2-8
2.1.6 Closing Data Files .................................................................2-10
2.2 Adjusting the Display Range
2.3 Organizing Windows
2.4 Manipulating Traces
2.4.1 Zooming, Centering, and Customizing a Trace ......................2-14
2.4.2 Duplicating a Trace ...............................................................2-15
2.4.3 Dividing the Active Trace ......................................................2-15
2.4.4 Adding Traces from
the Same Data File to a Window ...........................................2-16
2.4.5 Removing Traces ..................................................................2-21
2.4.6 Expanding and Linking Traces ..............................................2-21
2.4.7 Recalling and Rearranging Traces (Processing History) ........2-22
2.4.8 Overlaying Traces .................................................................2-24
2.4.9 Annotating Traces .................................................................2-28
2.4.10 Viewing Trace Labels ............................................................2-30
2.4.11 Printing Traces ......................................................................2-33
2.5 Working with Multiple Data Files
2.5.1 Working with Separate Data Files .........................................2-36
2.5.2 Copying Traces from Multiple Data Files to a Window ...........2-37
2.6 Saving, Opening, and Deleting .DAT Results
2.7 Exporting, Opening, and Deleting .RCD and .RSD Results Files
(Mariner Data Only)
2.8 Saving, Opening, and Deleting .SPC Results Files
(Mariner Data Only)
............................................................... 2-13
............................................................... 2-14
................................................................ 2-39
................................................................ 2-40
..................................................... 2-11
................................................ 2-36
................................ 2-38
iv Applied Biosystems
Page 5

Table of Contents
Chapter 3 Peak Detection and Labeling
3.1 Overview .............................................................................. 3-2
3.1.1 Default Peak Detection .......................................................... 3-2
3.1.2 The Resolution-Based Peak Detection Routine ...................... 3-3
3.2 Peak Detection
3.2.1 Strategy for Mariner Peak Detection ....................................... 3-6
3.2.2 Strategy for Voyager Peak Detection ...................................... 3-8
3.2.3 Setting Peak Detection Parameters ...................................... 3-11
3.2.4 Peak Detection Parameter Descriptions ................................3-19
3.2.5 Charge State
3.3 Peak List
3.3.1 Displaying the Peak List ........................................................3-37
3.3.2 Inserting Peaks in the Peak List ............................................3-39
3.3.3 Saving the Peak List .............................................................3-40
3.3.4 Sorting, Filtering, and Printing the Peak List .........................3-42
3.4 Deisotoping a Spectrum
3.5 Peak Labeling
3.5.1 Charge State Labels .............................................................3-53
3.5.2 Setting Chromatogram and Spectrum Peak Labels ...............3-54
3.5.3 Setting Custom Peak Labels .................................................3-61
3.6 Process that Occurs During Peak Detection,
Centroiding, and Integration
3.7 Default Peak Detection Settings
....................................................................... 3-6
Determination and Examples ................................................3-32
............................................................................. 3-37
.......................................................... 3-45
....................................................................... 3-52
..................................................... 3-67
................................................ 3-71
Data Explorer
™
Software User’s Guide v
Page 6

Table of Contents
Chapter 4 Examining Chromatogram Data
4.1 Overview .............................................................................. 4-2
4.2 Creating an Extracted Ion Chromatogram
4.2.1 Creating an Extracted Ion Chromatogram (XIC) ..................... 4-5
4.2.2 Creating a Constant Neutral Loss (CNL) Chromatogram ........ 4-9
4.3 Creating an Extracted Absorbance Chromatogram (XAC)
(Mariner Data Only)
4.4 Noise Filtering/Smoothing
4.5 Adding and Subtracting Raw Spectra Within a Data File
4.6 Displaying MS Method Data (Mariner Data Only)
4.7 Adjusting the Baseline
4.7.1 Using Baseline Offset ...........................................................4-27
4.7.2 Using Baseline Correction .....................................................4-29
................................................................ 4-13
........................................................ 4-17
............................................................ 4-27
..................................... 4-5
.................. 4-20
........................... 4-23
4.8 Using UV Trace Offset (Mariner Data Only)
.................................. 4-30
Chapter 5 Examining Spe ctrum Da ta
5.1 Overview .............................................................................. 5-2
5.2 Creating a Combined Spectrum
5.3 Manual Calibration
5.3.1 Overview of Manual Calibration ............................................. 5-5
5.3.2 Manually Calibrating .............................................................. 5-7
5.3.3 Creating or Modifying a Calibration Reference File (.REF) ....5-17
5.3.4 Reverting to Instrument Calibration .......................................5-22
5.3.5 Hints for Calibrating Mariner Data .........................................5-24
5.3.6 Hints for Calibrating Voyager Data ........................................5-25
5.4 Automatic Calibration
5.4.1 Overview of Automatic Calibration ........................................ 5-26
5.4.2 Importing and Specifying Automatic Calibration Settings .......5-29
5.4.3 Automatically Calibrating (Mariner Data Only) .......................5-34
.................................................................. 5-5
.............................................................. 5-26
................................................. 5-4
vi Applied Biosystems
Page 7

Table of Contents
5.5 Centroiding .......................................................................... 5-36
5.6 Mass Deconvolution (Mariner Data Only)
5.7 Noise Filtering/Smoothing
5.8 Adjusting the Baseline
5.8.1 Using Baseline Offset ...........................................................5-45
5.8.2 Using Baseline Correction .....................................................5-47
5.8.3 Using Advanced Baseline Correction ....................................5-48
........................................................ 5-42
............................................................ 5-45
..................................... 5-37
5.9 Truncating a Spectrum
5.10 Converting to a Singly Charged Spectrum (Mariner Data Only)
5.11 AutoSaturation Correction (Mariner Data Only)
5.12 Adding and Subtracting Raw or Processed Spectra from the Same
or Different Data Files (Dual Spectral Trace Arithmetic)
............................................................ 5-56
......... 5-59
............................. 5-62
................... 5-64
Chapter 6 Using Tools and Applications
6.1 Using the Elemental Composition Calcul ator ................................. 6-2
6.1.1 Determining Elemental Composition ...................................... 6-2
6.1.2 Setting Limits ......................................................................... 6-7
6.2 Using the Isotope Calculator
6.3 Using the Mass Resolution Calculator
6.4 Using the Signal-to-Noise Ratio Calculator
6.5 Using the Ion Fragmentation Calculator
6.6 Using the Elemental Targeting Application
6.7 Using the Macro Recorder
6.7.1 Before Using the Macro Recorder .........................................6-34
6.7.2 Recording a Macro ................................................................6-37
6.7.3 Assigning Macros to Buttons .................................................6-38
6.7.4 Running a Macro ..................................................................6-39
..................................................... 6-13
......................................... 6-20
................................... 6-23
....................................... 6-25
................................... 6-31
....................................................... 6-34
6.7.5 Deleting a Macro ...................................................................6-41
6.7.6 Advanced Macro Editing .......................................................6-42
6.7.7 Importing or Exporting Macros in DATAEXPLORER.VB6 ......6-43
6.7.8 Running Macros Automatically
When Opening and Closing Files ..........................................6-45
™
Data Explorer
Software User’s Guide vii
Page 8

Table of Contents
Chapter 7 Data Explorer Examples
7.1 Mariner Data Examples............................................................ 7-2
7.1.1 Improving Signal-To-Noise Ratio ............................................ 7-2
7.1.2 Deconvoluting and Evaluating
Unresolved Chromatographic Peaks .......................................7-4
7.1.3 Determining if a Peak is Background Noise ............................ 7-8
7.2 Voyager Data Examples
7.2.1 Detecting and Labeling Partially Resolved Peaks ..................7-11
7.2.2 Processing Before Calibrating to Optimize Mass Accuracy ...7-14
7.2.3 Detecting Peaks from Complex Digests ................................7-18
.......................................................... 7-11
Chapter 8 Viewing Voyager PSD D ata
8.1 Displaying PSD Data ............................................................... 8-2
8.2 Applying Fragment Labels
8.3 Calibrating a PSD Spectrum
8.3.1 Checking Peak Detection ......................................................8-11
8.3.2 Calibrating ............................................................................8-12
8.3.3 Creating PSD Calibration (.CAL) Files and
Applying to Other Data Files .................................................8-20
8.3.4 Creating PSD Calibration Reference (.REF) Files .................8-21
8.3.5 Changing the Precursor Mass ...............................................8-23
......................................................... 8-8
..................................................... 8-10
Chapter 9 Troubleshooting
9.1 Overview .............................................................................. 9-2
9.2 General Troubleshooting
9.3 Processing, Tools, and Applications Troubleshooting
9.4 Calibration Troubleshooting
9.5 Printing Troubleshooting
9.6 Peak Detection and Labeling Troubleshooting
.......................................................... 9-3
....................... 9-6
...................................................... 9-10
.......................................................... 9-14
............................... 9-15
viii Applied Biosystems
Page 9

Table of Contents
Appendix A Warranty Information ........................................ A-1
Appendix B Overview of Isotopes........................................ B-1
Appendix C Data Explorer Toolbox
(Visual Basic Macros)
......................................................... C-1
C.1 Overview
C.2 Preparing Data Before Accessing Macros
C.3 Accessing the Macros
C.4 Using the Ladder S equencing Toolbox
C.5 Using the Peptide Fragmentation Toolbox
C.6 Using the Polymer Analysis Toolbox
C.7 Using MS Fit/MS Tag Toolbox
.............................................................................. C-2
.............................................................. C-4
.................................................. C-18
Index
..................................... C-3
......................................... C-5
..................................... C-9
.......................................... C-15
Data Explorer
™
Software User’s Guide ix
Page 10

Table of Contents
x Applied Biosystems
Page 11
How to Use This Guide
How to Use This Guide
1
Purpose of
this guide
The Applied Biosystems Data Explorer ™ Software User’s
Guide describes processing and analyzing data with the
Data Explorer software. You can use the Data Explorer
software to analyze data collected on:
• Mariner
software
• Voyager
Version 5.0 and later software
™
Workstations with Version 3.0 and later
™
-DE Biospectrometry™ Workstations with
Audience This guide is intended for novice and experienced Mariner
or Voyager workstation users who are analyzing
biomolecules.
Structure of
this guide
Chapter/Append ix Content
Chapter 1, Data Explorer™ Basics Describes file formats, file management, the
The Applied Biosystems Data Explorer ™ Software User’s
Guide is organized into chapters and appendixes. Each
chapter page is marked with a tab and a header to help
you find information.
The table below describes the material covered in each
chapter and appendix.
parts of the Data Explorer window, and how
to customize the Data Explorer software.
Chapter 2, Using Chromatogram
and Spectrum Windows
Chapter 3, Peak Detection
and Labeling
Chapter 4, Examining
Chromatogram Data
Describes window and trace handling. Also
describes saving results.
Provides background information on peak
detection, centroiding, and integration.
Describes peak detection, peak labeling,
and peak deisotoping.
Describes processing and analyzing
chromatographic data.
™
Data Explorer
Software User’s Guide xi
Page 12
1
How to Use This Gu ide
Chapter/Append ix Content
Chapter 5, Examining Spectrum
Data
Chapter 6, Using Tools and
Applications
Chapter 7, Data Explorer Examples Includes specific examples for Mariner data
Chapter 8, Viewing Voyager PSD
Data
Chapter 9, Troubleshooting Includes symptoms and possible causes of,
Describes processing and analyzing mass
spectral data.
Describes how to generate results using
several tools and applications: the Centroid
calculator, Elemental Composition
calculator, Isotope calculator, Mass
Resolution calculator, Ion Fragmentation
calculator and Signal-to-Noise calculator.
Also describes using the Macro Recorder
and the Elemental Targeting Application.
and Voyager data. Examples include how to
improve the signal-to-noise ratio for
reserpine, deconvolute unresolved peaks in
cyctochrome c (Mariner data), and label
partially resolved peaks (Voyager data).
Describes how to view, label, and calibrate
PSD data.
and corrective actions for potential system
problems.
Appendix A, Warranty Provides warranty and service information.
Appendix B, Overview of Isotopes Includes background information you need
for understanding isotopes.
®
Appendix C, Data Explorer Toolbox
(Visual Basic Macros)
xii Applied Biosystems
Describes loading Visual Basic
preparing data, and running the macros.
macros,
Page 13

How to Use This Guide
Conventions This guide uses the following conventions to make text
easier to understand.
• Bold indicates user action. For example:
Type 0 and press Enter for the remaining
fields.
• Italic text denotes new or important words, and is also
used for emphasis. For example:
Before analyzing, always prepare fresh matrix.
1
Notes, Cautions,
Warnings, and
Hints
A note provides important information to the operator. For
example:
NOTE: If you are prompted to insert the boot diskette
into the drive, insert it, then press any key.
A caution provides information to avoid damage to the
system or loss of data. For example:
CAUTION
Do not touch the lamp. This may damage the lamp.
A warning provides information essential to the safety of
the operator. For example:
WARNING
CHEMICAL HAZARD. Wear appropriate personal
protection and always observe safe laboratory practices
when operating your system.
A hint provides helpful suggestions not essential to the
use of the system. For example:
Hint: To avoid complicated file naming, use Save First
to Pass or Save Best Only modes.
™
Data Explorer
Software User’s Guide xiii
Page 14

