Page 1

VINEO™Brettanomytest PCR Kit
Ref.: 354-8101
Instructions
Test to detect and quantify Brettanomyces bruxellensis
using real-time PCR
Page 2

TABLE OF CONTENTS
1 - INTRODUCTION
2 - PRINCIPLE BEHIND THE VINEO™ Brettanomytest PCR Kit
3 - KIT COMPOSITION
4 - VALIDITY AND PRESERVATION
5 - REQUIRED EQUIPMENT THAT IS NOT PROVIDED
6 - PRECAUTIONS AND RECOMMENDATIONS
7 - SAMPLING AND TRANSPORT OF SAMPLES
8 - DNA EXTRACTION
9 - REAL-TIME PCR
10 - ANALYSIS OF THE DATA
11 - INTERPRETATION OF THE RESULTS
12 - CONFIRMATION PROTOCOL FROM ISOLATED COLONIES
13 - TEST PERFORMANCE AND VALIDATIONS
APPENDIX A - PCR AMPLIFICATION REAGENT MIXTURE
PREPARATION TABLE
APPENDIX B - PLATE SETUP FOR THE Chromo4™ OR THE CFX96™
THERMOCYCLERS
APPENDIX C - PLATE SETUP FOR THE MiniOpticon™
THERMOCYCLER
2
© 2010 Bio-Rad
Page 3

1 - INTRODUCTION
Brettanomyces bruxellensis is a type of yeast that is responsible for the
presence of 4-ethyl Phenol and 4-ethylguaiacol in wine-type drinks, fruit
juices and beers that involves significant economic losses. In wines, its
presence is difficult to detect using classic culture methods, which take a
long time and are non-specific.
Rapid, specific and early detection of this yeast in the wine production would,
however, allow the oenologist and the producer to take preventive
measurements and to eliminate it before the appearance of the phenolated
character that is characteristic of changes induced by this yeast. Compared
with the traditional microbiological method, molecular biology provides high
speed, high sensitivity and high specificity detection solutions.
Diagnosis of the presence of this yeast must make it possible to determine at
what risk level the wine is and how this risk develops over time. This risk is
variable according to the contamination level of the wine. Therefore, an
adapted quantification tool is used to determine whether (i) the population is
low and therefore that risk is low or even controlled, (ii) regular monitoring is
required and the wine can be processed, in intermediate or critical quantities
in the event that the wine must be supervised (iii) and finally whether the
population is very high and in the event that the risk of producing volatile
phenols is also high, an immediate and urgent action is required for the wine.
In all cases, regular monitoring is recommended. It is important to monitor
the development Brettanomyces bruxellensis of in the wine.
VINEO™ Brettanomytest PCR Kit is a quantitative test used for specific
detection of Brettanomyces bruxellensis in wines and grape musts in
fermentation using the real-time polymerase chain technique (RT-PCR). A
specific sequence of Brettanomyces bruxellensis is amplified and detected
simultaneously by means of a fluorescent probe. Implementation of this
test is used to obtain a quantitative result less than 3 hours after extracting
the DNA (VINEO™ Extract DNA Kit, 354-8100). The analysis software
adapted to these tests, Opticon Monitor™ or CFX Manager™ Industrial
Diagnostic Edition, make it possible to measure the Brettanomyces
bruxellensis risk in the analysed wine sample by an automatic and
quantitative analysis. An interpretation of the risk level is proposed to the
user in this way. It is linked to the number of CFU.ml-1 (Colony-forming
unit) detected in the wine:
3
© 2010 Bio-Rad
Page 4

• Negative
• Low population, controlled risk
• Critical population to be monitored
• Very high population, risk of production of volatile Phenols
The VINEO™ Brettanomytest PCR Kit can also be used in a qualitative
mode to confirm isolated colonies from a culture medium.
2 - PRINCIPLE BEHIND THE VINEO™ Brettanomytest PCR Kit
This test is based on amplification, detection and quantification of DNA
sequences using the real-time PCR technique. It uses primers and a DNA
probe that are specific to Brettanomyces bruxellensis. Detection and
analysis of the results are optimised for use with a thermocycler for
Bio-Rad real-time PCR such as MiniOpticon™, Chromo4™ or CFX96™.
During the PCR reaction, the primers will be hybridised in the target region
and will then - catalysed by the polymerase - lengthen in the 5'-3'
direction using the desoxynucleotide triphosphate (dNTPs) present in the
reagent mixture, thereby creating a complementary DNA sequence, called
an amplicon.
During PCR, oligonucleotidic probes that are specific to the target
sequence will be hybridised with the amplicons. These probes, which are
marked by fluorophores, only emit fluorescence when hybridisation takes
place. The probe that is linked to the target sequence of Brettanomyces
bruxellensis is marked by a specific fluorophore. Intensity of fluorescence
increases proportionally to the increase in quantity of amplification
products in the PCR tube. The fluorescence that is generated in this way
is directly measured by the optical module of the thermocycler.
A synthetic DNA called the "Internal control" is added to each reaction. It
is amplified at the same time as the target sequences of Brettanomyces
bruxellensis but detected by a probe marked with a second fluorophore. It
is used to bring out any reaction inhibiting phenomenon.
The software associated with the device calculates the relation between
the intensity of the fluorescence and the amplification cycle automatically.
This relation shows the presence or absence of Brettanomyces
bruxellensis in the sample.
The results obtained by amplification of this DNA are analysed by the
Opticon Monitor™ or CFX Manager™ IDE software that have been
programmed to carry out an automatic and quantitative analysis. An
interpretation of the risk linked to the presence of Brettanomyces
bruxellensis is then proposed to the user.
4
© 2010 Bio-Rad
Page 5
This test is used to detect and quantify Brettanomyces bruxellensis in the
DNA extracted from fermented drinks produced from grape and
fermenting grape musts.
3 - KIT CONTENTS
This kit contains the quantity of reagent required for 96 reactions.
4 - VALIDITY AND PRESERVATION
On reception, the kit must be stored at temperatures between +2°C and
+8°C. Each reagent stored between +2°C and +8°C can be used until the
expiry date indicated on the tube.
NB: Do not freeze the reagents.
5 - REQUIRED EQUIPMENT THAT IS NOT PROVIDED
Equipment:
• Industrial Diagnostic CFX96™ real-time PCR detection system, 96
wells, ref. Bio-Rad :359-3990
• CFX Manager™ Software, Industrial Diagnostic Edition, ref. Bio-Rad :
359-3893
• MiniOpticon™ system for real-time PCR, reaction block of 48 wells,
ref. Bio-Rad: 359-3995
• Vortex
• Optionally: centrifuge with rotor for plates or strips (max. 2000 x G)
• 20 µl, 200 µl and 1000 µl micropipettes
• Multi-distributor such as combitip pipettes
Note: We recommend using an uninterruptible power supply with the
thermocycler.
5
© 2010 Bio-Rad
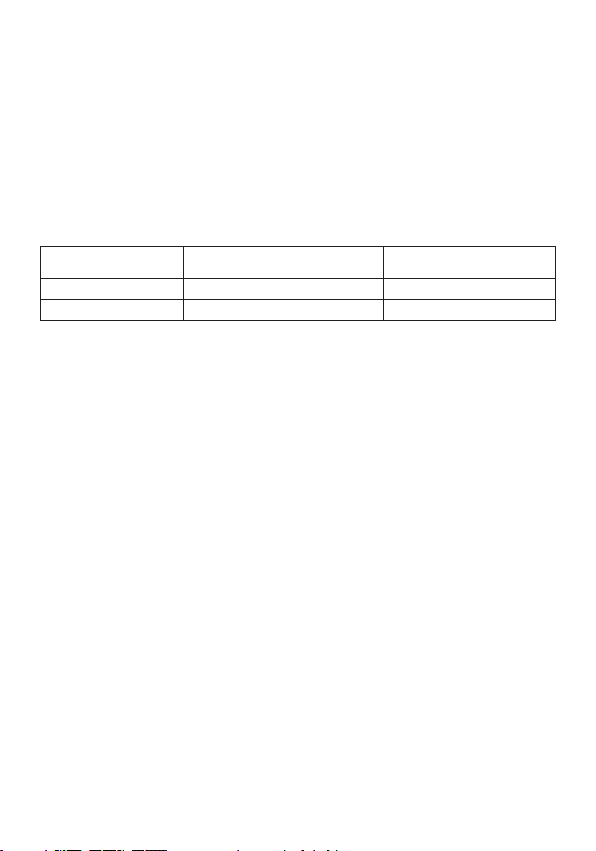
Stoppers Designation Reagent Volume
Pink Fluorescent probes Fluorescent probes 1 tube (0.55 mL)
White Amplification mix Amplification mix 2 tubes (2 x 2.2 mL)
Green Negative PCR control Negative PCR control 1 tube (0.5 mL)
Red Positive Qs PCR control Positive Qs PCR control 1 tube (0.5 mL)
TAKING
THE SAMPLE
DNA EXTRACTION
VINEO™ Extract
DNA Kit
(Ref.: 354-8100)
DNA
AMPLIFICATION by
PCR IN REAL TIME
VINEO™
Brettanomytest PCR
Kit (Ref.: 354-8101)
ANALYSIS &
INTERPRETATION
Opticon Monitor™
or
CFX Manager™ IDE
Page 6