1
How to Use This Gu ide
Related
documentation
The related documents shipped with your system include:
• Mariner
document to learn detailed information about the
Mariner Workstation.
• Voyager
User’s Guide—Use this document to learn detailed
information about the Voyager-DE Workstation.
• Printer documentation (depends on the printer you
purchase)—Use this documentation to set up and
service your printer.
• Microsoft
related documents—Use this guide to learn detailed
information about the Microsoft Windows NT user
interface.
™
Workstation User’s Guide—Use this
™
-DE Biospectrometry Workstation
®
Windows NT® User’s Guide and
Send us your
comments
We welcome your comments and suggestions for
improving our manuals. You can send us your comments
in two ways.
• On the web at:
www.appliedbiosystems.com/about/contact.html
• By e-mail at:
T echPubs@appliedbiosystems.com
xiv Applied Biosystems
Page 15
1 Data Explorer™
Basics
This chapter contains the following sections:
1.1 Overview................................................... 1-2
1.2 File Formats and Types ............................. 1-5
1.3 Parts of the Data Explorer Window.......... 1-11
1.4 Customizing the Data Explorer Window... 1-17
1.5 Setting Graphic Options .......................... 1-23
1.6 Managing Files........................................ 1-30
Chapter
1
Data Explorer™ Software User’s Guide 1-1
Page 16
Chapter 1 Data Explorer™ Basics
1
1.1 Overview
Description The Data Explorer
graphical software that you use to analyze, calibrate, and
report data. You can use the Data Explorer software to
analyze data collected on:
• Mariner
• Voyager
Features Data Explorer software includes a suite of tools and
processing options to allow you to graphically and interactively
manipulate chromatogra phi c and mas s spec tr al data. For
example, you can:
• Smooth and noise-filter data.
™
Version 4.0 processing software is
™
Workstations
™
-DE Biospectrometry™ Workstations
NOTE: Application systems that automatically acquire
and process data, such as Mariner High Throughput
Analysis Option (CombiSolve
Solution 1
Explorer software.
™
Option, require specific versions of Data
™
) and Proteomics
1-2 Applied Biosystems
• Automatically and manually calibrate spectral data.
• Set peak detection parameters and custom labels for
regions of the trace. Detected peaks can be evaluated
for charge-state determination according to
user-defined parameters.
• Determine elemental composition, theoretical isotope
distributions, resolution, signal-to-noise ratio, and
fragment ions.
• Perform target compound analysis (elemental
targeting).
• Customize windows, toolbars, and traces.
• Create scripts and macros to automate your work using
the Macro Recorder and Visual Basic
®
Editor.
Page 17
Overview
Starting and
exiting the
software
To start the Data Explorer software from the Windows NT
desktop, double-click the Data Explorer icon on the desktop.
The Data Explorer window opens.
The Data Explorer window is blank with only a few menus
displayed until you open a data file.
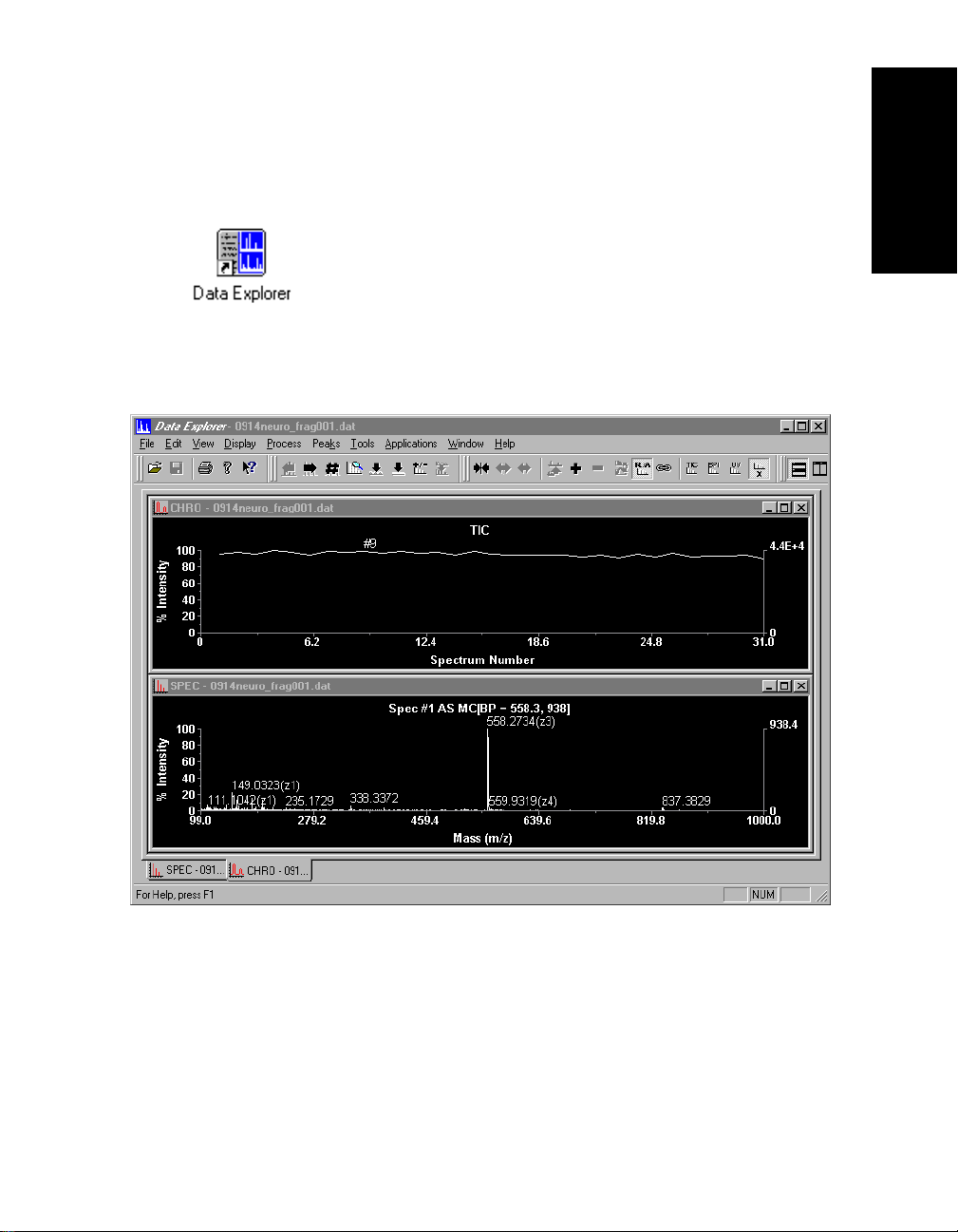
Figure 1-1 shows the Data Explorer main window with a
Mariner data file open. Figure 1-2 shows the Data Explorer
main window with a Voyager data file open.
To exit th e so ftw are, se lect Exit from the File menu in the Data
Explorer window. The Data Explorer software closes.
1
Figure 1-1 Data Explorer Window with Mariner Data
™
Data Explorer
Software User’s Guide 1-3
Page 18
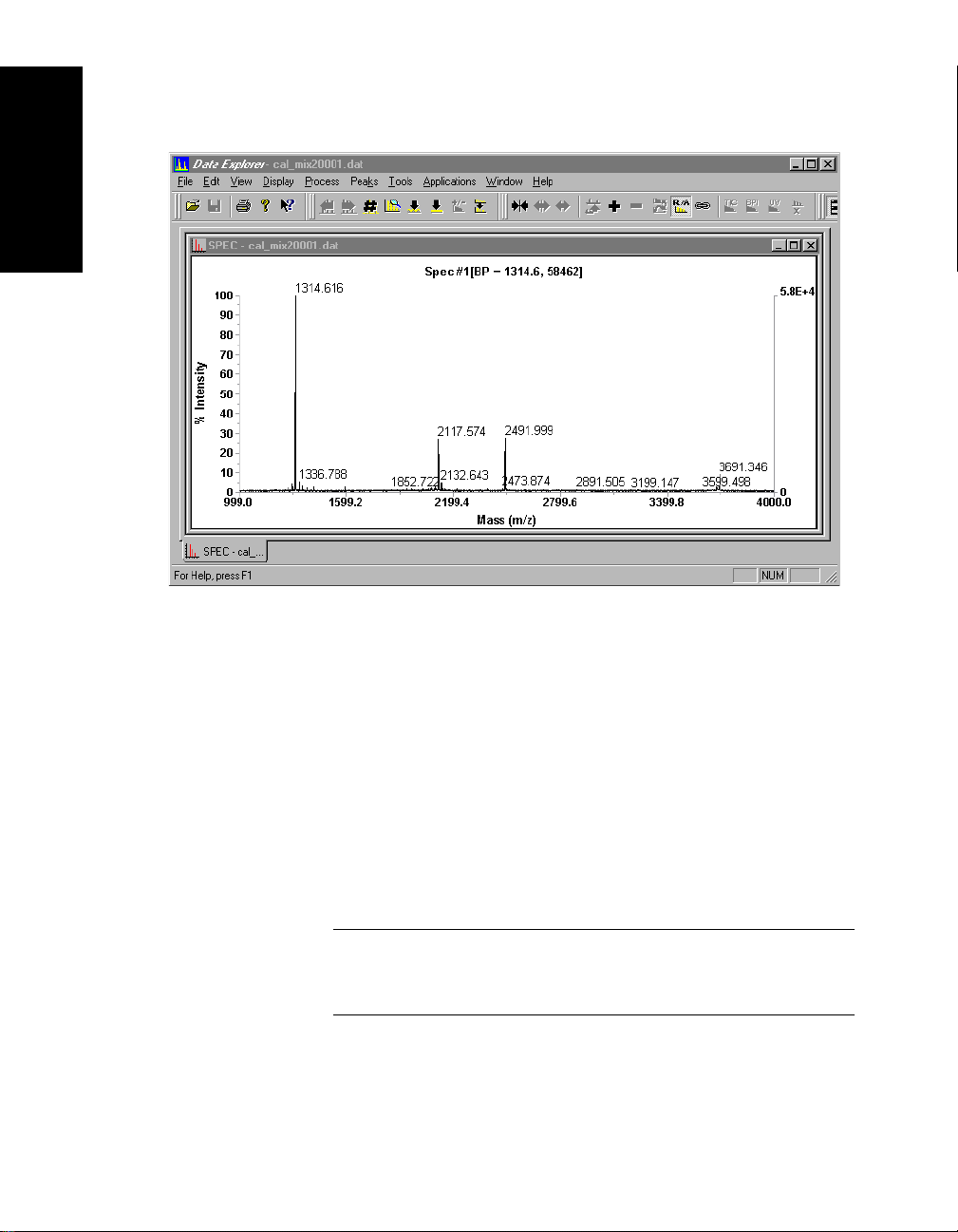
1
Chapter 1 Data Explorer™ Basics
Figure 1-2 Data Explorer Window with Voyager Data
Default colors The default colors are different for Mariner and Voyager:
• Mariner—Black background, yellow traces, and green
labels
• Voyager—White background, blue traces, and red
labels
You can customize the default colors as needed. See
Section 1.5.1, Changing Background Color.
NOTE: For consistency, all Mariner and Voyager screen
examples in the following sections of this User’s Guide are
shown with a white background.
1-4 Applied Biosystems
Page 19
File Formats and Types
1.2 File Formats and Types
This section describes:
• Software applications compati bility
• Data (.DAT) file format
1.2.1 Software Applications Compatibility
You can use the Data Explorer Macro Recorder function to
create Visual Basic scripts to automate tasks. You can also
use the Visual Basic Editor directly to create more complex
programs customized to suit your needs. For more
information, see Section 6.7, Using the Macro Recorder.
Additionally, you can convert data to ASCII format for import
into other software applications or import ASCII results. For
more information, see Section 1.6.3, Converting to and
Exporting ASCII Data, and Section 1.6.4, Importing a Trace in
ASCII Format.
1
1.2.2 Data (.DAT) File Format
.DAT file format Data generated by Mariner and Voyager systems is stored in
.DAT file format. The .DAT file format incorporates all
information about how a data file was acquired and processed
into a single file. This format improves data processing and
data storage efficiency.
Data files can contain spectra from a single acquisition or from
multiple acquisitions (for example, multiple spectral data from
a Voyager acquisition or multiple injection results from a
Mariner CombiSolv run).
™
Data Explorer
Software User’s Guide 1-5
Page 20
Chapter 1 Data Explorer™ Basics
1
Mariner .SPC file
format
Voyager .MS file
format
Extracting
information from
.DAT files
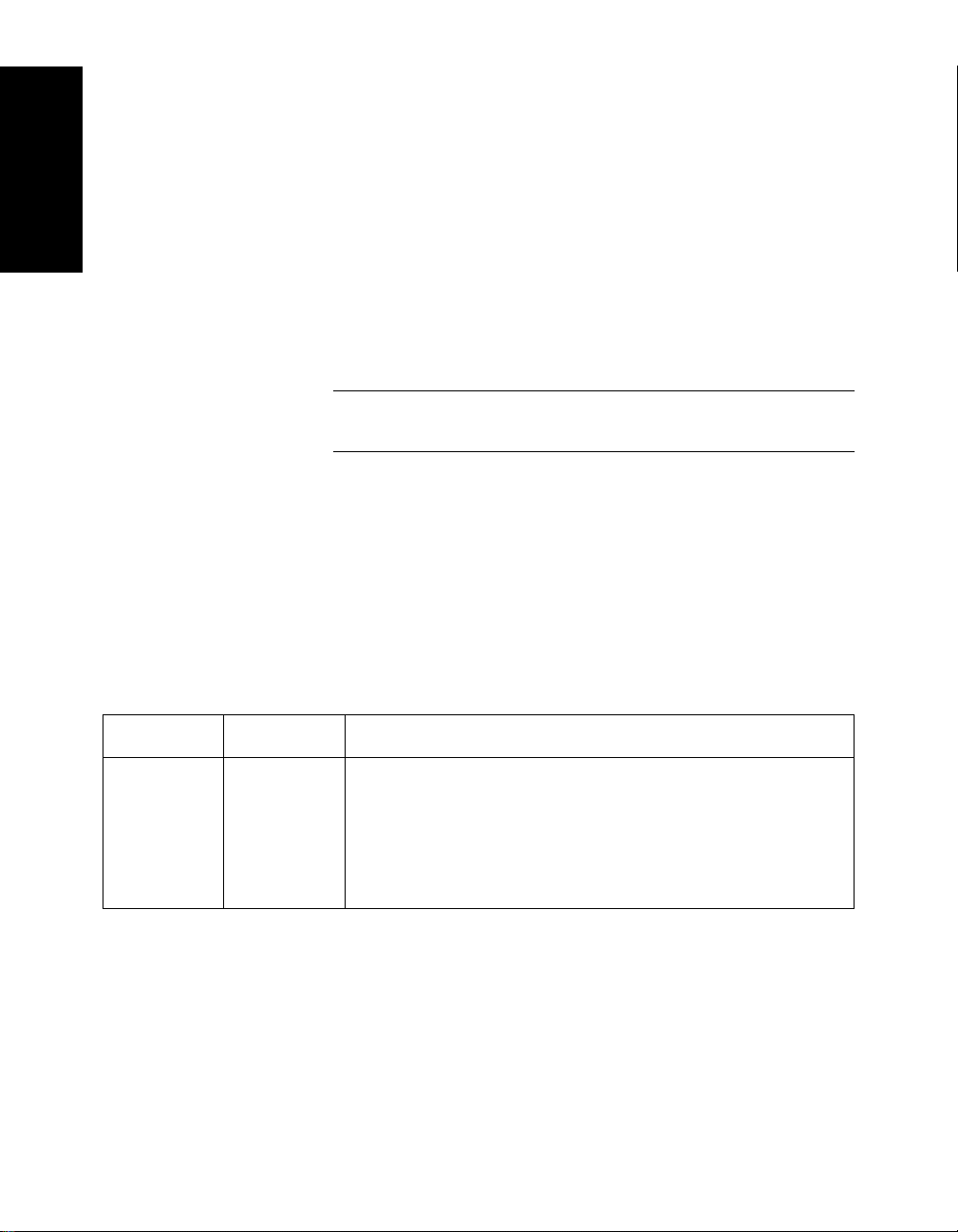
Table 1-1 Information Stored In a .DAT File
In Mariner software versions earlier than version 3.0, data files
are stored in .SPC format. You can view and process .SPC
files in Data Explorer, or you can convert these files to .DAT
format. For information about the differences between the
.DAT and the .SPC formats, see the Mariner
User’s Guide.
In Voyager software versions earlier than version 5.0, data
files are stored in .MS, .MSF, .MSA, and .MSB format. You can
view and process .MS, .MSF, .MSA, and .MSB files in Data
Explorer. You cannot convert them to .DAT format.
NOTE: Voyager .SPC format files are not s upported in the
Data Explorer software.
You can also store parameter settings in separate files by
extracting information from a .DAT file as needed for use with
other files. For more information, see Section 1.6.5, Extracting
and Saving Information from .DAT, .RSD, and .RCD Files.
The types of information stored in a .DAT file are described
below.
™
Workstation
Category File Type File Content
Settings .BIC Instrument settings for controlling the operation of the
mass analyzer. For more information, see the:
• Mariner Workstation User’s Guide
• Voyager Biospectrometry Workstation User’s
Guide
1-6 Applied Biosystems
Page 21
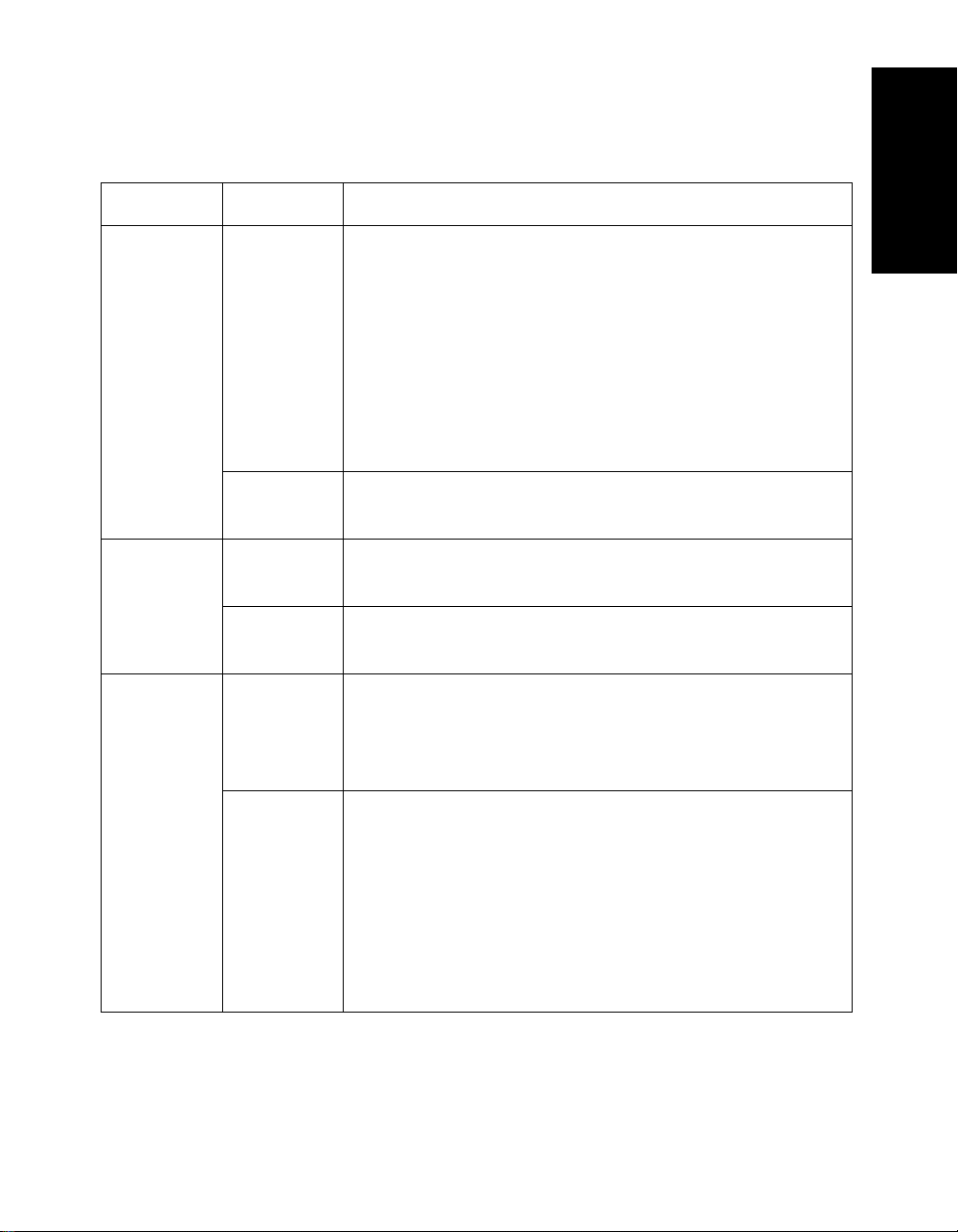
Table 1-1 Information Stored In a .DAT File (Continued)
Category File Type File Content
File Formats and Types
1
Settings
(continued)
Display .LBC Chromatogram label information. See Section 3.5.3,
Process .CAL Calibration constants generated by mass calibration.
.MSM
(Mariner
only)
.SET Graphic and processing settings. See Section 1.4.2,
.LBS Spectrum label information. See Section 3.5.3, Setting
MS Method settings, if data was acquired using an
.MSM file.
NOTE: To access the instrument settings used to
acquire each spectrum in an MS Method, you must first
extract the .MSM file from the data file, then export the
.BIC files from the .MSM file using the Export button in
the MS Method editor. For more information on
exporting a .BIC file from an .MSM file, see the Mariner
Workstation User’s Guide.
Customizing Processing and Graphic Settings (.SET).
Setting Custom Peak Labels.
Custom Peak Labels.
For more information, see “Exporting .BIC, .MSM, and
.CAL files” on page 1-36, and “Applying new constants
to additional files” on page 5-16.
.CTS Processed trace that you access by selecting
Processing History from the Display menu. For
information, see Section 2.4.7, Recalling and
Rearranging Traces (Processing History).
NOTE: You select the name of the trace from the
Processing History menu. You do not directly select a
.CTS file. To purge or disable .CTS files, see “Setting
Processing History options” on page 2-23.
™
Data Explorer
Software User’s Guide 1-7
Page 22
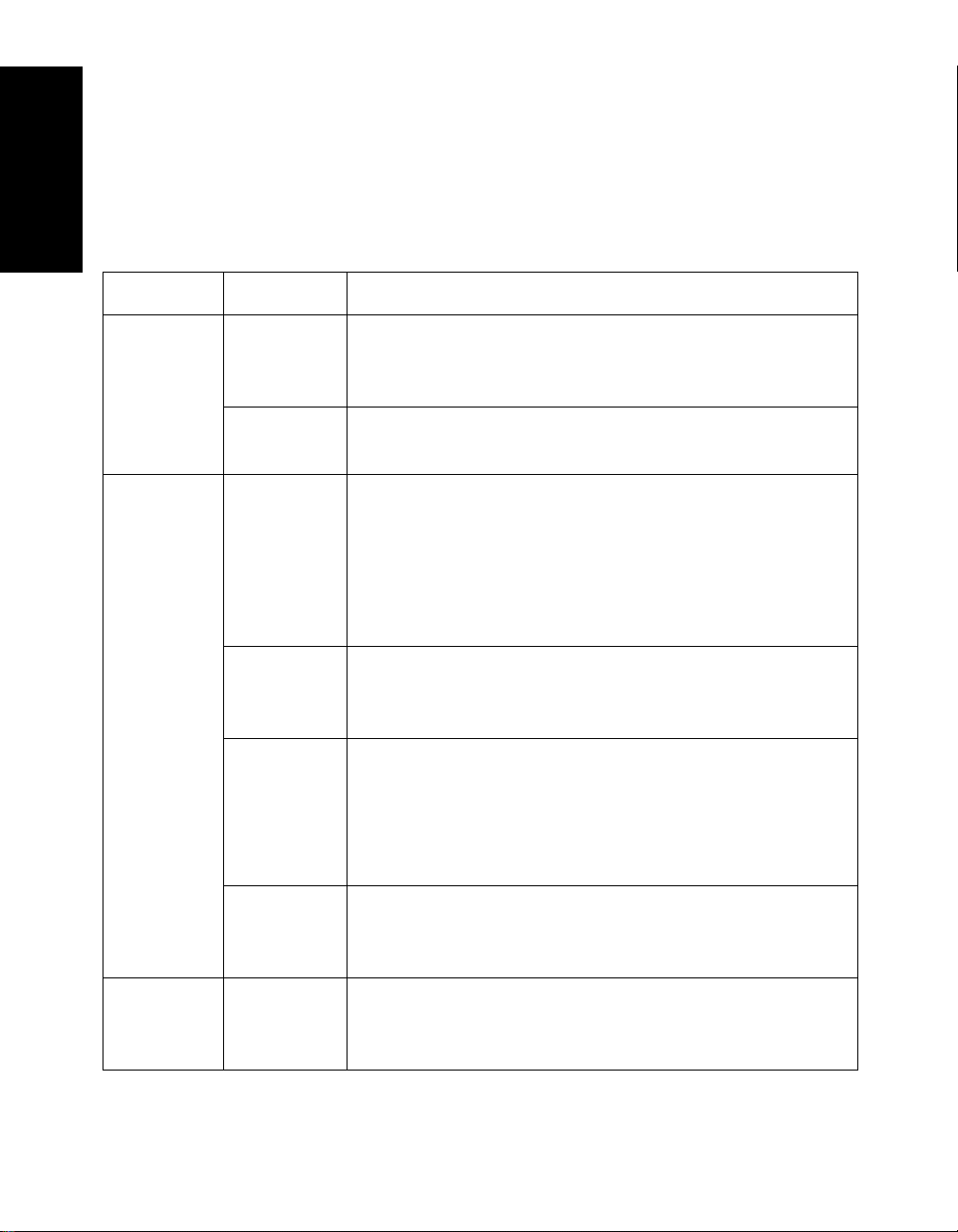
Chapter 1 Data Explorer™ Basics
1
Additional files
types
Category File Type File Content
Data .PKT Text file containing a chromatogram or a spectrum peak
.TXT Data file exported to an ASCII text file. See
Data .SPC
(Mariner
only)
.MS
(Voyager
only)
Additional file types you may see on your system are
described below.
Table 1-2 Additional File Types
list that you can save from the Output window. See
“Output window” on page 1-15.
Section 1.6.3, Converting to and Exporting ASCII Data.
Data file format for files acquired before Version 3.0 of
the Mariner Instrument Control Panel.
NOTE: Voyager data files in .SPC format have a file
structure different from Mariner data files in .SPC
format, and are not supported in the Data Explorer
software.
Data file format for files acquired before Version 5.0 of
the Voyager Instrument Control Panel.
.MSA and
.MSF
(Voyager
only)
.MSB
(Voyager
only)
Procedure .TUN
(Mariner
only)
1-8 Applied Biosystems
PSD data file format for composite and fragment files
acquired before Version 5.0 of the Voyager Instrument
Control Panel.
NOTE: The Data Explorer software cannot generate
composite spectra from .MSF fragment files.
Baseline-corrected data file format for composite and
fragment files acquired before Version 5.0 of the
Voyager Instrument Control Panel.
AutoTune method. See the Mariner Workstation User’s
Guide.
Page 23
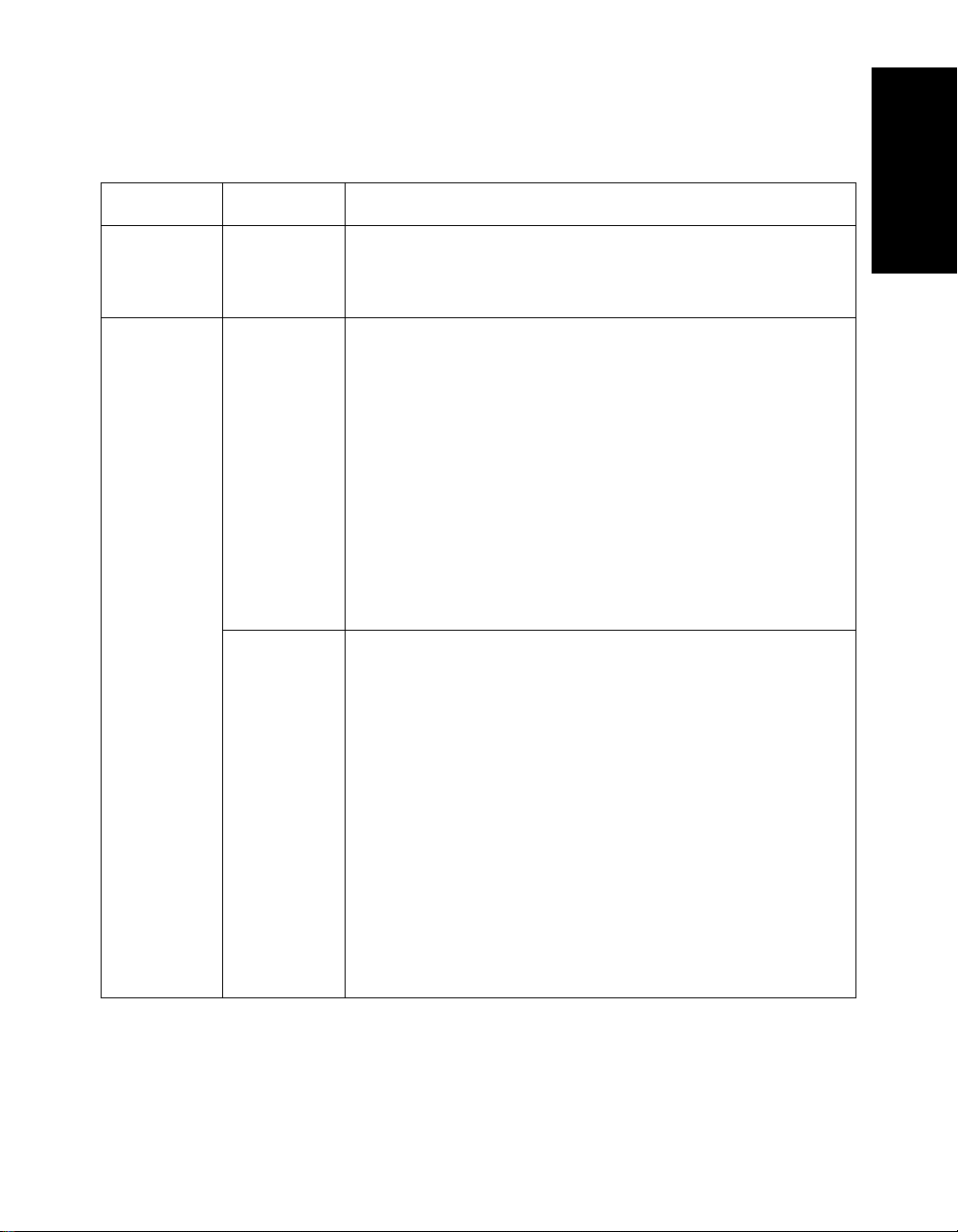
File Formats and Types
Table 1-2 Additional File Types (Continued)
Category File Type File Content
Reference .REF List of masses to select from during calibration. See
“Creating and saving a calibration reference file” on
page 5-18.
1
Process .RCT
(Mariner
only)
.RST Results file saved from:
Results file saved from:
• Mariner .SPC format data file (versions earlier
than 3.0) in the Data Explorer software after a
chromatogram is manually processed.
See Section 2.8, Saving, Opening, and Deleting
.SPC Results Files (Mariner Data Only).
• Mariner .DAT format data file (version 3.0 and later)
in the Mariner Instrument Control Panel for a
snapshot of chromatogram data.
NOTE: Results for a .DAT file are stored within the .DAT
file, not as a separate file.
• Mariner .SPC format data file (versions earlier
than 3.0) in the Data Explorer software after a
spectrum is manually processed.
See Section 2.8, Saving, Opening, and Deleting
.SPC Results Files (Mariner Data Only).
• Voyager .MS format data file in the Data Explorer
software after a spectrum is manually processed.
• Mariner .DAT format data file (version 3.0 and later)
in the Mariner Instrument Control Panel for a
snapshot of spectrum data.
NOTE: Results for a .DAT file are stored within the .DAT
file, not as a separate file.
Data Explorer
™
Software User’s Guide 1-9
Page 24
1
Chapter 1 Data Explorer™ Basics
Table 1-2 Additional File Types (Continued)
Category File Type File Content
Process
(continued)
.RCD Chromatogram results file exported from .DAT files. See
Section 2.7, Exporting, Opening, and Deleting .RCD and
.RSD Results Files (Mariner Data Only).
.RSD Spectr um results file exported from .DAT files. See
Section 2.7, Exporting, Opening, and Deleting .RCD and
.RSD Results Files (Mariner Data Only).
1-10 Applied Biosystems
Page 25

1.3 Parts of the
Data Explorer Window
This section describes:
• Overview
• Toolbar
• Chromatogr am and Spe ctru m windows
• Tabs for data files
• Data file names
• Output window
Overview Figure 1-3 shows the Data Explorer window with Mariner data.
Toolbar
Chromatogram
window
Parts of the Data E xplorer Window
1
Data file name
Spectrum
window
Tabs for
open data files
Output
window
Figure 1-3 Parts of the Data Explorer Window
™
Data Explorer
Software User’s Guide 1-11
Page 26
1
Chapter 1 Data Explorer™ Basics
Toolbar The toolbar contains buttons that access Data Explorer
functions.
For a description of a toolbar button, place the cursor on the
button. A brief description of the button (ToolTip) is displayed
below the button.
For information on adding or removing toolbar buttons, see
“Customizing toolbars” on page 1-21.
Chromatogram
and Spectrum
windows
Table 1-3 Mariner Data Displayed in Chromatogram and Spectrum Windows
Window Mariner Data
CHRO Displays:
• Total Ion Chromatogram—Includes the entire mass range saved in
the data file.
• Extracted Ion Chromatogram (XIC) (optional)—Includes only the
signal response from a mass window or range
• Constant Neutral Loss Chromatogram (CNL) (optional)—Extr a c ts
only the response from peaks that are separated by a selected mass
difference.
Optionally displays the following from Diode Array data (DAD):
• Total Absorbance Chromatogram (TAC)
• Channel (Ch)
• Extracted Absorbance Chromatogram (XAC)
Can be displayed as % Intensity versus Retention Time or Spectrum
number by selecting Traces from the Display menu, then selecting
X Axis In, then selecting Spectrum Number or Time.
Refer to the following tables for descriptions of the types of
data that you can display in the Chromatogram (CHRO) and
Spectrum (SPEC) windows:
• Mariner data—Table 1-3
• Voyager data—Table 1-4
1-12 Applied Biosystems
Page 27
Parts of the Data E xplorer Window
Table 1-3 Mariner Data Displayed in Chromatogram and Spectrum Windows
(Continued)
Window Marin er D ata
SPEC Displays the spectrum for the selected time in the TIC or TAC trace. By
default, displays spectrum #1. The trace label includes “DAD” for spectra
selected from TAC.
Indicates Base Peak (BP) mass and intensity for the tallest peak in the
spectrum. Displayed as % Intensity versus Mass-to-Charge (m/z). The
right axis is scaled to the intensity of the base peak.
Table 1-4 Voyager Data Displayed in Chromatogram and Spectrum Windows
Window Voyager Data
CHRO Window is not displayed by default.
Optionally displays Total Ion Current (TIC) for multiple spectra .DA T files if
you select Restore Chromatogram from the View menu.
NOTE: DAD functions are not supported for Voyager data.
1
SPEC Depends on the type of data file you open:
• Single spectrum files—Displays the spectrum and labels the plot as
spectrum #1.
• Multiple spectrum files—Displays the spectrum for the selected
time in the TIC trace (if displayed). By default, displays spectrum #1.
• PSD files—Displays the composite spectrum and labels it as
Stitched PSD. You can display segment spectra by clicking
and .
Indicates Base Peak (BP) mass and intensity for the tallest peak in the
spectrum. Displayed as % Intensity versus Mass-to-Charge (m/z). The
right axis is scaled to the intensity of the base peak.
Data Explorer
™
Software User’s Guide 1-13
Page 28