Consumables:
• For the plates, PCR tubes and stop caps:
- White plates of 96 "Low-profile" wells, ref. Bio-Rad: MLL-9651
- White plates of 48 "Low-profile" wells, ref. Bio-Rad: MLL-4851
- Strips of 8 tubes of 200l "Low-profile", ref. Bio-Rad: TLS-0851
- Strips of 8 flat optical stop caps for tubes or plates of 0.2 mL wells,
120 strips ref. Bio-Rad: TCS-0803
• Cones for Combitip or multi-distributor, sterile when packaged
individually
• Cones with sterile filters that are adaptable to 10 µL, 20 µL, 100 µL,
200 µL and 1000 µL micro-pipettes
• 2mL and 5mL sterile tubes
• Non-powdered latex or nitrile gloves
• Sterile distilled water
• 5% Chlorine Bleach
• Decontaminating agent such as DNA AWAY
®
or RNAse AWAY
®
6 - PRECAUTIONS AND RECOMMENDATIONS
• This trial must be carried out by staff who have had adequate
training.
• Result quality depends on scrupulous compliance with Laboratory
Good Practice, in particular in the area of PCR:
- The equipment (pipettes, tubes etc.) must no go from one work
station to another.
- It is indispensable to use a positive and a negative control for each
series of amplification reactions.
- Do not use the reagents beyond their expiry date.
- Subject the reagents in the kit to a vortex movement using the
apparatus provided for this purpose before using them to work
with homogeneous solutions.
- Check the exactness and the precision of the pipettes as well as
the proper functioning of the instruments.
- Change gloves regularly and as soon as you suspect that they may
have been contaminated.
- Clean work surfaces regularly with a 5% solution of chlorine bleach
and another agent such as DNA AWAY
®
or RNase AWAY®.
- Wear non-powdered gloves so as not to leave fingerprints on the
optical film used to seal the micro-plates. Do not write on the PCR
tube stoppers. In both cases, recording the data by the device can
be disturbed.
6
© 2010 Bio-Rad
Page 7

We strongly recommend that you read the complete protocol before
beginning the trial.
Follow the suggested protocol scrupulously.
7 - SAMPLING AND TRANSPORT OF SAMPLES
Wine samples are collected under aseptic conditions in sterile glass or
polyethylene recipients or recipients made out of similar material.
The samples must be submitted to the laboratory as quickly as possible,
preferably within 24 and not more than 48 after taking the sample. If the
samples are transported and analysed within 24 h, transport and storage take
place at ambient temperature (+18°C to +30°C). If the samples are
transported and analysed within 48 h, transport and storage take place at
between +2°C to +8°C.
8 - DNA EXTRACTION
DNA extraction must be carried out using the kit developed by Bio-Rad to
extract DNA from wine sample: VINEO™ Extract DNA Kit (ref. Bio-Rad
354-8100).
9 - REAL-TIME PCR
1. Starting up the PCR apparatus
Switch the thermocyler and the computer on in that order and start up the
Opticon Monitor software. For more information and for the definition of
software parameters, see the user guide for the Bio-Rad thermocycler for the
VINEO™ Brettanomytest PCR Kit.
2. Preparing PCR reactions
2.1 Prepare the VINEO™ Brettanomytest PCR Kit reagent mixture by mixing
(40 µL/sample) the amplification solution (tube with white stopper) and
(5 µL/sample) the fluorescent probes (tube with pink stopper). Carry out
the reagent mixture on the basis of the number of samples and controls
to be analysed (at least 1 duplicate (2) of the positive Qs PCR control and
a negative control must be used per plate). The PCR amplification
reagent mixture preparation table in Appendix A indicates the required
quantities of each reagent to be mixed on the basis of the number of
samples to be analysed. Do not forget to include the 2 control points.
Note: do not mix reagent batches.
7
© 2010 Bio-Rad
Page 8

2.2 After preparation, the reagent mix (amplification and probe solutions)
must be used immediately, or can be stored for 2 hours between +2°C
and +8°C.
2.3 Distribute 45 μl of this reagent mixture per well according to the defined
plate surface. Appendices B and C propose plate surfaces to be used
for the Chromo4™/CFX96™ and MiniOpticon™ thermocyclers
respectively.
Add 5 μl of sample or negative control or Qs PCR positive control and
seal the wells on the plate or the strips hermetically. It is important to
insist on sealing the wells and the strips to avoid any evaporation
phenomenon during the PCR reaction.
It is important to avoid the presence of bubbles at the bottom of the wells
by pipetting cautiously. To eliminate bubbles after sealing the plate or
closing the PCR strips, you may centrifuge the plate of PCR strips briefly.
The plate can be store at +2°C to +8°C for 2 hours.
2.4 Put the plate or the PCR strips into the thermocycler. Make sure that they
are correctly oriented (well A1 at the top left).
Close the reaction module.
3. Starting the amplification reaction
To restart the PCR, refer to the user guide for the Bio-Rad thermocycler for
the VINEO™ Brettanomytest PCR Kit.
10 - ANALYSIS OF THE DATA
Data analysis can be carried out directly at the end of the amplification
reaction or later by re-opening the data file. To open the data files and
analyse the results of the PCR, refer to the user guide for the Bio-Rad
thermocycler for the VINEO™ Brettanomytest PCR Kit.
11 - INTERPRETATION OF THE RESULTS
To obtain the analysis results, you only have to read the values of Ct (Cycle
threshold): value of the amplification cycle from which fluorescence increases
significantly above background noise.
Manual analysis will only be used for a qualitative analysis (presence or
absence).
8
© 2010 Bio-Rad
Page 9
You must use the the Opticon Monitor™ software (version 3.3 and higher)
or the CFX Manager™ software Industrial Diagnostic Edition for an
automated and quantitative interpretation. A complete report may be printed
(cf. software instructions).
1. Controls
Before a final interpretation of the results, you have to check the results of the
negative and positive controls.
For the test to be valid, the results of the negative and positive controls must
be as follows:
* N/A means “Not Applicable”. The software indicates N/A for the Ct of a sample when the
fluorescence curve does not cross the threshold.
If the results of the positive and negative PCR Qs controls are different from
those described in the table below, you have to begin the PCR again.
2. Samples
A sample is considered to be positive for Brettanomyces bruxellensis if the
value of Ct ≥ 10 is obtained for the FAM fluorophore.
If no value is obtained for Ct
FAM
(Ct
FAM
= N/A), the interpretation of the result
depends on the value of the internal control:
- A sample is considered to be negative for Brettanomyces bruxellensis if
no value of Ct is obtained for the FAM fluorophore (Ct
FAM
= N/A and if the
Ct of the internal control is greater than or equal to 28. (Ct
HEX
≥ 28).
- An N/A value for the Ct
HEX
of the internal control indicates that an
inhibition phenomenon of the PCR reaction has probably occurred, if
the Ct
FAM
of the target is also N/A. In this case, the DNA sample must
be diluted (to 1/10
th
for example) in sterile distilled water and then
subjected to a new PCR.
- If the Ct
HEX
of the internal control is less than 28, it is impossible to
interpret the result. Check that the threshold was set correctly or that
the raw curve shows an aspect characteristic of exponential
amplification. If the observed curve is not correct, it will be necessary to
repeat the PCR test for this sample.
9
© 2010 Bio-Rad
Detection of Brettanomyces
bruxellensis (FAM)
Detection of the internal
control (Channel HEX)
Negative control Ct = N/A* 25 ≤ Ct ≤ 40
Positive Qs PCR control 27 ≤ Ct ≤ 37 Not significant
Page 10