1
Chapter 1 Data Explorer™ Basics
Labels in the chromatogram or spectrum title identify the
type of data displayed in the window. For a description of
labels, see Section 2.4.10, Viewing Trace Labels.
For more information on Chromatogram and Spectrum
windows, see Chapter 2, Using Chromatogram and Spectrum
Windows.
Context-sensitive
menus
The commands displayed on the menus in the Data Explorer
window depend on the window that is active when you select
the menu:
• Only Mariner-related commands are displayed on
menus when you open a Mariner .DAT file.
• Only Voyager-related comma nds are displ ay ed on
menus when you open a Voyager .DAT file.
• Only spectrum-related commands are displayed if you
select menus when the Spectrum window is active.
• Only chromatogram-related commands are displayed if
you select menus when the Chromatogram window is
active. For Voyager multiple spectra .DAT files, UV
functions are disabled.
Tabs for data files Chromatogram and Spectrum windows display a tab at the
bottom (see Figure 1-3 on page 1-11) that allow you to switch
between data files, or between Chromatogram and Spectrum
windows for a Mariner data file. See “Moving Between Open
Files” on page 2-8.
Data file names Data file names are displayed in the:
• Title bar of each window
• Tab for each window
1-14 Applied Biosystems
NOTE: If the full name of the data file does not fit in the tab,
part of the name is displayed followed by “...”. To display
the full name of the data file, place the cursor on the tab.
The full name of the data file is displayed below the tab.
Page 29

Parts of the Data E xplorer Window
Output window The Output window (see Figure 1-3 on page 1-11) displays
tabs at the bottom that you can click to switch between the
types of information displayed:
1
Output wind ow
tabs
• Result—Displays results generated using commands on
the Process, Tools, and Applications menus. For more
information on results, see:
• Section 5.3, Manual Calibration, or Section 5.4,
Automatic Calibration
• Section 5.6, Mass Deconvolution (Mariner Data
Only)
• Section 6.2, Using the Isotope Calculator
• Section 6.3, Using the
Mass Resolution Calculator
• Section 6.4, Using the Signal-to-Noise Ratio
Calculator
• Chro Peak List —Displays results of chromatogram
peak detection and integration. For more information,
see Section 3.3, Peak List.
• Spectrum Peak List—Displays results of spectrum
peak detection, integration, and centroiding. For more
information, see Section 3.3, Peak List.
• Sample Info—For data files, displays the:
• Software version used for acquisition
• Acquisition time and sample comments entered
when data was acquired
For result files (.RCT, .RST, .RCD, .RSD), displays any
processing functions that have been performed and
saved in the result file.
™
Data Explorer
Software User’s Guide 1-15
Page 30

1
Chapter 1 Data Explorer™ Basics
• Instrument Setting—Displays a list of instrument
settings used to obtain the data. The settings are taken
from instrument settings pages in the Instrument
Control Panel. Also displays segments, event numbers,
and event tags from Mariner MS Method acquisitions
and LC information if LCMS was acquired using
Mariner.
• Elemental Analysis—Displays results for the
Elemental Composition calc ula tor. For information, see
Section 6.1, Using the Elemental
CompositionCalculator.
• Elemental Targeting—Displays results for the
Elemental Targeting application. For information, see
Section 6.6, Using the Elemental Targeting Application.
Displaying,
clearing, and
closing
The Output window is automatically displayed when you
generate results (for example, when you calculate resolution).
To display the Output window manually, select Output
Window from the View menu.
To clear the Output window, right-click the Output window,
then select Clear Window.
To close the Output window, deselect Output Window from
the View menu, or right-click the Output window, then select
Hide.
Hint: To maximize the Chromatogram and Spectrum
windows after hiding the Output window, click or
in the toolbar.
1-16 Applied Biosystems
Page 31

Customizing the Data Explorer Window
1.4 Customizing the Data Explorer Window
This section includes:
• Setting default values
• Customizing Graphic and Processing settings
• Customizing toolbars
1.4.1 Setting Default Values
You can set defaults for most dialog boxes in the Data
Explorer software by setting a value or selecting a button,
closing the dialog box, then closing the data file you are
viewing.
The next time you open the dialog box, the last settings
specified are displayed.
1
1.4.2 Customizing Processing
and Graphic Settings (.SET)
This section includes:
• Overview of processing and graphic settings
• What settin gs contain
• Default processing and graphic settings
• Default graphic settings
• Customizing settings saved in a data file
• Making a copy of default .SET files before customizing
• Opening, customizing, and saving .SET files
• Applying a .SET file
™
Data Explorer
Software User’s Guide 1-17
Page 32

Chapter 1 Data Explorer™ Basics
1
Overview of
processing and
graphic settings
What settings
contain
Processing and graphic settings control how data is processed
and displayed in the Data Explorer software. The last settings
used are automatically saved in the data file when you close it.
The next time you open the data file, you can select any of the
following to apply:
• Settings from the data file—Described in Sect ion 2.1,
Opening and Closing Data Files.
• Default settings—Described below.
• Settings from a selected set file—You can also apply
processing and graphic settings that have been
exported as stand-alone .SET files from other data files.
See “Applying a .SET file” on page 1-20.
Graphic settings include the attributes you set in Graphic
Options, described in Section 1.5, Setting Graphic Options.
Processing settings include:
• Peak detection parameters, described in Section 3.2.4,
Peak Detection Parameter Descriptions
• Smoothing points, described in Section 5.7, Noise
Filtering/Smoothing
• Automatic calibration settings, described in Section 5.4,
Automatic Calibration
Default
processing and
graphic settings
1-18 Applied Biosystems
NOTE: You can open a .SET file in Microsoft Notepad to
view the complete file contents.
The following default settings files (stand-alone .SET files) that
contain both processing and graphic settings are provided on
your system in the C:\MARINER\PROGRAM\Set Files
directory or the C:\VOYAGER\PROGRAM\Set Files directory:
• MARINER.SET
• VOYAGERLINEAR.SET
• VOYAGERREFLECTOR.SET
• VOYAGERPSD.SET
For more information on Peak Detection settings, see
Section 3.7, Default Peak Detection Settings .
Page 33

Customizing the Data Explorer Window
Additional .SET files that have been developed for detection of
different types of data are included in the
C:\VOYAGER\PROGRAM\SET FILES directory . The names of
the .SET files indicate the type of data the files can be used
for.
The appropriate default settings for the type of data you open
are automatically applied to a data file the first time you open it
in Data Explorer. You can also manually apply these settings if
desired. For information, see “Applying a .SET file” on
page 1-20.
1
Default graphic
settings
Customizing
settings saved in
a data file
Making a copy of
default .SET files
before
customizing
Two additional default settings files (stand-alone .SET files)
that contain graphic settings only are also provided on your
system in the C:\MARINER\PROGRAM directory or the
C:\VOYAGER\PROGRAM directory:
• DEFAULTWHITE.SET
• DEFAULTBLACK.SET
These .SET files contain the default graphic settings
applied when you select White Backgr ound or Dark
Background (after selecting Default from the Display
menu). For information, see Section 1.5.1, Changing
Background Color.
You can customize settings saved in a data file by adjusting
graphic or processing settings in the Data Explorer window.
Settings are saved with a data file when you close the data
file, and are automatically applied the next time you open the
data file, if specified. For more information, see Section 2.1,
Opening and Closing Data Files.
You can also save the settings in a .SET file for use with other
data files, as described in “Saving .SET files” on page 1-37.
All .SET files are user editable. However, before you edit the
default pr oc es si ng / g ra ph ic . SE T fi l e s (s ee page 1-18), m ak e a
copy of the original .SET file, in case you need to reload the
settings for default peak detection. Copy the file using
Windows NT
®
Explorer.
Data Explorer
™
Software User’s Guide 1-19
Page 34

Chapter 1 Data Explorer™ Basics
1
Opening,
customizing, and
saving .SET files
To open, customize, and save .SET files:
1. If you are customizing a default .SET file, make a copy
of the original file before opening it. See “Making a copy
of default .SET files before customizing” on page 1-19.
2. Select Settings from the File Menu.
3. Select one of the foll owing:
• Restore Processing Settings
• Restore Graphic Settings
• Restore Graphic/Processing Settings
4. In the Restore dialog box, select or type the name of the
.SET file, then click OK.
5. Customize processing and graphic settings as needed.
For information on the contents of processing and graphic
settings, see “What settings contain” on page 1-18.
6. To save settings, select S ettin gs from the File menu,
then select one of the following:
• Save Processing Settings As
• Save Graphic Settings As
• Save Graphic/Processing Settings As
7. In the Save As dialog box, type the name of the .SET file,
then click OK.
Applying a .SET
file
1-20 Applied Biosystems
You can apply a .SET file two ways:
• Select Recent Processing Settings from the File
menu, then select a .SET file from the list of most
recently used files
NOTE: When you apply a .SET file from the Recent
Processing Settings menu, only the process settings
are applied.
• Select an option from the Settings command on the
File menu
Page 35

To use the Settings option:
Customizing the Data Explorer Window
1. Select Settings from the File menu.
2. Select one of the following:
• Restore Processing Settings
• Restore Gra ph ic Set tin gs
• Restore Graphic/Processing Settings
• Revert to the Last Saved Graphic/Processing
Settings
3. If you select a Restore Settings option, select or type the
name of the .SET file in the Restore dialog box, then
click OK.
Hint: You can also restore default processing and graphic
settings when you open a file or files. See Section 2.1,
Opening and Closing Data Files.
1.4.3 Customizing Toolbars
Customizing
toolbars
To customize the toolbar:
1. Select Customize Toolbar from the Tools menu to
display the Customize dialog box.
1
2. To display or hide a toolbar section, click the Toolbars
tab, then select or deselect a toolbar.
3. To add a button to a toolbar, click the Commands tab,
select the appropriate category, then click-drag the button
to any toolbar in the main toolbar.
Hint: To display a button description, click the button
within the Customize dialog box. You can add buttons
from any category to any toolbar. For example, you can
add buttons from the File category to the Graph toolbar.
™
Data Explorer
Software User’s Guide 1-21
Page 36

1
Chapter 1 Data Explorer™ Basics
4. To remove a button from a toolbar, click-drag the button
from the toolbar.
NOTE: The Customize dialog box must be displayed to
click-drag a button from a toolbar.
5. Click OK to close the Customize dialog box.
Undocking
toolbars
Creating
toolbars
The toolbar at the top of the Data Explorer window is divided
into sections. A section is preceded by a double vertical bar.
You can move (undock) each section of the toolbar within the
Data Explorer window by click-dragging the double bar at the
left side of the toolbar section.
To move the toolbar section back to the top of the window,
click-drag the toolbar back to the original position.
You can display or hide each section individually, add or
remove buttons on the toolbar, and rearrange the order of
buttons displayed. To do so, you must have the Customize
dialog box open, as described in the previous section.
You can create new toolbars by click-dragging buttons to a
window area where there is no toolbar. You can then add
buttons to the new toolbar as described in “Customizing
toolbars” on page 1-21.
1-22 Applied Biosystems
Page 37

Setting Graphic Options
1.5 Setting Graphic Options
This section includes:
• Changing back ground color
• Customizing options
• Reverting to previous graphic options
NOTE: Changes you make to Graphic Options are saved
with the data file.
1.5.1 Changing Background Color
1
White or dark
background
You can switch background color by selecting Default from
the Display menu, then selecting:
• White Background—Displays blue traces and red
labels by default. Default settings are contained in
DEFAULTWHITE.SET.
• Dark Background—Displays yellow traces and green
labels by default. Default settings are contained in
DEFAULTBLACK.SET.
NOTE: These .SET files contain graphic settings only. They
do not contain processing settings.
You can customize the graphic settings associated with
default settings if desired. For information, see Section 1.4.2,
Customizing Processing and Graphic Settings (.SET).
Data Explorer
™
Software User’s Guide 1-23
Page 38

1
Chapter 1 Data Explorer™ Basics
1.5.2 Customizing Graphic Options
This section includes:
• Accessing graphic option s
• Setting colors
• Setting line widths
• Setting data cursors
• Setting traces in line or bar mode
• Setting graphic compression
Accessing
graphic options
To access graphic options:
1. Display the trace of interest.
2. From the Display menu, select Graphic Options.
3. To use the graphic options settings for all traces, click
Use same settings for all traces in the View Setup tab.
4. Click a Graph Setup tab in the Graph and Plot Options
dialog box (Figure 1-4).
5. Set colors, line widths, data cursors, and graphic
compression as described in the following sections.
6. Click OK.
1-24 Applied Biosystems
Page 39

Line or vertical
bar traces
Line width
Data cursor
Setting Graphic Options
1
Peak
bounds
Graphic
compression
Figure 1-4 Graphic Options Dialog Box—
Graph Setup Tab
Setting colors You can set colors manually or automatically.
Manually To manually select the color of graph features (axis, peak
bounds, tick labels, data cursor) and plot features (traces,
peak labels):
1. Select Graphic Options from the Display menu.
2. Set colors in the Graph Setup tab (see Figure 1-4).
™
Data Explorer
Software User’s Guide 1-25
Page 40

1
Chapter 1 Data Explorer™ Basics
When you manually set colors, note:
• Selections set to white (or line widths set to 0) may not
print on certain printers.
• If you select different trace colors for multiple traces, only
the color for the active trace is saved when you close the
data file.
Automatically
using Auto Color
Setting line
widths
Automatically assigning trace colors is useful when overlaying
traces. To automatically assign trace colors:
1. Select Graphic Options from the Display menu.
2. Select Auto Color in the View Setup tab. The software
assigns and displays trace colors when the traces are
overlaid.
When you use Auto Color:
• The active trace color stays at its original setting.
• Other trace colors are set based on the active trace color.
For example, if the active trace is yellow, other traces are
assigned the colors pale blue, pale green, and medium
gray, which are the colors listed after yellow in the Trace
color list, excluding whit e.
NOTE: White is not used in Auto Color because white may
not print on certain printers.
Y ou can control trace appearance by setting line widths. To set
line widths:
1. Select Graphic Options from the Display menu.
1-26 Applied Biosystems
2. Set line width in the Plot Setup section of the Graph Setup
tab (see Figure 1-4 on page 1-25).
NOTE: Line Widths of 0 or 1 (or lines set to the color white)
may not print on certain printers. If traces do not print,
change the line width (or color).
Page 41

Setting Graphic Options
Setting data
Label
Type
cursors
To enable data cursors and set cursor labels and attributes:
1. Select Graphic Options from the Display menu.
2. In the Data Cursor section of the Graph Setup tab (see
Figure 1-4 on page 1-25), select Show Data Cursors,
then select one of the following cursors from the Type
drop-down list:
• X—Single vertical cursor
• Y—Single horizontal cursor
• X-Y—Vertical and horizontal cursors
• X-X—Two vertical cursors
• X-Y-X—Two vertical cursors and one horizontal cursor
NOTE: If data cursors are displayed, they are printed when
you print traces. To suppress data cursor printing, deselect
Show Data Cursors before printing.
To set the cursor mode, select the appropriate label type as
described below:
Options
1
Y Label • Absolute—Displays the number of counts.
• BP Relativ e —Displays the % Intensity value relative to the base
peak in the trace. Includes “BP%” marker on the cursor label.
• Display Relative—Displays the % Intensity value relative to the
largest peak in the current display range.
X-X
Label
• Absolute—Displays the Spectrum Number or Retention Time
(chromatogram) or the Mass (spectrum).
• X-Relative—Available if you have two X cursors displayed. For the
first cursor, displays the Spectrum Number or Retention Time
(chromatogram) or the Mass (spectrum). For the second cursor,
displays the appropriate value relative to the first cursor.
For example, if you place the first X cursor in a spectrum at 100 m/z
and the second X cursor at 80 m/z, the cursors are labeled
100 and –20.
™
Data Explorer
Software User’s Guide 1-27
Page 42

Chapter 1 Data Explorer™ Basics
1
Setting traces in
Line or Vertical
Bar mode
Setting graphic
compression
You can change the trace display from Line to Vertical Bars.
Each vertical bar represents one data point.
Vertical bar mode is useful when setting peak detection
parameters to determine the number of points across a peak.
NOTE: Graphic Compression mode is not saved as part of
graphic settings. When you close a data file, it is
automatically reset to the default Local Max setting.
By default, all data is compressed when it is displayed on your
computer screen. The degree of compression is determined
by the number of data points in the data file and the resolution
setting of your computer monitor.
For example, assume that a data file contains 10,000 data
points and needs compression to 1,000 data points to fit on
your computer screen. Every 10 data points will be
compressed into a single data point.
To set graphic compression settings:
1. Select Graphic Options from the Display menu.
2. In the Graph Setup tab of the Graph and Plot Options
dialog box (see Figure 1-4 on page 1-25), select a
Graphic Compression mode :
1-28 Applied Biosystems
• Local Max (default)—Uses the maximum data
point within the range of data points being
compressed (range of 10 in the example above).
• Average—Uses the average of the data points
within the range of data points being compressed
(range of 10 in the example above).
• Sum (Binning)—Uses the sum of the data points
within the range of data points being compressed
(range of 10 in the example above).
NOTE: Changing the graphic compression mode may
alter the displayed intensities of peaks in a trace.
Page 43

Setting Graphic Options
1.5.3 Reverting to Previous Graphic Options
You have two options to revert to previously used graphic
options:
• Revert to Last Saved Graphic Setting—Reverts to
the last graphic settings saved in the data file. Does not
affect processing settings.
To access, select Default from the Display menu, then
select Revert to Last Saved Graphic Setting.
• Revert to Last Saved Graphic/Processing
Settings—Reverts to the last graphic and processing
settings saved in the data file.
To access, select Settings from the File menu, then
select Revert to Last Saved Graphic/Processing
Settings.
Hint: Instead of applying the settings saved with a data file,
you can apply the default settings stored in the default .SET
file for your system (see page 1-18). To apply the default
settings, close the data file, open the data file again, then
select Use Default Settings in the Open dialog box. For
more information, see Section 2.1, Opening and Closing
Data Files.
1
Data Explorer
™
Software User’s Guide 1-29
Page 44

Chapter 1 Data Explorer™ Basics
1
1.6 Managing Files
This section describes:
• Converting .SPC file format to .DAT file format
(Mariner only)
• Converting data from profile to centroid (Mariner only)
• Converting to and exporting ASCII data
• Importing a trace in ASCII format
• Extracting and saving information from .DA T, .RSD, and
.RCD files
• Copying from data files
1.6.1 Converting .SPC File Format to .DAT File Format (Mariner Data Only)
This section includes:
• When to convert
• Before you begin
• Converting
• Viewing file properties
• Searching file properties
NOTE: You cannot convert Voyager .MS files to .DAT
format.
When to convert You are not required to convert Mariner .SPC file format (for
data acquired in software versions earlier than 3.0) to .DA T file
format. However, the .DAT format allows you to store all
information associated with the file (such as data, results,
settings) in one file, simplifying file management.
1-30 Applied Biosystems
Page 45

Managing Files
Before you begin Confirm that the .SPC and .CGM files are located in the
same directory. Use Windows NT
directory contents and to move the .SPC and .CGM files as
necessary. If the .SPC and .CGM files are not in the same
directory, when you open the .SPC file, a “Failed to open
chromatogram data” message is displayed.
To check that the .SPC and .CGM files are in the same
directory:
1. Select Open from the File menu.
The Open dialog box is displayed.
2. From the Files of type drop-down list, select All Files (*.*).
A list of all files contained in the directory is displayed.
3. Check that the .SPC and .CGM files are present.
®
Explorer to display the
Converting In the Data Explorer window:
1. Open or click the .SPC file to convert.
2. Select Convert from the File menu.
3. Select New Dat a Fo rmat.
The Convert to .DAT Format dialog box is displayed
(Figure 1-5).
1
Figure 1-5 Convert to .DAT Format Dialog Box
4. To add file property information (for example, Title, Author,
or Keywords) to the file, click Set Property, enter the
appropriate information, then click OK. For more
information, see “Viewing file properties” on page 1-32.
™
Data Explorer
Software User’s Guide 1-31
Page 46

1
Chapter 1 Data Explorer™ Basics
The Convert to .DAT Format dialog box reappears.
5. Click OK.
A message box is displayed, showing the file name of
the newly created .DAT file.
6. Close the .SPC file and open the .DAT file before
processing. If desired, manually delete the .SPC file and
associated files.
Viewing file
properties
Searching file
properties
File properties are accessible in Windows NT Explorer and
provide general information about a data file without opening
the file in the Data Explorer software.
To view file properties:
1. In Data Explorer, close the file of interest.
2. In Windows NT Explorer, select the file, then right-click.
3. Select Properties from the menu.
4. Click the Summary tab.
NOTE: If the Summary tab is not available, the file may
be open in Data Explorer. Close the file and repeat the
steps above.
To search for a file based on file properties:
1. In Windows NT Explorer, select Find from the Tools
menu, then select Files or Folders.
The Find: All Files dialog box is displayed.
2. In the Name & Location tab, type or select a directory in
the Look in text box.
1-32 Applied Biosystems
3. Select the Advanced tab.
4. Select Data Explore Document from the Of type
drop-down list.
5. Type the information you are searching for (for example, a
title or keyword) in the Containing text box, then click Find
Now.
Page 47

Managing Files
1.6.2 Converting Data from
Profile to Centroid (Mariner Data Only)
Overview You can convert an entire data file from profile to centroid
format. Centroid format files are smaller than profile format
files.
NOTE: Profile data is not automatically deleted when you
convert to centroid data. You can delete the profile data file
using Windows NT Explorer.
Before you begin If the file to convert is in .SPC format, confirm that the .SPC
and .CGM files are located in the same directory. For
information see “Before you begin” on page 1-31. If the
.SPC and .CGM files are not in the same directory, when
you open the.SPC file, a “Failed to open chromatogram
data” message is displayed.
1
Converting to
centroid
To convert from profile to centroid format:
1. Open or activate the .DAT or .SPC file to convert.
2. Select Convert from the File menu, then select Centroid.
The Save As dialog box is displayed. The software
appends a “-CT” suffix to the file name. This is the
default file name. You can enter a new file name for the
centroid file.
3. Click Save.
NOTE: The m/z range in a data file that is converted
from profile to centroid is determined by the peak
detection range set in Data Explorer, not the m/z range
in the original data file. For more information, see
Section 3.2, Peak Detection.
™
Data Explorer
Software User’s Guide 1-33
Page 48

1
Chapter 1 Data Explorer™ Basics
1.6.3 Converting to and
Exporting ASCII Data
This section describes:
• Converting a data file to ASCII format
• Exporting a trace to ASCII format
Converting a data
file to ASCII
format
Exporting a trace
to ASCII format
Y ou can convert a data file for use in a spreadsheet or another
application and export and import single traces in ASCII
format.
To convert an entire active data file to an ASCII text file:
1. Open the data file to convert.
2. Fr om the File menu, select Convert, then select ASCII
text.
A Save As dialog box is displayed.
3. Specify the name and destination for the file to be
exported. By default, the software assigns a .TXT
extension to the file.
4. Click OK.
You can export a selected trace to ASCII format for display in
Data Explorer software or for use in another application.
To export trace data to ASCII format:
1. With a data file open, select the trace window.
2. Fr om the File menu, select Export, then select
Chromatogram or Spectrum (the menu item is context
sensitive).
1-34 Applied Biosystems
A Save As dialog box is displayed.
3. Specify the name and destination of the file to be
exported. By default, the software assigns a .TXT
extension to the file.
4. Click OK.
Page 49

Managing Files
1.6.4 Importing a Trace in ASCII Format
You can import trace data in ASCII format. If the file you are
importing was originally exported using the Data Explorer
software, you can import a spectrum trace only into the
Spectrum window and a chromatogram trace only into the
Chromatogram window. However, if the data is from another
source, the software does not know the data type and you
must make sure to import the data into the correct window
type.
1
Importing a Trace
in ASCII Format
To import an ASCII trace:
1. Open a data file and activate a Chromatogram or
Spectrum window.
NOTE: You must open a data file before you can import
ASCII data, even if the data file is unrelated to the
imported data.
2. Select Add/Remove Traces from the File menu, then set
the Replace Mode by selecting one of the following:
• Replace the Active Trace
• Add a New Trace
3. From the File menu, select Import, then select ASCII
Chromatogram or ASCII Spectrum (the menu item is
context sensitive to the selected window).
An Open dialog box appears.
4. Select the ASCII file to import.
5. Click OK.
The imported trace is displayed in the active window.
Data Explorer
™
Software User’s Guide 1-35
Page 50

Chapter 1 Data Explorer™ Basics
1
CAUTION
An imported ASCII format trace contains only the data
points for the trace. The Sample Info and Instrument
settings tabs in the Output window display data from the
data file you opened in step 1. These tabs do not include
information about the imported trace.
1.6.5 Extracting and Saving Information from .DAT, .RSD, and .RCD Files
Overview You can extract the following information from a .DA T data file,
.RSD spectrum results file, and .RCD chromatogram results
file, then save the information as a stand-alone file for use with
other files:
• Instrument settings (.BIC)
• MS Method (.MSM) (Mariner data only)
• Calibration constants (.CAL)
• Processing/graphic settings (.SET)
• Spectrum or chromatogram peak labels (.LBS or .LBC)
• ASCII Spectrum and ASCII Chromatogram (.TXT)
Exporting .BIC,
.MSM, and .CAL
files
1-36 Applied Biosystems
To export Instrument settings (.BIC), MS Method settings
(.MSM), or calibration constants (.CAL) from a .DAT or results
file:
1. Open or activate the data (.DAT) or results (.RSD or
.RCD) file.
2. Fr om the File menu, select Export, then select:
• Calibration—To export the last applied
calibration constants in a .CAL file.
Page 51