Interpretation of results obtained for the samples:
**: In the event of a null value for a sample and its internal control (Ct = N/A), the analysis must be done
again with the diluted DNA sample (diluted to 1/5thor to 1/10th for example).
***: A value less than 10 may be obtained; check whether the curve as raw data shows an aspect that
is characteristic of exponential amplification (flat base line and regular increase in fluorescence followed
by a plateau). If the observed curve is correct, the sample for the presence of Brettanomyces
bruxellensis may be considered to be positive. Otherwise, the interpretation of the result depends on
the value of the internal control, as is explained in the previous paragraphs.
12 - CONFIRMATION PROTOCOL FROM ISOLATED COLONIES
You can use the VINEO™ Brettanomytest PCR Kit to confirm isolated
colonies on agarose gel.
1. Puncture an isolated colony, whether it comes from a selective medium or
not, using a toothpick or a 1 µl piercing instrument, or any other
consumable adapted to this purpose (a pipette cone for example).
2. Resuspend the colony in 100 l of R2 in an Eppendorf tube (it is possible
to resuspend the colony in steril physiological water, PCR efficiency can
then be affected). Homogenise the suspension by means of a vortex.
3. Put 5 µl of suspension in 45 µl of PCR reagent mixture (cf. part 9.2
Real-time PCR) and follow the rest of the VINEO™ Brettanomytest PCR
Kit method to obtain and interpret results. Note: only a qualitative
analysis is made from the PCR carried out on this suspension. The
signal obtained by PCR is often quite high because the quantity of cells
sampled and analysed by PCR using this method is quite significant.
13 - TEST PERFORMANCE AND VALIDATIONS
The VINEO™ Brettanomytest PCR Kit is specific for detecting
Brettanomyces bruxellensis. You can detect the yeast from:
• 500 CFU/mL of wine using the DNA extraction protocol described in
the VINEO™ Extract DNA Kit: for 1.8 mL for fermenting grape must or
a heavy wine during the wine-making or maturing process and
• 10 CFU/mL of wine according to the protocol starting from 45 mL for
slightly heavy wine, close to bottling or already bottled, the analysis of
which requires a high level of analytic sensitivity.
10
© 2010 Bio-Rad
Detection of Brettanomyces
bruxellensis (FAM)
Detection of the internal
control
Interpretation
Ct ≥ 10*** Not significant Positive
Ct = N/A
Ct
HEX
≥ 28
Negative
Ct = N/A Ct = N/A Inhibition**
Page 11
11
© 2010 Bio-Rad
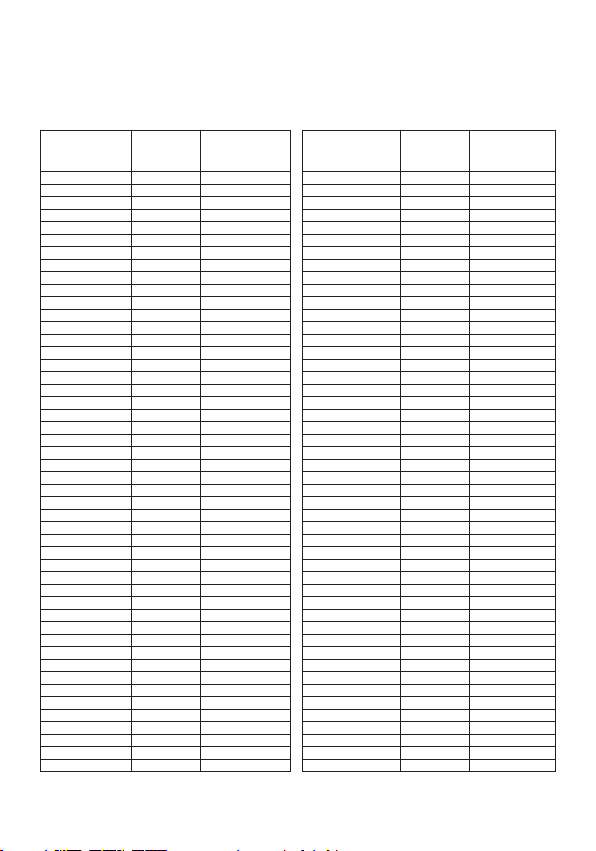
APPENDIX A - PCR AMPLIFICATION REAGENT MIXTURE PREPARATION TABLE
Use this table to determine the required quantities of fluorescent probes and amplification
solution to prepare the PCR reagent mixture. Calculation of the number of samples
must include the 2 positive Qs PCR controls and the negative control.
Number
of samples
Fluorescent
probes (μL)
(Pink stopper)
Amplification
solution (μL)
(White stopper)
Number
of samples
Fluorescent
probes (μL)
(Pink stopper)
Amplification
solution (μL)
(White stopper)
1 5 40 49 265 2117
2 11 86 50 270 2160
3 16 130 51 275 2203
4 22 173 52 281 2246
5 27 216 53 286 2290
6 32 259 54 292 2333
7 38 302 55 297 2376
8 43 346 56 302 2419
9 49 389 57 308 2462
10 54 432 58 313 2506
11 59 475 59 319 2549
12 65 518 60 324 2592
13 70 562 61 329 2635
14 76 605 62 335 2678
15 81 648 63 340 2722
16 86 691 64 346 2765
17 92 734 65 351 2808
18 97 778 66 356 2851
19 103 821 67 362 2894
20 108 864 68 367 2938
21 113 907 69 373 2981
22 119 950 70 378 3024
23 124 994 71 383 3067
24 130 1037 72 389 3110
25 135 1080 73 394 3154
26 140 1123 74 400 3197
27 146 1166 75 405 3240
28 151 1210 76 410 3283
29 157 1253 77 416 3326
30 162 1296 78 421 3370
31 167 1339 79 427 3413
32 173 1382 80 432 3456
33 178 1426 81 437 3499
34 184 1469 82 443 3542
35 189 1512 83 448 3586
36 194 1555 84 454 3629
37 200 1598 85 459 3672
38 205 1642 86 464 3715
39 211 1685 87 470 3758
40 216 1728 88 475 3802
41 221 1771 89 481 3845
42 227 1814 90 486 3888
43 232 1858 91 491 3931
44 238 1901 92 497 3974
45 243 1944 93 502 4018
46 248 1987 94 508 4061
47 254 2030 95 513 4104
48 259 2074 96 518 4147
Page 12

12
© 2010 Bio-Rad
APPENDIX B
PLATE SETUP FOR THE Chromo4™/CFX96™ THERMOCYCLERS
The first 3 wells in column 1 may be used to analyse the 3 control points. The other
wells in the plate will be used for the samples to be analysed.
NC: Negative PCR control
QS: Positive Qs PCR control
APPENDIX C
PLATE SETUP FOR THE MiniOpticon™ THERMOCYCLER
The first 3 wells in column 1 may be used to analyse the 3 control points. The other
wells in the plate will be used for the samples to be analysed.
NC: Negative PCR control
QS: Positive Qs PCR control
1 2 3 4 5 6 7 8 9 10 11 12
A QS
B QS
C NC
D Sample
E Sample
F …
G
H
1 2 3 4 5 6
A QS
B QS
C NC
D Sample
E Sample
F …
G
H
Page 13

13
© 2010 Bio-Rad
NOTICE OF CONCERN TO THE PURCHASER: LIMITED LICENSE
Use of this product is covered by one or more of the following US patents and the corresponding patent
claims outside the United States: 5 079 352, 5 789 224, 5 618 711, 6 127 155, 5 677 152 (claims 1 to
23 only) and 5 773 258 (claims 1 and 6 only) and by the claims outside the United States that correspond
to US patent n° 4 889 818. The purchase of this product includes limited immunity from legal proceedings
that may not be transferred according to the claims of previous patents when this quantity of product is
solely used for alimentary analysis, environmental and industrial microbiology purposes including
publication of the results of the purchaser's activities in return for payment or other commercial
counterpart, and when it is also used for the purchaser's own research purposes. No right according to
any patent claim (such as the claims of method 5'-nuclease in US patents n° 5 210 015 and 5 487 972)
is not expressly transferred whether by implication or preclusion. Further information on the purchase of
licenses may be obtained by contacting the Director of Licenses, Applied Biosystems, 850 Lincoln Centre
Drive, Foster City, California 94404, United States.
Page 14

14
© 2010 Bio-Rad
Page 15

VINEO™Brettanomytest PCR Kit
Réf. : 354-8101
Notice d’utilisation
Test pour la détection et quantification par PCR
en temps réel de Brettanomyces bruxellensis
15
© 2010 Bio-Rad
Page 16

SOMMAIRE
1 - INTRODUCTION
2 - PRINCIPE DU VINEO™ Brettanomytest PCR Kit
3 - COMPOSITION DU KIT
4 - VALIDITE ET CONSERVATION
5 - EQUIPEMENT ET MATERIEL NECESSAIRES NON FOURNIS
6 - PRECAUTIONS ET RECOMMANDATIONS
7 - PRELEVEMENT ET TRANSPORT DES ECHANTILLONS
8 - EXTRACTION DE L’ADN
9 - PCR EN TEMPS REEL
10 - ANALYSE DES DONNEES
11 - INTERPRETATION DES RESULTATS
12 - PROTOCOLE DE CONFIRMATION A PARTIR DE COLONIES ISOLEES
13 - PERFORMANCES DU TEST ET VALIDATIONS
ANNEXE A - TABLEAU DE PREPARATION DU MELANGE
REACTIONNEL D’AMPLIFICATION PCR
ANNEXE B - PLAN DE PLAQUE A UTILISER SUR LES
THERMOCYCLEURS Chromo4™ OU CFX96™
ANNEXE C - PLAN DE PLAQUE A UTILISER SUR LE
THERMOCYCLEUR MiniOpticon™
16
© 2010 Bio-Rad
Page 17

1 - INTRODUCTION
Brettanomyces bruxellensis est une levure responsable de la présence
des 4-ethyl Phénol et 4-ethyl Guaiacol dans les boissons de type vins, jus
de fruits et bières impliquant des pertes économiques importantes. Dans
les vins, sa présence est difficile à détecter par les méthodes de culture
classiques qui sont longues et non spécifiques.
La détection rapide, spécifique et précoce de cette levure dans le
processus d’élaboration des vins permettrait cependant à l’œnologue et
au producteur de prendre des mesures de prévention et de l’éliminer
avant l’apparition du caractère phénolé, caractéristique de l’altération par
cette levure. Face à la méthode microbiologique traditionnelle, la biologie
moléculaire apporte des solutions de détection à de hauts niveaux de
rapidité, de sensibilité et de spécificité.
Le diagnostic de la présence de cette levure doit permettre de déterminer à
quel niveau de risque se trouve le vin et comment évolue ce risque dans le
temps. Ce risque est variable selon le niveau de contamination du vin. Ainsi
un outil de quantification adapté permet de déterminer si (i) la population est
en faible quantité et donc que le risque est faible voire maîtrisé, (ii) en quantité
intermédiaire ou critique, dans ce cas le vin doit être surveillé, un suivi régulier
s’impose, le vin peut être traité (iii) et enfin si la population est très importante,
et dans ce cas le risque de production des phénols volatils l’est aussi, une
action immédiate et urgente s’impose sur le vin.
Dans tous les cas un suivi régulier est préconisé. Il est important de suivre
l’évolution des Brettanomyces bruxellensis au sein du vin.
VINEO™ Brettanomytest PCR Kit est un test quantitatif permettant la
détection spécifique de Brettanomyces bruxellensis dans les vins et les
moûts en fermentation par la technique de polymérisation en chaîne en
temps réel (RT-PCR). Une séquence d’ADN spécifique de Brettanomyces
bruxellensis est amplifiée et détectée simultanément grâce à une sonde
fluorescente. La mise en œuvre de ce test permet l'obtention d'un résultat
quantitatif moins de 3h après l’extraction d’ADN (VINEO™ Extract DNA Kit,
354-8100). Les logiciels d’analyse adaptés à ce test, Opticon Monitor™ ou
CFX Manager™ Industrial Diagnostic Edition, permettent par une analyse
automatique et quantitative, de mesurer le risque Brettanomyces
bruxellensis dans l’échantillon de vin analysé. Une interprétation du niveau
de risque est ainsi proposée à l’utilisateur. Elle est liée au nombre
d’UFC.mL-1 (Unité Formant Colonie) détecté dans le vin :
17
© 2010 Bio-Rad
Page 18