Managing Files
NOTE: To export calibration constants used to
acquire the data, select Mass Calibration from
the Process menu, then select the Revert to
Instrument Calibration before exporting. For
more information, see Section 5.3.4, Reverting to
Instrument Calibration.
• Configuration—To export .BIC or .MSM files
(.MSM—Mariner data only).
NOTE: To access the instrument settings for each
spectrum in a data file acquired using a Mariner
MS Method, you must first extract the .MSM file
from the data file as described above. Then export
the .BIC files from the .MSM file using the Export
button in the MS Method editor. For more
information on exporting a .BIC file from .MSM,
see the Mariner Wo rks tati on U ser’s Guide.
3. In the Save As dialog box, type a name for the exported
file, then click Save.
1
Saving .SET files To sa ve proces sing and graphic s ettings ( .SET) from a .DAT or
results file:
1. Open or activate the .DAT, .RSD or .RCD file.
2. From the File menu, select Settings, then select one of
the following:
• Save Processing Settings As
• Save Graphic Settings As
• Save Graphic/Processing Settings As
3. In the Save As dialog box, type a name for the .SET file,
then click OK.
For information, see Section1.4.2, Customizing Processing
and Graphic Settings (.SET).
™
Data Explorer
Software User’s Guide 1-37
Page 52

Chapter 1 Data Explorer™ Basics
1
Saving .LBS
and .LBC files
To save spectrum (.LBS) or chromatogram (.LBC) peak label
files from a .DAT, .RSD, or .RSC file, see Section 3.5.3,
Setting Custom Peak Labels.
1.6.6 Copying from Data Files
Overview You can copy the following types of data from data and result
files to the Windows clipboard:
• Trace Image—Copies the graphic image of the trace in
the active window.
• Trace Data—Copies the raw data for the trace
displayed in the active window. You can also use this
command by right-clicking, then selecting Copy Trace
Data. For more information, see Section 2.5.2, Copying
Traces from Multiple Data Files to a Window.
• Displayed Peaks—Copies peak list entries for the
peaks displayed in the active window.
• All Peaks—Copies peak list entries for all peaks in the
active view.
• Mass List—Copies all centroid or apex masses fro m
the peak list for the active Spectrum window.
Copy trace image To copy the trace as it is displayed in the active window to the
Windows clipboard:
1. Select the trace window to copy. Zoom and adjust the
trace as needed.
2. From the Edit menu, select Copy, then select Trace
Image.
3. Paste the image into an application that handles Windows
Metafile format images, for example Microsoft
PowerPoint
1-38 Applied Biosystems
®
.
Page 53

Managing Files
NOTE: If you paste the image into an application that
does not handle Windows Metafile format images, for
example Microsoft Paint, images are distorted.
Copy trace data To copy raw data (x,y pairs) for the peaks displayed in the
active trace to the Windows clipboard:
1. Select the trace window to copy. Zoom and adjust the
trace as needed.
NOTE: Only raw data for the set of peaks displayed in
the trace window is copied.
2. From the Edit menu, select Copy, then select Trace Data.
3. Paste the data into an appropriate application, for
example, another Data Explorer trace window or Microsoft
Excel.
When pasted into a Data Explorer trace window, the
trace appears with the filename in parentheses and the
trace label from the copied trace.
CAUTION
A trace pasted into an active Data Explorer window
contains only the data points for the trace. The Sample
Info and Instrument settings tabs display data from the
original data file. These tabs do not include information
about the copied trace.
1
Copy displayed
peaks
Use this method to copy the section of the peak list pertaining
to the peaks displayed in the active trace window. To copy the
peak list for all peaks in the active trace, see “Copy all peaks”
on page 1-40.
™
Data Explorer
Software User’s Guide 1-39
Page 54

Chapter 1 Data Explorer™ Basics
1
NOTE: Copy Displayed Peaks copies all fields and
headings. However, some data applications may not work
correctly if headings are present because the first row
contains text and not data. For information on copying the
peaks list without headings, see Section 3.3.3, Saving the
Peak List.
To copy the peak list for the displayed peaks to the Windows
clipboard:
1. Set peak detection as needed to create a peak list. See
Section 3.2, Peak Detection.
2. Select the trace window to copy. Zoom and adjust the
trace as needed.
NOTE: Only peak list information for the set of peaks
displayed in the trace window is copied.
3. From the Edit menu, select Copy, then select Displayed
Peaks.
4. Paste the data into an appropriate application, for
example Microsoft Excel.
Copy all peaks Use this method to copy the peak list for all peaks in the active
trace. T o copy only the section of the peak list pertaining to the
peaks displayed in the active trace window, see “Copy
displayed peaks” on page 1-39.
NOTE: Copy All Peaks copies all fields and headings.
However, some data applications may not work correctly if
headings are present because the first row contains text
and not data. For information on copying the peaks list
without headings, see Section 3.3.3, Saving the Peak List.
To copy the peak list for all peaks in the active trace to the
Windows clipboard:
1. Set peak detection as needed to create a peak list. See
Section 3.2, Peak Detection.
1-40 Applied Biosystems
Page 55

2. Select the trace window to copy.
Managing Files
3. Display the peak list by selecting Output Window from
the Display menu, then clicking the Chro Peak List or
Spec Peak List tab.
4. Sort the peak list as needed. See “Sorting the peak list”
on page 3-42.
5. From the Edit menu, select Copy, then select All Peaks.
6. Paste the data into an appropriate application, for
example, Microsoft Excel.
Copy mass list This function copies only the masses for the spectrum and is
useful for pasting data directly into spreadsheets or other
applications without having to delete header information.
To copy all centroid or apex mass data from the spectrum
peak list to the clipboard:
1. Select the Spectrum window to copy from.
2. Select Peak Label from the Peaks menu.
3. Specify the Mass Type by selecting Apex or Centroid.
For more information, see Section 3.5.2, Setting
Chromatogram and Spectrum Peak Labels.
4. Click OK.
1
5. From the Edit menu, select Copy, then select Mass List.
Hint: If you activate the Spec Peak List tab, you can
copy the mass list by right-clicking, then selecting Copy
Mass List from the popup menu.
The mass list is copied to the clipboard with a header
indicating Apex Mass or Centroid Mass.
6. Paste the data into an appropriate application, for
example, Microsoft Excel.
™
Data Explorer
Software User’s Guide 1-41
Page 56

1
Chapter 1 Data Explorer™ Basics
1-42 Applied Biosystems
Page 57

2 Using Chromatogram
and Spectrum
Windows
This chapter contains the following sections:
2.1 Opening and Closing Data Files......................................... 2-2
2.2 Adjusting the Display Range............................................ 2-11
2.3 Organizing Windows........................................................ 2-13
2.4 Manipulating Traces......................................................... 2-14
2.5 Working with Multiple Data Files ...................................... 2-36
2.6 Saving, Opening, and Deleting .DAT Results ................... 2-38
2.7 Exporting, Opening, and Deleting .RCD
and .RSD Results Files (Mariner Data Only) .................... 2-39
2.8 Saving, Opening, and Deleting .SPC Results Files
(Mariner Data Only) ......................................................... 2-40
Chapter
2
Data Explorer™ Software User’s Guide 2-1
Page 58

2
Chapter 2 Using Chro matogr am and Spectrum Windows
2.1 Opening and Clos ing Data Files
This section includes:
• Opening data files
• Displaying Mariner UV traces
• Displaying Voyager chromatograms
• Viewing read-only files
• Moving between open files
• Closing data files
2.1.1 Opening Data Files
This section includes:
• Opening a recently opened file
• Opening data files with File Open
• Automatically running macros
Opening a
recently opened
file
Opening data files
with File Open
2-2 Applied Biosystems
The File menu lists the last several opened files (up to a
maximum of nine). To quickly open one of these files, select it
from the list.
NOTE: If you access Data Explorer software from the
Voyager Instrument Control Panel, the most recently saved
Voyager data file is displayed in the Data Explorer window.
This data file and other recently acquired data files are
listed in the Data Explorer File menu the next time you open
Data Explorer.
You can open up to eight data files using the File Open dialog
box:
1. Select Open from the File menu.
The Select Files dialog box (Figure 2-1) appears.
Page 59

Opening and Closing Data Files
.
2
Displaying
acquisition
comment
Figure 2-1 Select Files Dialog Box
2. Click the down arrow to display the Files of type list,
then select the file extension to display.
3. Select up to eight data files to open, then click Add or
Add All. Add All is not active if the number of selected
files exceeds eight.
NOTE: You can also select files by double-clicking the
file name in the file list.
The files are listed in the Files Selected box.
4. To display acquisition comments before opening the data
file, right-click a file name in the top pane of the dialog
box, select Properties, then click the Summary tab.
™
Data Explorer
Software User’s Guide 2-3
Page 60

2
Chapter 2 Using Chro matogr am and Spectrum Windows
Specifying settings 5. Select a Restoring Graphics and Processing Settings
option to apply to new files you are opening (settings are
not applied to files in the list that are already open):
• Use Settings from Data File—Applies the last
settings used on the data.
• Use Default Settings—Applies settings from
the appropriate default .SET file for your
system. See “Default processing and graphic
settings” on page 1-18.
• Use Selected Set File—Opens the Restore
Graphics and Processing Settings dialog box,
where you select the .SET file to open.
For information on customizing a .SET file, see
Section 1.4.2, Customizing Processing
and Graphic S ettings (.SET).
6. Click Finish to open the selected files.
The Data Explorer window displays the selected data
files with the processing and graphic settings you
selected (Figure 2-2 and Figure 2-3).
2-4 Applied Biosystems
Page 61

Opening and Closing Data Files
Figure 2-2 Data Explorer Window with Four Mariner Data Files Open (Each .DAT
File Displays a Chromatogram and a Spectrum Trace)
2
Figure 2-3 Data Explorer Window with Four Voyager Data Files Open (Spectrum
Traces Only Displayed by Default)
™
Data Explorer
Software User’s Guide 2-5
Page 62

2
Chapter 2 Using Chro matogr am and Spectrum Windows
The data is labeled accordingly:
Type of Data Spectrum Trace Labels
Mariner data If you open a data file you previously calibrated in Data Explorer,
all spectra in the data file are calibrated and displayed with an
MC or AC trace label.
If AutoSaturation Correction is turned on, all spectra in the data
file are corrected and displayed with an ASC trace label. For
more information, see Section 5.11, AutoSaturation Correction
(Mariner Data Only).
Voyager data If you open a data file you previously calibrated in Data Explorer,
the spectrum is calibrated and displayed with an MC trace label.
For multispectrum data files, you can display chromatograms.
For more information, see Section 2.1.3, Displaying Voyager
Chromatograms.
Automatically
running macros
You can set macros to automatically run when you open or
close files. For information, see “Running Macros
Automatically When Opening and Closing Files” on page 6-45.
2.1.2 Displaying Mariner DAD Traces
When you use a diode array detector (DAD) to acquire
Mariner data, you can display the following types of
DAD traces:
• DAD chromatogram—With the Chromatogram
window active, select Traces from the Display menu,
then select DAD TAC or DAD Channel.
• DAD spectrum—Double click a DAD chromatogram.
For more information, see “Types of Mariner data” on page 4-2
and “Creating an Extracted Absorbance Chromatogram (XAC)
(Mariner Data Only)” on page 4-13.
2-6 Applied Biosystems
Page 63

Opening and Closing Data Files
2.1.3 Displaying Voyager Chromatograms
To display chromatograms for multispectrum
Voyager.DAT files:
1. Open the .DAT files as desc ribed in “Opening Data
Files” on page 2-2.
Spectrum traces for the .DAT files are displayed.
2. To display a Chromatogram window, click a Spectrum
window to activate it, then select Restore
Chromatogram from the View menu.
A Chromatogram window is displayed for the data file.
NOTE: Diode array support and UV display are disabled
for Voyager chromatogram traces.
2.1.4 Viewing Read-Only Files
2
For quick scanning of archived data files, you can view
read-only files using the File Open dialog box. When you open
a read-only file, a message indicates that the file is
read-only and prompts you to open the file. All functions that
write information to disk are disabled when you view a
read-only file.
CAUTION
Although you can process read-only files, you cannot save
any changes you make. When you close a read-only file
any changes you made are lost.
™
Data Explorer
Software User’s Guide 2-7
Page 64

2
Chapter 2 Using Chro matogr am and Spectrum Windows
2.1.5 Moving Between Open Files
Y ou can have more than one file open at a time. You can move
between open files in three ways:
• Select the open file to view from the Window menu
• Use tabs
• Use the Activate File dialog box
Using tabs To move between open files using tabs, click the tab at the
bottom of the Data Explorer window (Figure 2-4) to activate
the file.
Click a tab to
select an open data file
Using Activate
File
Double-click
an active title bar
to maximize a window
Figure 2-4 Tabs for Open Data Files
If the window for the tab you click is already open, clicking the
tab activates the window.
If the window for the tab you click is minimized, clicking the tab
activates the title bar for the window. Double-click the active
title bar to display the window.
To move between Spectrum windows of open data files using
Activate File:
1. Select Activate File from the Windows menu.
2. Select a file from the Current Data File list.
2-8 Applied Biosystems
Page 65

Opening and Closing Data Files
Figure 2-5 Select File to Activate Dialog Box
3. Select:
• Maximize—To maximize the Spectrum window
of the selected file
2
• Activate—To activate the Spectrum window of
the selected file
™
Data Explorer
Software User’s Guide 2-9
Page 66

2
Chapter 2 Using Chro matogr am and Spectrum Windows
2.1.6 Closing Data Files
You can close files in the following ways:
• Select Close from the File menu to close the active file.
• Select Close All Files from the File menu to close all
files.
• In the Open File dialog box, select any open files from
the Files Selected list, click Remove, then click Finish
to close the selected files.
• Select Exit from the File menu to close all files and exit
the software.
If you enabled Processing History and selected the Show
Save History option, a dialog box is displayed. Click Save or
Purge. For more information, see “Setting Processing History
options” on page 2-23.
2-10 Applied Biosystems
Page 67

Adjusting the Display Range
2.2 Adjusting the Display Range
To set the display range:
1. Click the Chromatogram or Spectrum window to
activate it.
2. From the Display menu, select Range.
3. Select X Range to set the x-axis range. The scaling units
depend on the window you are scaling:
• Chromatog ra m —Scales in the same units
currently displayed in the Chromatogram
window (Spectrum Number or Time [Mariner
data only])
• Spectrum—Scales in m/z units
4. Select Y Range to scale the y-axis range.
The Y Axis Setup dialog box (Figure 2-6) is displayed.
2
Figure 2-6 Y Axis Setup Dialog Box
™
Data Explorer
Software User’s Guide 2-11
Page 68

Chapter 2 Using Chro matogr am and Spectrum Windows
5. Set the parameters described below as needed:
Parameter Description
Scaling Mode
Display Relative Autoscales the trace to the largest peak in the selected range.
2
Base Peak
Relative
Absolute Value Sets the trace to the Y Display Range you enter in the Y Display
Display Min/Max S ets the trace display to the minimum and maximum Y values.
Minimum Absolute Max Y
Use Limit Sets the minimum value for Y-axis scaling. Useful to maintain
Y Display Range
From/To Sets the range for scaling. If Display Relative or Base Peak
Autoscales the trace to the base peak in the entire range, not the
selected range. Displays a right-axis label with the base peak
intensity.
NOTE: To turn off the right axis, select Graphic Options from
the Display menu, click the Graph #1 Setup tab, then deselect
the Show Right Y-Axis check box.
Range From/To boxes.
relative scaling of peaks and to prevent autoscaling noise to full
scale.
Relative is selected, range is in percent.
If Absolute Value is selected, range is in counts.
2-12 Applied Biosystems
6. Click OK.
Page 69

Organizing Windows
2.3 Organizing Windows
Linking views Linking Chromatogram or Spectrum windows in different data
files allows you to zoom on multiple data files.
NOTE: When different data files are linked, zooming
functions performed on one data file are applied to all linked
files. Processing and peak centering functions are applied
to the active file only.
To link data files:
1. Open the data files you want to link.
2. In the first data file, click the window (Chromatogram or
Spectrum) that you want to link to another data file, then
select Link View from the View menu.
2
Organizing
windows
NOTE: Clicking in the toolbar links traces, not
views.
3. Repeat step 2 if you want to link both windows.
4. Repeat step 2 and step 3 for the remaining data files.
NOTE: You must select Link View for each window and
each data file you want to link.
You can organize all open windows by clicking buttons in the
toolbar:
• Tile Horizontal
• Tile Vertical
• Cascade Windows
You can also move and resize windows by click-dragging.
™
Data Explorer
Software User’s Guide 2-13
Page 70

2
Chapter 2 Using Chro matogr am and Spectrum Windows
2.4 Manipulating Traces
This section includes:
• Zooming centering, and customizing a trace
• Duplicating a trace
• Dividing the active trace
• Adding traces from the same data file to a window
• Removing traces
• Expanding and linking traces
• Recalling and rearranging traces (Processing History)
• Overlaying traces
• Annotating traces
• Viewing trace labels
• Printing traces
2.4.1 Zooming, Centering,
and Customizing a Trace
Zooming and
unzooming
2-14 Applied Biosystems
Y ou can expand (zoom) an area of a trace by click-dragging a
box around the area of interest.
You can also click buttons in the toolbar to:
• Zoom in
NOTE: Display data cursors, then click the point you
want to zoom on before clicking this button. See
“Setting data cursors” on page 1-27.
• Zoom out to the previous zoom
• Full Unzoom
Page 71

Manipulating Traces
Centering a peak
in the trace
Customizing a
trace
To center a peak in the trace window:
1. Display the trace containing the peak of interest.
2. Click the Spec Peak List or Chro Peak List tab in the
Output window.
3. Double-click the peak to center.
The peak is centered in the active trace.
See Section 1.5, Setting Graphic Options, for information on
customizing trace parameters such as line widths and data
compression.
2.4.2 Duplicating a Trace
To make a duplicate of a trace:
1. Click the trace.
2. From the Display menu, select Duplicate Active Trace.
The active trace is duplicated and displayed in another
trace position.
2
2.4.3 Dividing the Active Trace
You can divide the active trace into equal segments with one
command. This is useful to expand the x-axis range to better
see spectral and chromato graphic features.
To divide a trace:
1. Click the trace.
2. From the Display menu, select Divide Active Trace, then
select the number of segments to divide the trace into.
The active trace is automatically split into the selected
number of traces, with the range evenly divided among
the traces.
™
Data Explorer
Software User’s Guide 2-15
Page 72

Chapter 2 Using Chro matogr am and Spectrum Windows
For example, if you select Divide Active Trace to Four
when the active trace has a range of 0.0 to 20.0, the
active trace is divided into four traces:
• First trace represents the range from 0 to 5
• Second trace represents the range from 4 to 10
• Third trace represents the range from 9 to 16
• Fourth trace represents the range from 15 to 20
2
NOTE: To restore the display to a single trace, select
Remove Inactive Traces from the Display menu.
2.4.4 Adding Traces from the Same Data File to a Window
This section describes:
• Overview
• Setting the Replace mode
• Adding a trace
Overview By default, the Data Explorer software displays:
Data Type Window
Mariner Chromatogram and Spectrum.
Voyager single
spectrum
Voyager
multispectrum
Spectrum only.
Spectrum only. Can also optionally
display chromatogram.
2-16 Applied Biosystems
Chromatogram and Spectrum windows each contain
one trace (see Figure 1-3 on page 1-11).
Page 73

Manipulating Traces
When you perform certain functions (f or example,
smoothing), a new trace is created. You can set the Replace
mode to add to or replace the active trace.
Y ou can add up to seven new traces to a window to allow you
to keep original data displayed when you generate new
traces.
Setting the
Replace mode
To set the Replace mode:
1. From the Display menu, select Add/Remove Traces.
The Display Trace dialog box is displayed (Figure 2- 7).
2
Figure 2-7 Display Trace Dialog Box
™
Data Explorer
Software User’s Guide 2-17
Page 74

Chapter 2 Using Chro matogr am and Spectrum Windows
2. Select the Replace Mode:
• Replace the Active Trace (default)—Replaces
the active trace with the newly created trace.
• Add a New Trace—Adds the newly created
trace to the window. The original trace remains
displayed and accessible, allowing visual
comparison of the traces.
2
NOTE: If Replace Mode is set to Add a New Trace and
eight traces are present when you perform a function,
the active trace is replaced when a new trace is
generated.
Hint: A toolbar button is available for switching
between Replace and Add mode. See “Customizing
toolbars” on page 1-21 for information. The button
is located in the Graph category.
3. If desired, you can also add traces by selecting the trace
to add from the Select Traces to Display section.
4. Click OK.
Adding a trace To add a new trace to a window:
1. Click the Chromatogram or Spectrum window.
2. Click in the toolbar.
NOTE: You can also add traces by selecting Add/
Remove Traces from the Display menu.
2-18 Applied Biosystems
The software adds a trace to the window. You can add
up to seven traces (Figure 2-8).
Hint: Resize the window to view all added traces.
Page 75

Original
trace
Manipulating Traces
Added
traces
Figure 2-8 Adding Traces (Four Traces Shown, up to Four More Can Be Added)
When you perform a function that adds a new trace, the label
of the trace changes from Not Used to the label for the type of
trace created (Figure 2-9).
2
Data Explorer
™
Software User’s Guide 2-19
Page 76

2
Chapter 2 Using Chro matogr am and Spectrum Windows
Figure 2-9 shows the original trace and three added traces
that now contain a smoothed spectrum (SM), a centroided
spectrum (CT), and a baseline offset spectrum (BO).
Original
trace
Added
traces
Figure 2-9 Added Traces Containing Data
2-20 Applied Biosystems
For a description of trace labels, see “Viewing Trace Labels”
on page 2-30.
Page 77

2.4.5 Removing Traces
Manipulating Traces
Removing the
active trace
Removing
inactive traces
To remove the active trace from a window:
1. Click the trace to remove.
2. Click in the toolbar, or right-click the trace, then
select Remove Trace from the menu.
The trace is removed.
To quickly remove all inactive traces from the window:
1. Click the trace to keep displayed to make it the active
trace.
2. Select Remove Inactive Traces from the Display menu.
2.4.6 Expanding and Linking Traces
When you have more than one trace displayed in a window,
you can:
• Expand traces—Click a trace, then click in the
toolbar to expand the selected trace for closer
examination. To display all traces, click again.
2
• Link traces—Click in the toolbar to link all traces.
Any zooming actions you perform on one trace affect all
traces in that window. To unlink traces, click
again.
™
Data Explorer
Software User’s Guide 2-21
Page 78

2
Chapter 2 Using Chro matogr am and Spectrum Windows
2.4.7 Recalling and Rearranging Traces (Processing History)
Overview The Chromatogram and Spectrum windows can display up to
8 traces at a time for a data file. However, up to 16 traces are
held in memory.
You can recall traces previously displayed for an open data
file, or rearrange the order of traces, using the Processing
History command. You can also set Processing History
Options to automatically delete history or to disable the
function completely.
Recalling or
rearranging
Removing t rac es
from the history
list
To recall or rearrange previously displayed traces:
1. Click the trace position in which you want to recall or
rearrange a trace.
2. From the Display menu, select Processing History.
A submenu is displayed, listing the last 16 processed
traces viewed in the window.
NOTE: Unprocessed traces and theoretical traces
generated using commands on the Applications menu
are not listed.
3. Select the trace to display.
The selected trace is displayed in the active trace
position.
To remove traces from the Processing History list:
1. From the Display menu, select Processing History.
2. Select Remove History.
3. Select the traces to remove from the list, then click OK.
2-22 Applied Biosystems
Page 79

Manipulating Traces
Setting
Processing
History options
To set Processing History options:
1. From the Tools menu, select Processing History
Options.
The Processing History Options dialog box
(Figure 2-10) is displayed.
2
Figure 2-10 Processing History Options Dialog Box
2. Turn Processing History on or off. If you turn on, specify:
• Purge processing history—Records all
processing functions performed. Stores the
history log in a .CTS file and purges the history
log when you close the data file.
Purging a history file does not affect the data
contained in the data file. It clears the contents
of the .CTS file that contains the processing
history.
• Save processing history—Records al l
processing functions performed. Stores the
history log in a .CTS file and maintains the
history log when you close the data file.
If you save processing history, .CTS files can
become very large. You can periodically delete
older .CTS files to clear disk space.
3. If you want a reminder dialog box to appear when you
close a data file that prompts you to selectively save or
purge the history file, select Show Save History dialog.
4. C lick OK.
™
Data Explorer
Software User’s Guide 2-23
Page 80

Chapter 2 Using Chro matogr am and Spectrum Windows
2.4.8 Overlaying Traces
This section includes:
• Overlaying traces from different data files
• Overlaying traces in a single data file
• Changing the active trace
• Sequentially activating overlaid spectra
• Setting over lay attribu tes
2
Overlaying traces
from different
data files
To overlay traces from different data files:
1. Copy chromatogram or spectrum traces you want to
overlay into a trace window. For more information, see
Section 2.5.2, Copying Traces from Multiple Data Files
to a Window.
Hint: The copied traces display the original trace label
and filename.
2. To use settings other than defaults, set attributes for
the overlay if needed. See “Setting overlay attributes”
on page 2-26.
3. Click the trace of interest to activate it.
NOTE: Only the active trace in an overlay is affected by
processing tools. However, all traces are affected by
zooming functions.
4. From the Display menu, select Overlay Traces.
2-24 Applied Biosystems
Hint: A toolbar button is available to toggle between
Overlay and Undo Overlay mode. See “Customizing
toolbars” on page 1-21 for information. The button
is located in the Graph category.
The traces are overlaid. The trace names are listed in
the trace label.
Page 81

Manipulating Traces
NOTE: When saving results, only the results for the
active trace are saved.
Overlaying traces
in a single data
file
To overlay traces in a single data file:
1. Display the chromatogram or spectrum traces you want
to overlay. For more information, see Section 2.4.4,
Adding Traces from the Same Data File to a Window
2. To use settings other than defaults, set attributes for the
overlay if needed. See “Setting overlay attributes” on
page 2-26.
3. Click the trace of interest to activate it.
NOTE: Only the active trace in an overlay is aff ected by
processing tools. However, all traces are affected by
zooming functions.
4. From the Display menu, select Overlay Traces.
Hint: A toolbar button is available to toggle between
Overlay and Undo Overlay mode. See “Customizing
toolbars” on page 1-21 for information. The button
is located in the Graph category.
2
The traces are overlaid. The trace names are listed in
the trace label.
NOTE: When saving results, only the results for the
active trace are saved.
™
Data Explorer
Software User’s Guide 2-25
Page 82

Chapter 2 Using Chro matogr am and Spectrum Windows
2
Changing the
active trace
Sequentially
activating
overlaid spectra
Setting overlay
attributes
To change the active trace in an overlay:
1. From the Display menu, deselect Overlay.
2. Click the trace to activate.
3. From the Display menu, select Overlay.
When you display overlaid traces, you can sequentially
activate each trace by clicking and .
NOTE: If you overlay processed spectra, and then display a
different spectrum number, the processed spectrum is lost.
For example, assume you smooth Spectrum #3 in a trace,
then overlay it with another trace. If you select Spectrum
#3, then click to display Spectrum #2, then click
to return to Spectrum #3, Spectrum #3 is no longer
smoothed.
To set overlay attributes:
1. Display the individual traces to overlay.
2. Select Graphic Options from the Display menu.
The Graph and Plot Options dialog box is displayed.
2-26 Applied Biosystems
NOTE: If traces are overlaid when you select Graphic
Options, you can set attributes for the active trace only.
3. Click View Setu p.
4. To apply the graphic options to all traces, select the Use
same settings for all traces check box.
NOTE: If the Use same settings for all traces check box
is selected when traces are overlaid, the attributes are
applied to the active trace only. When you display
individual traces again, settings are applied to all
traces.
Page 83

Manipulating Traces
NOTE: You must select the Use same settings for all
traces check box before selecting options for traces. If
you do not, settings are applied only to the active trace.
5. In View Setup, select Overlay Trace scaling:
• Display Relative—Autoscales each trace to the
base peak in the display range
• BP Relative —Autoscales each trace to the base
peak in the trace
• Absolute Value—Maintains scaling of individual
traces
6. Set trace colors as needed:
• If you selected Use same settings for all traces,
click Autocolo r to allow the software to
automatically assign trace colors.
2
NOTE: The active trace color stays at its original
setting. Other trace colors are set based on the
active trace color. For example, if the active trace
is yellow, other traces are assigned the colors pale
blue, pale green and medium gray, which are the
colors listed after yellow in the Trace color list,
excluding white. White is not used in Autocolor,
because white may not print on certain printers.
• If you did not select Use same settings for all
traces, click Graph Setup, then select trace
colors. See “Setting colors” on page 1-25 for
information.
7. C lick OK.
™
Data Explorer
Software User’s Guide 2-27
Page 84

Chapter 2 Using Chro matogr am and Spectrum Windows
2.4.9 Annotating Traces
You can add text annotation to traces by:
• Copying a line of results from the Output window (or
copying any ASCII text from any source), then pasting
the copied information on the trace
• Typing text on the trace
2
Copying results To copy results:
1. Generate results as desired by selecting commands
from the Tools or Applications menu. For more
information, see:
• Section 5.3, Manual Calibration, or Section 5.4,
Automatic Calibration
• Section 5.6, Mass Deconvolution (Mariner Data
Only)
• Section 6.2, Using the Isotope Calculator
• Section 6.3, Using the
Mass Resolution Calculator
• Section 6.4, Using the Signal-to-Noise Ratio
Calculator
2. Click the Results tab in the Output window.
3. Select the line of text to copy, then right-click and select
Copy from the menu displayed.
NOTE: If you select more than one line of text, only the
first line is pasted when you annotate the trace.
2-28 Applied Biosystems
Page 85