• Négatif
• Population faible, risque maîtrisé
• Population critique à surveiller
• Population très élevée, risque de production de Phénols volatils
Le VINEO™ Brettanomytest PCR Kit peut également être utilisé sur un
mode qualitatif pour la confirmation de colonies isolées à partir d’un milieu
de culture.
2 - PRINCIPE DU VINEO™ Brettanomytest PCR Kit
Ce test repose sur l’amplification, la détection et la quantification de
séquences d’ADN par la technique de PCR en temps réel. Il utilise des
amorces et une sonde d’ADN spécifiques de Brettanomyces bruxellensis.
La détection et l’analyse des résultats sont optimisées pour l’utilisation
avec un thermocycleur pour la PCR en temps réel Bio-Rad, tel que le
MiniOpticon™, le Chromo4™ ou le CFX96™.
Au cours de la réaction de PCR, les amorces vont s’hybrider à la région
cible puis, catalysées par la polymérase, s’allonger dans le sens 5’-3’ en
utilisant les désoxynucléotides triphosphates (dNTPs) présents dans le
mélange réactionnel, créant ainsi une séquence complémentaire d’ADN,
appelée amplicon.
Pendant la PCR, des sondes oligonucléotidiques, spécifiques de la
séquence cible, vont s’hybrider aux amplicons. Ces sondes, marquées
par des fluorophores, émettent de la fluorescence uniquement quand
l’hybridation a lieu. La sonde qui se lie à la séquence cible de
Brettanomyces bruxellensis est marquée par un fluorophore spécifique.
L’intensité de fluorescence augmente proportionnellement à
l’augmentation de la quantité des produits d’amplification dans le tube de
PCR. La fluorescence ainsi générée est mesurée directement par le
module optique du thermocycleur.
Un ADN synthétique appelé « Contrôle interne » est ajouté à chaque
réaction. Il est amplifié en même temps que les séquences cibles de
Brettanomyces bruxellensis, mais détecté par une sonde marquée avec
un deuxième fluorophore. Il permet de mettre en évidence un éventuel
phénomène d’inhibition de la réaction.
Le logiciel associé à l’appareil calcule automatiquement la relation entre
l’intensité de la fluorescence et le cycle d’amplification. Cette relation indique
la présence, ou l’absence, de Brettanomyces bruxellensis dans l’échantillon.
Les résultats obtenus pour l’amplification de ces ADN sont analysés par les
logiciels Opticon Monitor™ ou CFX Manager™ IDE software programmés
18
© 2010 Bio-Rad
Page 19

pour réaliser une analyse automatique et quantitative. Une interprétation
concernant le risque lié à la présence de Brettanomyces bruxellensis est
alors proposée à l’utilisateur.
Ce test permet la détection et la quantification de Brettanomyces
bruxellensis dans les ADN extraits à partir de boissons fermentées issues
de raisin et des moûts en fermentation.
3 - COMPOSITION DU KIT
Ce kit contient la quantité de réactif nécessaire pour 96 réactions.
4 - VALIDITE ET CONSERVATION
Dès réception, le kit doit être conservé entre +2°C et +8°C. Chaque réactif
conservé entre +2°C et +8°C peut être utilisé jusqu'à la date de
péremption indiquée sur le tube.
NB : Ne pas congeler les réactifs.
5 - EQUIPEMENT ET MATERIEL NECESSAIRES NON FOURNIS
Equipements :
• Industrial Diagnostic CFX96™ pour la PCR en temps réel, bloc
réactionnel de 96 puits, réf. Bio-Rad: 359-3990
• CFX Manager™ Software, Industrial Diagnostic Edition, réf. Bio-Rad :
359-3893
• Système MiniOpticon™ pour la PCR en temps réel, bloc réactionnel
de 48 puits, réf. Bio-Rad: 359-3995
• Vortex
• En option: centrifugeuse avec rotor pour plaque ou barrette (max.
2000 x G)
19
© 2010 Bio-Rad
PRÉLÈVEMENT DE
L’ÉCHANTILLON
EXTRACTION ADN
VINEO™ Extract
DNA Kit
(Réf. : 354-8100)
AMPLIFICATION ADN
par PCR EN TEMPS
REEL
VINEO™
Brettanomytest PCR
Kit (Réf. : 354-8101)
ANALYSE &
INTERPRETATION
Opticon Monitor™
ou
CFX Manager™ IDE
Bouchons Désignation Réactif Volume
Rose Fluorescent probes Sondes fluorescentes 1 tube (0,55 mL)
Blanc Amplification mix Solution d’amplification 2 tubes (2 x 2,2 mL)
Vert Negative PCR control Contrôle négatif PCR 1 tube (0,5 mL)
Rouge Positive Qs PCR control Contrôle positif Qs PCR 1 tube (0,5 mL)
Page 20

• 20 µl, 200 µl and 1000 µl micropipettes
• Multi-distributeur, tel que combitip pipettes
Remarque : Nous recommandons l’utilisation d’un onduleur électrique
avec le thermocycleur.
Consommables :
• Pour les plaques, tubes PCR et capuchons :
- Plaques blanches de 96 puits “Low-profile”, réf. Bio-Rad: MLL-9651
- Plaques blanches de 48 puits “Low-profile”, réf. Bio-Rad: MLL-4851
- Barrettes de 8 tubes de 200 l “Low-profile”, réf. Bio-Rad: TLS-0851
- Barrettes de 8 capuchons optiques plats, pour tubes ou plaques de
puits de 0,2 mL, 120 barrettes Réf. Bio-Rad : TCS-0803
• Cônes pour Combitip ou multi-distributeur, stériles à emballage individuel
• Cônes à filtre stériles, adaptables sur les micropipettes de 10 µl,
20 µl, 100 µl, 200 µl et 1000 µl
• Tubes stériles de 2 mL et de 5 mL
• Gants non-poudrés latex ou nitrile
• Eau distillée stérile
• Eau de javel 5%
• Agent décontaminant tel que DNA AWAY
®
ou RNAse AWAY
®
6 - PRECAUTIONS ET RECOMMANDATIONS
• Cet essai doit être réalisé par des personnes ayant reçu une formation
adéquate.
• La qualité des résultats dépend du respect scrupuleux des Bonnes
Pratiques de Laboratoire, en particulier en matière de PCR :
- Le matériel (pipettes, tubes etc...) ne doit pas circuler d'un poste de
travail à l'autre.
- Il est indispensable d'utiliser un contrôle positif et un contrôle négatif
pour chaque série de réactions d'amplification.
- Ne pas utiliser les réactifs au-delà de leur date de péremption.
- Vortexer les réactifs du kit avant leur utilisation pour travailler avec
des solutions homogènes.
- Vérifier l'exactitude et la précision des pipettes ainsi que le bon
fonctionnement des instruments.
- Changer de gants régulièrement et dès que vous soupçonnez qu’ils
peuvent être contaminés.
- Nettoyer les surfaces de travail régulièrement avec de l’eau de javel
5% et autre agent tel que DNA AWAY
®
ou RNase AWAY®.
- Porter des gants non-poudrés pour ne pas laisser des traces de
20
© 2010 Bio-Rad
Page 21

doigts sur le film optique utilisé pour sceller les microplaques. Ne pas
écrire sur les bouchons des tubes PCR. Dans les deux cas,
l’enregistrement des données par l’appareil peut être perturbé.
Nous vous recommandons vivement de lire l'ensemble du protocole
avant de commencer l'essai.
Respecter scrupuleusement le protocole proposé.
7 - PRELEVEMENT ET TRANSPORT DES ECHANTILLONS
Les échantillons de vins sont collectés, en conditions aseptiques, dans des
récipients stériles en verre, en polyéthylène ou en matériel similaire.
Les échantillons doivent être remis au laboratoire le plus rapidement possible,
de préférence dans les 24 heures et pas plus de 48 heures après le
prélèvement. Si les échantillons sont transportés et analysés dans les 24
heures, le transport et le stockage ont lieu à température ambiante (+18°C à
+30°C). Si les échantillons sont transportés et analysés dans les 48 heures, le
transport et le stockage ont lieu entre +2°C et +8°C.
8 - EXTRACTION DE L’ADN
L’extraction de l’ADN doit être réalisée en utilisant le kit développé par
Bio-Rad pour l’extraction d’ADN à partir d’échantillons de vin VINEO™
Extract DNA Kit (réf. Bio-Rad 354-8100).
9 - PCR EN TEMPS REEL
1. Mise en marche appareil PCR
Allumer dans l’ordre le thermocycleur, l’ordinateur puis ouvrir le logiciel
Opticon Monitor. Pour plus d’informations et pour la définition des
paramètres du logiciel consulter le manuel utilisateur du thermocycleur
Bio-Rad pour le kit VINEO™ Brettanomytest PCR Kit.
2. Préparation des réactions PCR
2.1 Préparer le mélange réactionnel VINEO™ Brettanomytest PCR Kit en
mélangeant (40 µL/échantillon) de la solution d’amplification (tube à
bouchon blanc) et (5 µL/échantillon) des sondes fluorescentes (tube à
bouchon rose). Réaliser le mélange réactionnel en fonction du nombre
d’échantillons et des contrôles à analyser (au minimum 1 duplicat (2) du
contrôle positif Qs PCR et un contrôle négatif doivent être utilisés par
plaque). Le tableau de préparation du mélange réactionnel d’amplification
PCR présent en Annexe A indique les quantités nécessaires de chaque
réactif à mélanger en fonction du nombre d’échantillons à analyser. Ne
pas oublier de comptabiliser les 3 points de contrôles.
Note : ne pas mélanger les lots de réactifs.
21
© 2010 Bio-Rad
Page 22