Manipulating Traces
Annotating the
trace
To annotate the trace:
1. Click the trace at the location where you want to insert
text.
2. Right-click, then select:
• Paste text—If you copied results.
• Add text annotation—If you want to type in
text. Type in text as needed.
The text is added to the trace, and remains in the
Spectrum window until you delete it.
NOTE: The text is associated with the x-coordinate. If
you display another spectrum, the text remains in view.
If you zoom on a different region of the trace and the xcoordinate moves out of view, the annotated text also
moves out of view. If you annotate overlaid traces, only
the text associated with the active trace is displayed.
NOTE: To move the text, click-drag the text to the
desired position.
2
3. To customize the appearance of the annotated text, see
Section 1.5, Setting Graphic Options.
4. To delete an annotation, do either of the following:
• Select the text, right-click, then select the
appropriate delete or cut option.
• Right-click the trace, then select Delete All
Text.
NOTE: Text annotations are included when you print
the spectrum. They are not saved with the spectrum
when you close the data file.
™
Data Explorer
Software User’s Guide 2-29
Page 86

Chapter 2 Using Chro matogr am and Spectrum Windows
2.4.10 Viewing Trace Labels
The Data Explorer software includes a label in the trace
header to identify the type of data displayed.
NOTE: Trace labels are applied by the software and cannot
be removed.
2
Chromatogram
trace labels
Chromatogram Trace
Label
BC Baseline corrected
BO Baseline offset
BP Base Peak mass and intensity
CNL Constant neutral loss chromatogram
EF Event filtered chromatogram (MS Method data only)
Mass xxx:yyy Extracted ion chromatogram for a mass range where xxx
NFX Noise-filtered trace, where X is the applied Correlation
NRX Noise-removed trace, where X is the number of standard
SMX Smoothed trace, where X is the number of smoothing
Labels in the chromatogram title identify the following types of
displayed data:
Description
is the starting mass and yyy is the ending mass
Factor
deviations of noise removed
points applied
TAC Total absorbance chromatogram
+ TAC Realigned total absorbance chromatogram
XAC E xtracted absorbance chromatogram
2-30 Applied Biosystems
Page 87

Manipulating Traces
Chromatogram Trace
Label
+ XAC Realigned extracted absorbance chromatogram
XIC xxx ± yyy Extracted ion chromatogram for a selected mass where
xxx is the center mass and yyy is the specified window
NOTE: Extracted ion chromatograms were previously
labeled with “Mass” instead of “XIC”.
Figure 2-11 illustrates an extracted ion chromatogram with a
“XIC” chromatogram trace label.
Trace label
Figure 2-11 Chromatogram Trace Label
Spectrum trace
labels
Labels in the spectrum title identify the following types of data
displayed:
Description
2
Spectrum Trace Label Type of Processing
+, /, or –mmm:nnn
(Mariner data only)
AC Automatically mass calibrated
AdvBC Advanced baseline correction
ASC (Mariner data only) AutoSaturation corrected trace
BC Baseline corrected
BO Baseline offset
Added, accumulated, or subtracted spectrum from
spectrum #mmm to #nnn
™
Data Explorer
Software User’s Guide 2-31
Page 88

Chapter 2 Using Chro matogr am and Spectrum Windows
Spectrum Trace Label Type of Processing
BPI Base peak intensity
CT Centroid
DAD Diode array data
2
DECONV
(Mariner data only)
DI Deisotoped trace
ISO Isotope
MC Manually mass calibrated
NF Noise-filtered trace
NRX Noise-removed trace, where X is the number of standard
RSMX (does not apply
to Voyager PSD data)
SC (Mariner data only) Conver ted to single-charge
SMX Smoothed, where X is the number of smoothing points
Stitched PSD Composite PSD spectrum
Zero charge deconvoluted trace
deviations of noise removed
Default smoothing applied, where X is the resolution
value (from peak detection) used to calculate the
optimum number of smoothing points to apply at every
mass point
applied
TR Truncated spectrum
2-32 Applied Biosystems
Page 89

Manipulating Traces
Figure 2-12 illustrates a smoothed spectrum with an “SM5”
spectrum trace label.
Trace label
Figure 2-12 Spectrum Trace Label
2.4.11 Printing Traces
Printing traces To print traces:
1. Display the traces to print.
To obtain a clear printout, you can set the Trace Color
and other attributes to dark colors before printing traces
by selecting Default from the Display menu, then
selecting White Background.
NOTE: If you previously modified the colors associated
with this command as described in Section 1.5.1,
Changing Background Color, selecting this command
may not set a white background and black traces.
When you manually set colors, note the following:
• Selections set to white (or line widths set to 0)
may not print on certain printers.
2
• If you select different trace colors for multiple
traces, only the color for the active trace is saved.
2. Click the window (Chromatogram or Spectrum) to print.
™
Data Explorer
Software User’s Guide 2-33
Page 90

Chapter 2 Using Chro matogr am and Spectrum Windows
3. To print with the x-axis along the longest length of the
paper, select Print Setup from the File menu, then select
Landscape orientation.
NOTE: The Landscape printing orientation you set in
Data Explorer is lost when you close Data Explorer. To
permanently set the printer, see “Dedicating a printer to
landscape orientation” on page 2-35.
2
4. From the File menu, select a print option:
• Print File with Instrument Settings—Pri nts
displayed trace windows from the active file
along with the settings used to obtain the data.
• Print Spectrum or Chromatogram Trace—
Prints the active trace.
• Print Spectrum or Chromatogram View—
Prints all traces in the Spectrum or
Chromatogram window.
• Print All Views—Prints traces for all open data
files.
NOTE: If you select Print All Views when more
than two data files are open, certain printers may
not print the data file name. To ensure data file
names are printed, print views individually, or only
open two data files before you select Print All
Views.
2-34 Applied Biosystems
NOTE: To print a trace that is displayed as Vertical
Bars, change the Line Width to 1. If Line Width is set to
0, Vertical Bar traces may not print on certain printers.
See Section 1.4, Customizing the Data Explorer
Window, for information.
Page 91

Manipulating Traces
NOTE: Line Widths of 0 or 1 (or lines set to the color
white) may not print on certain printers. If traces do not
print, change the line width (or color).
Dedicating a
printer to
landscape
orientation
To dedicate the printer to landscape orientation:
1. From the Windows desktop, click Start, then select
Settings.
2. C lick Printers.
3. Select the printer name in the displayed list.
4. C lick File, then select Document Defaults.
5. In the Page Setup tab, select Landscape orientation.
NOTE: If you cannot select Landscape orientation, you
do not have access permission. See your administrator.
Print Setup The Print Setup function allows you to select a printer and set
printer options. For more information on Print Setup and on
connecting printers to your computer, refer to the
documentation provided with your computer.
2
Data Explorer
™
Software User’s Guide 2-35
Page 92

2
Chapter 2 Using Chro matogr am and Spectrum Windows
2.5 Working with Multiple Data Files
When you have multiple data files open, you can:
• Work with the data files separately to view, zoom, and
print
• Copy traces from one data file to another to compare or
combine data
2.5.1 Working with Separate Data Files
To view, zoom, and print multiple data files:
1. Open the data files as described in Section 2.1,
Opening and Closing Data Files.
2. Set the Y Scaling mode to Display Relative, as described
in Section 2.2, Adjusting the Display Range.
3. Link the data files by selecting the Spectrum window for
each data file, then selecting Link View from the View
menu.
2-36 Applied Biosystems
You must select Link View for each data file.
NOTE: Clicking in the toolbar links traces, not
views.
4. Organize the Spectrum windows for the data files by
clicking and in the toolbar.
5. Zoom and manipulate traces as needed.
6. To print all data files, select Print All Views from the File
menu.
Page 93

Working with Multiple Data Files
NOTE: If you select Print All Views when more
than two data files are open, certain printers may
not print the data file name. To ensure data file
names are printed, print views individually, or only
open two data files before you select Print All
Views.
2.5.2 Copying Traces from Multiple Data Files to a Window
You can copy up to seven traces from open data files to a
different trace window:
1. Select the trace to copy.
2. From the Edit menu, select Copy, then Trace Data, or
right-click the trace, then select Copy Trace Data.
3. Activate the window in which to paste the trace.
NOTE: The Add/Replace Trace state, as set in the
Display Trace dialog box, determines whether the
copied trace replaces or is added below the active
trace. See “Setting the Replace mode” on page 2-17.
4. From the Edit menu, select Paste, then Trace Data, or
right-click the window, then select Paste Trace Data.
The copied trace is added to the active window. The
original trace label is preceded by the name of the file
from which you copied the trace.
2
Y ou can repeat step 1 through step 4 until a maximum of eight
traces are displayed in a trace window. If eight traces are
displayed and you copy a new trace to the window, the active
trace is replaced.
™
Data Explorer
Software User’s Guide 2-37
Page 94

Chapter 2 Using Chro matogr am and Spectrum Windows
2
Comparing
copied traces
After you copy a trace to another trace window, you can
compare traces by overlaying (see Section 2.4.8, Overlaying
Traces) or by using trace arithmetic (see Section 5.12, Adding
and Subtracting Raw or Processed Spectra from the Same or
Different Data Files (Dual Spectral Trace Arithmetic).
2.6 Saving, Opening, and
Deleting .DAT Results
Saving results for
.DAT files
To save results for .DAT files:
1. Process a data file to generate results as needed.
2. Fr om the File menu, select Result Spectrum or Result
Chromatogram, then select Save As.
3. Use the default title, or enter a name (up to 31 characters)
for the results in the Title text box, then click OK.
Only the results for the active trace are saved.
NOTE: Results are stored within the .DAT file, not as
separate files. Title is an identifier you can use to recall
the results. It is not a file name.
Opening results
for .DAT
files
Deleting results
for .DAT files
2-38 Applied Biosystems
To open results for .DAT files:
1. From the File menu, select Result Spectrum or Result
Chromatogram, then select Open.
2. Select the title of the previously saved results to open,
then click OK.
3. Click the Sample Info tab in the Output window to display
information for the result.
To delete results for .DAT files:
1. From the File menu, select Result Spectrum or Result
Chromatogram, then select Delete.
2. Select the title of the results to delete, then click OK.
Page 95

Exporting, Opening, and Deleting .RCD and .RSD Results Files (Mariner Data Only)
2.7 Exporting, Op ening, and
Deleting .RCD and .RSD Results
Files (Mariner Data Onl y)
Exporting results
for .RCD and
.RSD files
Opening results
for .RCD and
.RSD files
To export results to a .RSD or .RCD file:
1. With a data file open in Data Explorer, select a
spectrum or chromatogram.
2. From the File menu, select Export, then Result
Chromatogram or Result Spectrum.
A Save As dialog box appears.
3. Specify the name and destination of the exported file. By
default, the software assigns a .RCD extension for a
chromatogram or a .RSD extension for a spectrum.
4. C lick OK.
To open results from .RCD and .RSD files:
1. Select Open from the File menu.
The Select Data File to Open dialog box is displayed.
2. From the Files of Type drop-down list, select Result
Spectrum Files (*.RS*) or Result Chromatogram Files
(*.RC*).
3. Select the .RCD or .RSD file to open, then click OK.
4. Click the Sample Info tab in the Output window to display
the following information for the result file:
• Name of the original raw data file the result file
was generated from
2
• Processing functions that were performed and
saved in the result file
™
Data Explorer
Software User’s Guide 2-39
Page 96

Chapter 2 Using Chro matogr am and Spectrum Windows
2
Deleting results
for .RCD and .RSD
Use Windows NT Explorer to delete .RCD and .RSD result
files.
files
2.8 Saving, Opening, and Deleting
.SPC Results Files
(Mariner Data Only)
Saving results for
.SPC files
To save results for .SPC files (Mariner data only):
1. Process a data file to generate results as needed.
2. Fr om the File menu, select Result Spectrum or Result
Chromatogram, then select Save As.
3. Enter a file name for the result file in the File Name text
box.
Spectrum files are automatically named with an .RST
extension. Chromatogram files are automatically
named with an .RCT extension.
Opening results:
for .SPC files
2-40 Applied Biosystems
4. Click OK.
NOTE: You can also save Mariner .RCT and .RST files
from .DAT format data files in the Instrument Control
Panel. For more information, see the Mariner
Workstation User’s Guide.
To open results for .SPC files:
1. Select Open from the File menu.
The Select Data File to Open dialog box is displayed.
2. From the Files of Type drop-down list at the bottom of the
dialog box, select Result Spec Files (*.RS*) or Result
Chro Files (*.RC*).
Page 97

Saving, Opening, and Deleting .S PC R esults Files (Mariner Data Only)
3. Select the .RST or .RCT file to open, then click OK.
NOTE: Saturation Correction is not applied to Mariner
.RST files. For more information, see Section 5.11,
AutoSaturation Correction (Mariner Data Only).
4. Click the Sample Info tab in the Output window to display
the following information for the result file:
• Name of the original raw data file from which the
result file was generated
• Processing functions that were performed and
saved in the result file
2
Deleting results
for .SPC files
Use Windows NT Explorer to delete .RST and .RCT result files
generated from .SPC files.
Data Explorer
™
Software User’s Guide 2-41
Page 98

2
Chapter 2 Using Chro matogr am and Spectrum Windows
2-42 Applied Biosystems
Page 99

3 Peak Detection
and Labeling
This chapter contains the following sections:
3.1 Overview ......................................................... 3-2
3.2 Peak Detection ................................................ 3-6
3.3 Peak List ....................................................... 3-37
3.4 Deisotoping a Spectrum ................................. 3-45
3.5 Peak Labeling................................................ 3-52
3.6 Process that Occurs During Peak Detection,
Centroiding, and Integration .......................... 3-67
Chapter
3
3.7 Default Peak Detection Settings .................... 3-71
Data Explorer™ Software User’s Guide 3-1
Page 100

3
Chapter 3 Peak Detecti on and Labeling
3.1 Overview
This section includes:
• Default peak detection
• The resolution-based peak detection routine
3.1.1 Default Peak Detection
Overview When you open a data file, it is automatically peak detected.
For:
• Chromatographic data—The software uses default
settings that have been optimized to yield acceptable
peak detection for many sample types. For more
information, see Section 3.7, Default Peak Detection
Settings.
• Spectral data—The software uses a resolution-based
peak detection routine to calculate peak detection values
that provide optimum peak detection for most sample
types. For more information, see Section 3.1.2, The
Resolution-Based Peak Detection Routine.
3-2 Applied Biosystems
For many applications, the default peak detection settings and
settings calculated by the resolution-based peak detection
routine provide acceptable peak detection. If default settings
do not provide acceptable peak detection, you can adjust the
settings as described in:
• Section 3.2.1, Strategy for Mariner Peak Detection
• Section 3.2.2, Strategy for Voyager Peak Detection
NOTE: If peak detection settings do not detect desired
peaks, you can manually insert peaks. See Section 3.3.2,
Inserting Peaks in the Peak List.
 Loading...
Loading...