2.2 Après préparation, le mélange réactionnel (Solutions d’amplification et de
sondes) doit être utilisé immédiatement, ou il peut être conservé pendant
2 heures de +2°C à +8 °C.
2.3 Répartir 45 μl de ce mélange réactionnel par puits, selon le plan de plaque
défini. Les Annexes B et C proposent des plans de plaque à utiliser
respectivement sur les thermocycleurs Chromo4™/CFX96™ et
MiniOpticon™.
Ajouter 5 μl d’échantillon ou de contrôle négatif ou de contrôle positif Qs
PCR et sceller de façon hermétique les puits de la plaque ou des
barrettes. Il est important d’insister sur la fermeture des puits et des
barrettes pour éviter tout phénomène d’évaporation au cours de la
réaction de PCR.
Il est important d’éviter la présence de bulles au fond des puits en
pipetant précautionneusement. Pour éliminer des bulles, après avoir
scellé la plaque ou avoir fermé les barrettes PCR, il est possible de
centrifuger brièvement la plaque ou les barrettes PCR. La plaque peut
être conservée pendant 2 heures de +2°C à +8 °C.
2.4 Placer la plaque ou les barrettes PCR dans le thermocycleur. S’assurer
de leur bonne orientation (puits A1 en haut à gauche).
Fermer le module réactionnel.
3. Lancement de la réaction d’amplification
Pour le lancement de la PCR, consulter le manuel utilisateur du
thermocycleur Bio-Rad pour le kit VINEO™ Brettanomytest PCR Kit.
10 - ANALYSE DES DONNEES
L’analyse des données peut être réalisée directement à la fin de la réaction
d’amplification ou ultérieurement en ré-ouvrant le fichier de données. Pour
ouvrir des fichiers de données et analyser les résultats de la PCR, consulter le
manuel utilisateur du thermocycleur Bio-Rad pour le kit VINEO™
Brettanomytest PCR Kit.
11 - INTERPRETATION DES RESULTATS
Pour obtenir les résultats de l’analyse, il suffit de lire les valeurs de Ct (Cycle
treshold) : valeur du cycle d’amplification à partir duquel la fluorescence
s’élève significativement au-dessus du bruit de fond.
L’analyse manuelle permettra uniquement une analyse qualitative (présence
ou absence).
22
© 2010 Bio-Rad
Page 23

Il est nécessaire d’utiliser les logiciels Opticon Monitor™ (version 3.3 et plus)
ou CFX Manager™ Industrial Diagnostic Edition pour une interprétation
automatisée et quantitative. Un rapport complet pourra être imprimé (Cf.
notice d’utilisation des logiciels).
1. Contrôles
Avant l’interprétation finale des résultats il est nécessaire de vérifier les
résultats des contrôles négatif et positif.
Pour que le test soit valide, les résultats des contrôles négatif et positif
doivent être les suivants :
* N/A signifie « Non Applicable ». Le logiciel indique N/A pour le Ct d’un échantillon quand la courbe de
fluorescence ne croise pas le seuil.
Si les résultats des contrôles positif Qs PCR et négatif sont différents de ceux
décrits dans le tableau ci-dessus, il est nécessaire de recommencer la PCR.
2. Echantillons
Un échantillon est considéré positif pour Brettanomyces bruxellensis si une
valeur de Ct ≥ 10 est obtenue pour le fluorophore FAM.
Si aucune valeur n’est obtenue pour le Ct
FAM
(Ct
FAM
= N/A), l’interprétation
du résultat dépend de la valeur du contrôle interne :
- Un échantillon est considéré négatif pour Brettanomyces bruxellensis si
aucune valeur de Ct n’est obtenue pour le fluorophore FAM (Ct
FAM
=N/A)
et si le Ct du contrôle interne est supérieur ou égal à 28. (Ct
HEX
≥ 28).
- Une valeur N/A pour le Ct
HEX
du contrôle interne indique, lorsque le
Ct
FAM
de la cible est également N/A, qu’un phénomène d’inhibition de
la réaction de PCR a probablement eu lieu. Dans ce cas, l’échantillon
d’ADN doit être dilué (au 1/10
e
par exemple) en eau distillée stérile, puis
soumis à une nouvelle PCR.
- Si le Ct
HEX
du contrôle interne est inférieur à 28 il n’est pas possible
d’interpréter le résultat. Vérifier que le seuil a été correctement placé ou
que la courbe brute montre un aspect caractéristique d’amplification
exponentielle. Si la courbe observée n’est pas correcte il sera
nécessaire de répéter le test PCR pour cet échantillon.
23
© 2010 Bio-Rad
Détection de Brettanomyces
bruxellensis (FAM)
Détection du contrôle interne
(Canal HEX)
Contrôle négatif Ct = N/A* 25 ≤ Ct ≤ 40
Contrôle positif Qs PCR 27 ≤ Ct ≤ 37 Non significatif
Page 24

Interprétation des résultats obtenus sur les échantillons :
** : En cas de valeur nulle pour un échantillon et son contrôle interne (Ct = N/A), l’analyse doit être
renouvelée avec l’échantillon d’ADN dilué (au 1/5
e
ou au 1/10epar exemple).
*** : Il peut arriver qu’une valeur inférieure à 10 soit obtenue, vérifier que la courbe en tant que donnée
brute montre un aspect caractéristique d’amplification exponentielle (ligne de base plate, puis
augmentation régulière de la fluorescence, suivi par un plateau). Si la courbe observée est correcte, on
peut considérer l’échantillon positif pour la présence de Brettanomyces bruxellensis. Dans le cas
contraire, l’interprétation du résultat dépend de la valeur du contrôle interne, comme il est expliqué
dans les paragraphes ci-dessus.
12 - PROTOCOLE DE CONFIRMATION A PARTIR DE COLONIES ISOLÉES
Il est possible d’utiliser le test VINEO™ Brettanomytest PCR Kit pour la
confirmation de colonies isolées sur gélose.
1. Piquer une colonie isolée, à partir d’un milieu sélectif ou non, à l’aide d’un
cure-dent ou d’une œse de 1 µl, ou autre consommable adapté (cône
pipette par exemple).
2. Resuspendre la colonie dans 100 µl de R2 dans un tube de type Eppendorf
(il est possible de resuspendre la colonie dans de l'eau physiologique stérile,
l'efficacité PCR peut alors être affectée). Bien homogénéiser la suspension à
l’aide d’un vortex.
3. Déposer 5 µl de la suspension dans 45 µl du mélange réactionnel PCR (Cf.
partie 9.2 PCR en temps réel) et suivre le reste de la méthode VINEO™
Brettanomytest PCR Kit pour l’obtention et l’interprétation des résultats.
Note : seule une analyse qualitative est réalisée à partir de la PCR
effectuée sur cette suspension. Le signal obtenu en PCR est souvent
très élevé car la quantité de cellules prélevées et analysées par PCR
avec cette méthode est très importante.
13 - PERFORMANCES DU TEST ET VALIDATIONS
Le test VINEO™ Brettanomytest PCR Kit est spécifique pour la détection de
Brettanomyces bruxellensis. Il est possible de détecter la levure à partir de :
• 500 CFU/mL de vin avec le protocole d’extraction d’ADN décrit dans le
VINEO™ Extract DNA Kit : sur 1,8 mL pour un moût en fermentation
ou un vin chargé en cours de vinification ou d’élevage et
• 10 CFU/mL de vin selon le protocole partant de 45 mL pour un vin peu
chargé, proche de l’embouteillage ou déjà mis en bouteille dont
l’analyse nécessite un niveau de sensibilité analytique élevé.
24
© 2010 Bio-Rad
Détection de Brettanomyces
bruxellensis (FAM)
Détection du contrôle interne Interprétation
Ct ≥ 10*** Non significatif Positif
Ct = N/A
Ct
HEX
≥ 28
Négatif
Ct = N/A Ct = N/A Inhibition**
Page 25

25
© 2010 Bio-Rad
ANNEXE A - TABLEAU DE PREPARATION DU MELANGE REACTIONNEL
D’AMPLIFICATION PCR
Utiliser ce tableau pour déterminer les quantités nécessaires de sondes fluorescentes et
solution d’amplification pour préparer le mélange réactionnel de PCR. Le calcul du
nombre d’échantillons doit inclure les 2 contrôles positifs Qs PCR et le contrôle
négatif.
Nombre
d'échantillons
Sondes
fluorescentes
(μL)
(Bouchon rose)
Solution
d’amplification (μL)
(Bouchon blanc)
Nombre
d'échantillons
Sondes
fluorescentes
(μL)
(Bouchon rose)
Solution
d’amplification (μL)
(Bouchon blanc)
1 5 40 49 265 2117
2 11 86 50 270 2160
3 16 130 51 275 2203
4 22 173 52 281 2246
5 27 216 53 286 2290
6 32 259 54 292 2333
7 38 302 55 297 2376
8 43 346 56 302 2419
9 49 389 57 308 2462
10 54 432 58 313 2506
11 59 475 59 319 2549
12 65 518 60 324 2592
13 70 562 61 329 2635
14 76 605 62 335 2678
15 81 648 63 340 2722
16 86 691 64 346 2765
17 92 734 65 351 2808
18 97 778 66 356 2851
19 103 821 67 362 2894
20 108 864 68 367 2938
21 113 907 69 373 2981
22 119 950 70 378 3024
23 124 994 71 383 3067
24 130 1037 72 389 3110
25 135 1080 73 394 3154
26 140 1123 74 400 3197
27 146 1166 75 405 3240
28 151 1210 76 410 3283
29 157 1253 77 416 3326
30 162 1296 78 421 3370
31 167 1339 79 427 3413
32 173 1382 80 432 3456
33 178 1426 81 437 3499
34 184 1469 82 443 3542
35 189 1512 83 448 3586
36 194 1555 84 454 3629
37 200 1598 85 459 3672
38 205 1642 86 464 3715
39 211 1685 87 470 3758
40 216 1728 88 475 3802
41 221 1771 89 481 3845
42 227 1814 90 486 3888
43 232 1858 91 491 3931
44 238 1901 92 497 3974
45 243 1944 93 502 4018
46 248 1987 94 508 4061
47 254 2030 95 513 4104
48 259 2074 96 518 4147
Page 26

26
© 2010 Bio-Rad
ANNEXE B
PLAN DE PLAQUE A UTILISER SUR LES THERMOCYCLEURS
Chromo4™/CFX96™
Les 3 premiers puits de la colonne 1 peuvent être utilisés pour analyser les 3 points de
contrôles. Les autres puits de la plaque seront utilisés pour déposer les échantillons à
analyser.
NC : Contrôle négatif PCR
QS : Contrôle positif Qs PCR
ANNEXE C
PLAN DE PLAQUE A UTILISER SUR LE THERMOCYCLEUR MiniOpticon™
Les 3 premiers puits de la colonne 1 peuvent être utilisés pour analyser les 3 points de
contrôles. Les autres puits de la plaque seront utilisés pour déposer les échantillons à
analyser.
NC : Contrôle négatif PCR
QS : Contrôle positif Qs PCR
1 2 3 4 5 6 7 8 9 10 11 12
A QS
B QS
C NC
D Echantillons
E Echantillons
F …
G
H
1 2 3 4 5 6
A QS
B QS
C NC
D Echantillons
E Echantillons
F …
G
H
Page 27

27
© 2010 Bio-Rad
AVIS CONCERNANT L'ACHETEUR : LICENCE LIMITEE
L’utilisation de ce produit est couverte par un ou plusieurs des brevets US suivants et les revendications
de brevet correspondantes hors Etats-Unis : 5 079 352, 5 789 224, 5 618 711, 6 127 155, 5 677 152
(revendications 1 à 23 seulement) et 5 773 258 (revendications 1 et 6 seulement) et par les revendications
hors Etats-Unis correspondant au brevet US n° 4 889 818. L’achat de ce produit comprend une
immunité de poursuite restreinte et non transférable selon les revendications de brevet précédentes
lorsque cette quantité de produit est uniquement utilisée à des fins d’analyse alimentaire, d’analyse
environnementale et en microbiologie industrielle, y compris la publication des résultats des activités de
l’acheteur moyennant paiement ou toute autre contrepartie commerciale, et lorsqu’elle est également
utilisée aux fins des propres recherches de l’acheteur. Aucun droit selon une quelconque revendication de
brevet (comme les revendications de la méthode 5’-nucléase dans les brevets US n° 5 210 015 et 5 487
972) n’est expressément cédé, que ce soit par implication ou par préclusion. De plus amples informations
concernant l’achat de licences peuvent être obtenues en contactant le Directeur des Licences, Applied
Biosystems, 850 Lincoln Centre Drive, Foster City, Californie, 94404, Etats-Unis.
Page 28

28
© 2010 Bio-Rad
Page 29

VINEO™Brettanomytest PCR Kit
Ref.: 354-8101
Instrucciones de uso
Prueba para la detección y cuantificación mediante
PCR en tiempo real de Brettanomyces bruxellensis
29
© 2010 Bio-Rad
Page 30

ÍNDICE
1 - INTRODUCCIÓN
2 - PRINCIPIO DEL Kit VINEO™ Brettanomytest PCR Kit
3 - COMPOSICIÓN DEL KIT
4 - CADUCIDAD Y CONSERVACIÓN
5 - EQUIPO Y MATERIAL NECESARIOS NO SUMINISTRADOS
6 - PRECAUCIONES Y RECOMENDACIONES
7 - EXTRACCIÓN Y TRANSPORTE DE LAS MUESTRAS
8 - EXTRACCIÓN DEL ADN
9 - PCR EN TIEMPO REAL
10 - ANÁLISIS DE DATOS
11 - INTERPRETACIÓN DE LOS RESULTADOS
12 - PROTOCOLO DE CONFIRMACIÓN A PARTIR DE COLONIAS
AISLADAS
13 - RENDIMIENTOS DE LA PRUEBA Y VALIDACIONES
ANEXO A. TABLA DE PREPARACIÓN DE LA MEZCLA REACTIVA DE
AMPLIFICACIÓN POR PCR
ANEXO B. PLAN DE PLACA PARA LOS TERMOCICLADORES
Chromo4™ O CFX96™
ANEXO C. PLAN DE PLACA PARA EL TERMOCICLADOR
MiniOpticon™
30
© 2010 Bio-Rad
Page 31

1 - INTRODUCCIÓN
La Brettanomyces bruxellensis es una levadura responsable de la
presencia de 4-etil fenol y 4-etil guayacol en bebidas como los vinos, los
zumos de frutas y las cervezas que causan pérdidas económicas
importantes. Resulta difícil detectar su presencia en los vinos mediante los
métodos de cultivo clásicos, que son largos y no específicos.
Sin embargo, la detección rápida, específica y precoz de esta levadura en
el proceso de elaboración de los vinos permitiría al enólogo y al productor
tomar medidas de prevención y eliminarla antes de la aparición del
carácter fenolado característico de la alteración provocada por esta
levadura. Frente al método microbiológico tradicional, la biología
molecular ofrece soluciones de detección con elevados niveles de
rapidez, sensibilidad y especificidad.
El diagnóstico de la presencia de esta levadura debe permitir determinar
el nivel de riesgo en que se encuentra el vino y cómo evoluciona dicho
riesgo con el tiempo. El riesgo varía según el nivel de contaminación del
vino. Así pues, una herramienta de cuantificación adaptada permite
determinar si: a) existe una pequeña población y, por tanto, el riesgo es
bajo o está controlado; b) existe una población intermedia o crítica, en
cuyo caso el vino debe ser vigilado, es necesario hacer un seguimiento de
forma regular y puede tratarse el vino; o c) la población es considerable,
en cuyo caso también lo es el riesgo de producción de fenoles volátiles, y
se requiere actuar de forma inmediata y urgente sobre el vino.
En todos los casos se recomienda realizar un seguimiento regular. Es
importante seguir la evolución de las Brettanomyces bruxellensis en el vino.
El kit VINEO™ Brettanomytest PCR Kit es una prueba cuantitativa que permite
detectar de forma específica la Brettanomyces bruxellensis en los vinos y
mostos en fermentación mediante la técnica de reacción en cadena de la
polimerasa en tiempo real (RT-PCR). Se amplifica y detecta simultáneamente
una secuencia de ADN específica de la Brettanomyces bruxellensis gracias a
una sonda fluorescente. La aplicación de esta prueba permite obtener un
resultado cuantitativo en menos de 3 h tras la extracción de ADN (VINEO™
Extract DNA Kit, 354-8100). Los softwares diseñados para este análisis
Opticon Monitor™ o CFX Manager™ Industrial Diagnostic Edition permiten la
monitorización del riesgo derivado de la presencia de Brettanomyces
bruxellensis en la muestra de vino analizada. Además, se ofrece al usuario una
interpretación del nivel de riesgo en relación con el número de UFC/mL-1
(unidades formadoras de colonias) detectado en el vino:
31
© 2010 Bio-Rad
Page 32

• Negativo
• Población pequeña, riesgo controlado
• Población crítica que debe vigilarse
• Población muy elevada, riesgo de producción de fenoles volátiles
El kit VINEO™ Brettanomytest PCR Kit puede usarse asimismo de modo
cualitativo para confirmar colonias aisladas a partir de un medio de cultivo.
2 - PRINCIPIO DEL Kit VINEO™ Brettanomytest PCR Kit
Esta prueba se basa en la amplificación, detección y cuantificación de
secuencias de ADN mediante la técnica de PCR en tiempo real. Utiliza
cebadores y una sonda de ADN específicos de la Brettanomyces
bruxellensis. La detección y el análisis de los resultados se optimizan para
el uso con un termociclador para PCR en tiempo real de Bio-Rad, como
el MiniOpticon™, el Chromo4™ o el CFX96™.
Durante la reacción de la PCR, los cebadores se hibridan en la región
objetivo y después, catalizados por la polimerasa, se extienden en sentido
5’-3’ utilizando los desoxinucleótidos trifosfatos (dNTP) presentes en la
mezcla reactiva, creando de este modo una secuencia complementaria
de ADN llamada «amplicón».
Durante la PCR, se hibridan con los amplicones sondas oligonucleotídicas
específicas de la secuencia objetivo. Dichas sondas, marcadas mediante
fluoróforos, sólo emiten fluorescencia cuando se produce la hibridación.
La sonda que se une a la secuencia objetivo de Brettanomyces
bruxellensis se marca con un fluoróforo específico. La intensidad de la
fluorescencia aumenta en proporción a la cantidad de productos de
amplificación en el tubo de PCR. El módulo óptico del termociclador mide
directamente la fluorescencia generada de este modo.
A cada reacción se añade un ADN sintético llamado «control interno» que se
amplifica a la vez que las secuencias objetivo de Brettanomyces bruxellensis pero
es detectado por una sonda marcada con un segundo fluoróforo. Este control
interno permite evidenciar un posible fenómeno de inhibición de la reacción.
El software asociado al aparato calcula automáticamente la relación entre la
intensidad de la fluorescencia y el ciclo de amplificación. Esta relación indica la
presencia o ausencia de Brettanomyces bruxellensis en la muestra.
Los resultados obtenidos mediante amplificación de este DNA son analizados
por los softwares Opticon Monitor™ o CFX Manager™ IDE que han sido
programados para realizar análisis cuantitativos y automáticos. Entonces se
ofrece al usuario una interpretación del riesgo relacionado con la presencia de
Brettanomyces bruxellensis.
32
© 2010 Bio-Rad
Page 33

Esta prueba permite detectar y cuantificar la Brettanomyces bruxellensis
en los ADN extraídos a partir de bebidas fermentadas procedentes de la
uva y de mostos en fermentación.
3 - COMPOSICIÓN DEL KIT
Este kit contiene la cantidad de reactivos necesaria para 96 reacciones.
4 - CADUCIDAD Y CONSERVACIÓN
Desde el momento de su recepción, el kit debe conservarse entre +2 °C y
+8 °C. Cada reactivo conservado entre +2 °C y +8 °C puede ser utilizado
hasta la fecha de caducidad indicada en el tubo.
Nota: No congelar los reactivos.
5 - EQUIPO Y MATERIAL NECESARIOS NO SUMINISTRADOS
Equipo:
• Industrial Diagnostic CFX96™ para la PCR en tiempo real, bloque
reactivo de 96 pocillos, ref. Bio-Rad: 359-3990
• CFX Manager™ Software, Industrial Diagnostic Edition, ref. Bio-Rad:
359-3893
• Sistema MiniOpticon™ para la PCR en tiempo real, bloque reactivo
de 48 pocillos; ref. Bio-Rad: 359-3995
• Vórtex
• Opcional: centrífuga con rotor para placa o tira de tubos (máx. 2000 x g)
• Micropipetas de 20 µl, 200 µl y 1000 µl
• Dispensador múltiple, como pipetas Combitip
Observación: Recomendamos utilizar un ondulador eléctrico con el
termociclador.
33
© 2010 Bio-Rad
EXTRACCIÓN DE
LA MUESTRA
EXTRACCIÓN DE
ADN – VINEO™
Extract DNA Kit
(Ref.: 354-8100)
AMPLIFICACIÓN de
ADN PCR EN TIEMPO
REAL
VINEO™
Brettanomytest PCR
Kit (Ref.: 354-8101)
ANÁLISIS e
INTERPRETACIÓN
Opticon Monitor™
o
CFX Manager™ IDE
Tapones Denominación Reactivo Volumen
Rosa Fluorescent probes Sondas fluorescentes 1 tubo (0,55 mL)
Blanco Amplification mix Solución de amplificación 2 tubos (2 x 2,2 mL)
Verde Negative PCR control Control negativo PCR 1 tubo (0,5 mL)
Rojo Positive Qs PCR control Control positivo Qs PCR 1 tubo (0,5 mL)
Page 34

Consumibles:
• Para las placas, tubos PCR y tapones:
- Placas blancas de 96 pocillos de perfil bajo; ref. Bio-Rad: MLL-9651
- Placas blancas de 48 pocillos de perfil bajo; ref. Bio-Rad: MLL-4851
- Tiras de 8 tubos de 200l de perfil bajo, ref. Bio-Rad: TLS-0851
- Tiras de 8 tapones ópticos planos para tubos o placas de pocillos
de 0,2 mL, 120 tiras, ref. Bio-Rad : TCS-0803
• Conos para Combitip o dispensador múltiple, estériles y envasados
de forma individual
• Conos con filtro estériles, adaptables a micropipetas de 10 µl, 20 µl,
100 µl, 200 µl y 1000 µl
• Tubos estériles de 2 mL y de 5 mL
• Guantes de látex o nitrilo sin polvo
• Agua destilada estéril
• Lejía diluida al 5 %
• Agente descontaminante, como DNA AWAY
®
o RNAse AWAY
®
6 - PRECAUCIONES Y RECOMENDACIONES
• El ensayo debe ser realizado por personas que hayan recibido una
formación adecuada.
• La calidad de los resultados depende de que se cumplan
escrupulosamente las buenas prácticas de laboratorio, en especial
en lo referente a la PCR:
- El material (pipetas, tubos, etc.) no debe cambiarse de puesto de
trabajo.
- Es indispensable utilizar un control positivo y otro negativo para
cada serie de reacciones de amplificación.
- No utilizar los reactivos después de la fecha de caducidad.
- Agitar con vórtex los reactivos del kit antes de utilizarlos para
trabajar con soluciones homogéneas.
- Comprobar la exactitud y la precisión de las pipetas, así como el
correcto funcionamiento del instrumental.
- Cambiarse los guantes de forma regular y en cuanto se sospeche
que puedan estar contaminados.
- Limpiar regularmente las superficies de trabajo con lejía diluida al
5 % y otro agente limpiador, como DNA AWAY
®
o RNase AWAY®.
- Usar guantes sin polvo para no dejar huellas de dedos en la lámina
óptica empleada para sellar las microplacas. No escribir en los
tapones de los tubos para PCR. En ambos casos, puede dificultar
el registro de datos por parte del aparato.
34
© 2010 Bio-Rad
Page 35

Recomendamos encarecidamente leer el protocolo completo antes
de comenzar el ensayo.
Cumpla escrupulosamente el protocolo propuesto.
7 - EXTRACCIÓN Y TRANSPORTE DE LAS MUESTRAS
Las muestras de vino se recogen, en condiciones asépticas, en recipientes
estériles de vidrio, polietileno o de otro material parecido.
Las muestras deben enviarse al laboratorio lo más rápidamente posible,
preferentemente antes de 24 horas y no más de 48 horas tras su extracción.
Si las muestras se transportan y analizan en un plazo de 24 horas, el transporte
y almacenamiento se realizan a temperatura ambiente (entre +18 C y +30 C).
Si las muestras se transportan y analizan en un plazo de 48 horas, el transporte
y almacenamiento se realizan a una temperatura de entre +2 C y +8 C.
8 - EXTRACCIÓN DEL ADN
La extracción del ADN debe realizarse con el kit VINEO™ Extract DNA Kit
(ref. Bio-Rad 354-8100), desarrollado por Bio-Rad para la extracción de ADN
a partir de muestras de vino.
9 - PCR EN TIEMPO REAL
1. Puesta en marcha de la máquina de PCR
Encender, por este orden, el termociclador y el ordenador y, a continuación,
abrir el software Opticon Monitor. Encontrará más información y la definición
de los parámetros del software en el manual del usuario del termociclador
Bio-Rad para el kit VINEO™ Brettanomytest PCR Kit.
2. Preparación de las reacciones de PCR
2.1 Preparar la mezcla reactiva del kit VINEO™ Brettanomytest PCR Kit con
40 µL/muestra de la solución de amplificación (tubo con tapón blanco)
y 5 µL/muestra de sondas fluorescentes (tubo con tapón rosa). Hacer
mezcla reactiva en función del número de muestras y de controles que
se desee analizar, teniendo en cuenta que por cada placa deben usarse
como mínimo 1 duplicado (2) del control positivo Qs PCR y un control
negativo. La tabla de preparación de la mezcla reactiva de amplificación
por PCR, incluida en el Anexo A, indica las cantidades de cada reactivo
que deben mezclarse en función del número de muestras que se desee
analizar. No olvide contar los 3 puntos de control.
Nota: No mezclar lotes de reactivos.
35
© 2010 Bio-Rad
Page 36

2.2 Una vez preparada, la mezcla reactiva (soluciones de amplificación y
sondas) debe utilizarse inmediatamente, o bien puede conservarse
durante 2 horas entre +2 °C y +8 °C.
2.3 Repartir 45 μL de esta mezcla reactiva por pocillos, según el plan de
placa definido. En los Anexos B y C se proponen planes de placa para
los termocicladores Chromo4™/CFX96™ y MiniOpticon™,
respectivamente.
Añadir 5 μL de muestra, de control negativo o de control positivo Qs
PCR y sellar herméticamente los pocillos de la placa o de las tiras. Es
importante insistir en el cierre de los pocillos y de las tiras de pocillos para
evitar toda posible evaporación durante la PCR.
Es importante asimismo evitar la presencia de burbujas en el fondo de
los pocillos; para ello deberá pipetearse con cuidado. Para eliminar las
burbujas una vez sellada la placa o cerradas las tiras de pocillos para
PCR, éstas pueden centrifugarse brevemente. La placa puede
conservarse durante 2 horas entre +2 °C y +8 °C.
2.4 Colocar la placa o las tiras para PCR en el termociclador. Asegurarse de
que estén orientadas correctamente (pocillos A1 en la parte superior
izquierda).
Cerrar el módulo reactivo.
3. Inicio de la reacción de amplificación
Para iniciar la PCR, consulte el manual del usuario del termociclador Bio-Rad
para el kit VINEO™ Brettanomytest PCR Kit.
10 - ANÁLISIS DE DATOS
El análisis de los datos puede realizarse directamente al final de la reacción
de amplificación o posteriormente, reabriendo el fichero de datos. Para abrir
los ficheros de datos y analizar los resultados de la PCR, consulte el manual
del usuario del termociclador Bio-Rad para el kit VINEO™ Brettanomytest
PCR Kit.
11 - INTERPRETACIÓN DE LOS RESULTADOS
Para obtener los resultados del análisis, basta con leer los valores Ct (del
inglés «cycle threshold», ‘ciclo umbral’): valor del ciclo de amplificación a
partir del cual la fluorescencia se eleva significativamente por encima del
ruido de fondo.
36
© 2010 Bio-Rad
Page 37

El análisis manual permitirá únicamente un análisis cualitativo (presencia o
ausencia).
Para una interpretación automática es necesario utilizar el software Opticon
Monitor™ (versión 3.3 y posteriores) o el software CFX Manager™ Industrial
Diagnostic Edition. Será posible imprimir un informe completo (ver
instrucciones de software).
1. Controles
Antes de la interpretación final de los resultados es necesario verificar los
resultados de los controles positivo y negativo.
Para que la prueba sea válida, los resultados de los controles positivo y
negativo deberán ser los siguientes:
* N/A significa «no aplicable»: el software indica «N/A» para el Ct de una muestra cuando la curva de
fluorescencia no supera el umbral.
Si los resultados de los controles positivos Qs PCR y negativo son distintos
de los descritos en la tabla anterior, es necesario repetir la PCR desde el
principio.
2. Muestras
Una muestra se considera positiva en Brettanomyces bruxellensis si se
obtiene un valor Ct ≥ 10 para el fluoróforo FAM.
Si no se obtiene ningún valor para el Ct
FAM
(Ct
FAM
= N/A), la interpretación
del resultado depende del valor del control interno:
- Una muestra se considera negativa en Brettanomyces bruxellensis si no
se ha obtenido ningún valor de Ct para el fluoróforo FAM (Ct
FAM
=N/A) y el
Ct del control interno es igual o superior a 28 (Ct
HEX
≥ 28).
- Un valor N/A para el Ct
HEX
del control interno indica, cuando el Ct
FAM
del objetivo también es N/A, que probablemente se ha producido un
fenómeno de inhibición de la PCR. En este caso, la muestra de ADN
debe ser diluida (por ejemplo en relación 1:9) en agua destilada estéril y
para luego someterla a una nueva PCR.
37
© 2010 Bio-Rad
Detección de Brettanomyces
bruxellensis (FAM)
Detección del control interno
(Canal HEX)
Control negativo Ct = N/A* 25 ≤ Ct ≤ 40
Control positivo Qs PCR 27 ≤ Ct ≤ 37 No significativo
Page 38

- Si el Ct
HEX
del control interno es inferior a 28, no es posible interpretar el
resultado. Comprobar que se haya colocado el umbral correctamente o
que la curva bruta muestre un aspecto característico de amplificación
exponencial. Si la curva observada no es correcta, será necesario
repetir la prueba de PCR para esa muestra.
Interpretación de los resultados obtenidos con las muestras:
**: En caso de valor nulo en una muestra y su control interno (Ct = N/A), el análisis debe repetirse con
la muestra de ADN diluida (por ejemplo, en relación 1:4 o 1:9).
***: Puede ocurrir que se obtenga un valor inferior a 10. En ese caso, hay que comprobar que la curva
de datos brutos muestre un aspecto característico de amplificación exponencial (línea de base plana
seguida de aumento regular de la fluorescencia hasta llegar a una meseta). Si la curva observada es
correcta, puede considerarse que la muestra es positiva en Brettanomyces bruxellensis. En caso
contrario, la interpretación del resultado depende del valor del control interno, según lo explicado en los
párrafos anteriores.
12 - PROTOCOLO DE CONFIRMACIÓN A PARTIR DE COLONIAS
AISLADAS
Es posible utilizar la prueba VINEO™ Brettanomytest PCR Kit para confirmar
colonias aisladas en agar.
1. Pinchar la colonia aislada, ya sea a partir de un medio selectivo o no, con
ayuda de un palillo, un asa de siembra de 1 µL u otro consumible
adecuado (p. ej. un cono de pipeta).
2. Volver a suspender la colonia en 100 µl de R2 en un tubo Eppendorf (es
posible para volver a suspender la colonia en agua fisiológica estéril, la
eficiencia de PCR pueden ser afectadas). Homogeneizar bien la
suspensión en un vórtex.
3. Poner 5 µL de la suspensión en 45 µL de la mezcla reactiva de la PCR
(véase apartado 9.2 PCR en tiempo real) y seguir el resto del método del kit
VINEO™ Brettanomytest PCR Kit para obtener e interpretar los resultados.
Nota: Sólo se realiza un análisis cualitativo a partir de la PCR efectuada
con esta suspensión. A menudo, la señal obtenida con la PCR es muy
elevada debido a que la cantidad de células extraídas y analizadas
mediante PCR con este método es muy grande.
38
© 2010 Bio-Rad
Detección de Brettanomyces
bruxellensis (FAM)
Detección del control interno
Interpretación
Ct ≥ 10*** No significativo Positivo
Ct = N/A
Ct
HEX
≥ 28
Negativo
Ct = N/A Ct = N/A Inhibición**
Page 39

13 - RENDIMIENTOS DE LA PRUEBA Y VALIDACIONES
La prueba VINEO™ Brettanomytest PCR Kit es específica para la detección
de la Brettanomyces bruxellensis. La levadura puede detectarse a partir de:
• 500 UFC/mL de vino con el protocolo de extracción de ADN descrito
en el kit VINEO™ Extract DNA Kit: 1,8 mL para un mosto en
fermentación o un vino pesado durante la vinificación o la crianza.
• 10 UFC/mL de vino según el protocolo: 45 mL para un vino poco
pesado, próximo al embotellado o ya embotellado cuyo análisis precise
de un nivel elevado de sensibilidad analítica.
39
© 2010 Bio-Rad
Page 40

40
© 2010 Bio-Rad
ANEXO A
TABLA DE PREPARACIÓN DE LA MEZCLA REACTIVA DE AMPLIFICACIÓN POR
PCR
Utilizar esta tabla para determinar las cantidades de sondas fluorescentes y solución de
amplificación necesarias para preparar la mezcla reactiva de la PCR. En el cálculo del
número de muestras deben incluirse los 2 controles positivos Qs PCR y el control
negativo.
Número de
muestras
Sondas
fluorescentes
(μL)
(Tapón rosa)
Solución de
amplificación (μL)
(Tapón blanco)
Número de
muestras
Sondas
fluorescentes
(μL)
(Tapón rosa)
Solución de
amplificación (μL)
(Tapón blanco)
1 5 40 49 265 2117
2 11 86 50 270 2160
3 16 130 51 275 2203
4 22 173 52 281 2246
5 27 216 53 286 2290
6 32 259 54 292 2333
7 38 302 55 297 2376
8 43 346 56 302 2419
9 49 389 57 308 2462
10 54 432 58 313 2506
11 59 475 59 319 2549
12 65 518 60 324 2592
13 70 562 61 329 2635
14 76 605 62 335 2678
15 81 648 63 340 2722
16 86 691 64 346 2765
17 92 734 65 351 2808
18 97 778 66 356 2851
19 103 821 67 362 2894
20 108 864 68 367 2938
21 113 907 69 373 2981
22 119 950 70 378 3024
23 124 994 71 383 3067
24 130 1037 72 389 3110
25 135 1080 73 394 3154
26 140 1123 74 400 3197
27 146 1166 75 405 3240
28 151 1210 76 410 3283
29 157 1253 77 416 3326
30 162 1296 78 421 3370
31 167 1339 79 427 3413
32 173 1382 80 432 3456
33 178 1426 81 437 3499
34 184 1469 82 443 3542
35 189 1512 83 448 3586
36 194 1555 84 454 3629
37 200 1598 85 459 3672
38 205 1642 86 464 3715
39 211 1685 87 470 3758
40 216 1728 88 475 3802
41 221 1771 89 481 3845
42 227 1814 90 486 3888
43 232 1858 91 491 3931
44 238 1901 92 497 3974
45 243 1944 93 502 4018
46 248 1987 94 508 4061
47 254 2030 95 513 4104
48 259 2074 96 518 4147
Page 41

41
© 2010 Bio-Rad
ANEXO B
PLAN DE PLACA PARA LOS TERMOCICLADORES Chromo4™/CFX96™
Los 3 primeros pocillos de la columna 1 pueden usarse para analizar los 3 puntos de
control. Los demás pocillos de la placa se usarán para colocar las muestras que se
desee analizar.
CN: Control negativo PCR
QS: Control positivo Qs PCR
ANEXO C
PLAN DE PLACA PARA EL TERMOCICLADOR MiniOpticon™
Los 3 primeros pocillos de la columna 1 pueden usarse para analizar los 3 puntos de
control. Los demás pocillos de la placa se usarán para colocar las muestras que se
desee analizar.
CN: Control negativo PCR
QS: Control positivo Qs PCR
1 2 3 4 5 6 7 8 9 10 11 12
A QS
B QS
C NC
D Muestra
E Muestra
F …
G
H
1 2 3 4 5 6
A QS
B QS
C NC
D Muestra
E Muestra
F …
G
H
Page 42

AVISO AL COMPRADOR: LICENCIA LIMITADA
Este producto está protegido por una o más de las siguientes patentes de EE. UU. y las reivindicaciones
de patentes correspondientes fuera de Estados Unidos: 5 079 352, 5 789 224, 5 618 711, 6 127 155, 5
677 152 (sólo reivindicaciones de la 1 a la 23) y 5 773 258 (sólo reivindicaciones de la 1 a la 6) y las
reivindicaciones fuera de EE.UU. correspondientes a la patente n° 4 889 818 de EE. UU. La adquisición
de este producto incluye inmunidad judicial restringida e intransferible según las anteriores
reivindicaciones de patentes cuando esta cantidad de producto se use únicamente con fines de análisis
alimentario, análisis medioambiental y en microbiología industrial, incluida la publicación de los resultados
de las actividades del comprador por medio del pago o toda contrapartida comercial, cuando se utilice
para los fines de las propias investigaciones del comprador. No se cede de forma expresa ningún
derecho a cualquier reivindicación de patente (como las reivindicaciones del método de la 5’ nucleasa en
las patentes n° 5 210 015 y 5 487 972 de EE. UU.), ya sea por implicación o por preclusión. Puede
obtenerse más información respecto a la compra de licencias poniéndose en contacto con el Director de
Licencias, Applied Biosystems, 850 Lincoln Centre Drive, Foster City, California, 94404, EE. UU.
42
© 2010 Bio-Rad
Page 43

VINEO™Brettanomytest PCR Kit
Rif.: 354-8101
Istruzioni per l'uso
Test per la rilevazione e la quantificazione mediante
PCR in tempo reale di Brettanomyces bruxellensis
43
© 2010 Bio-Rad
Page 44

SOMMARIO
1 - INTRODUZIONE
2 - PRINCIPIO DEL VINEO™ Brettanomytest PCR Kit
3 - COMPOSIZIONE DEL KIT
4 - PERIODO DI VALIDITÀ E CONSERVAZIONE
5 - ATTREZZATURA E MATERIALE NECESSARI NON FORNITI
6 - PRECAUZIONI E RACCOMANDAZIONI
7 - PRELIEVO E TRASPORTO DEI CAMPIONI
8 - ESTRAZIONE DEL DNA
9 - PCR IN TEMPO REALE
10 - ANALISI DEI DATI
11 - INTERPRETAZIONE DEI RISULTATI
12 - PROTOCOLLO DI CONFERMA A PARTIRE DA COLONIE ISOLATE
13 - PRESTAZIONI DEL TEST E VALIDAZIONI
ALLEGATO A - TABELLA DI PREPARAZIONE DELLA MISCELA DI
REAZIONE DI AMPLIFICAZIONE PCR
ALLEGATO B - PIANO DI PIASTRA DA UTILIZZARE SUI
TERMOCICLATORI Chromo4™ O CFX96™
ALLEGATO C - PIANO DI PIASTRA DA UTILIZZARE SUL
TERMOCICLATORE MiniOpticon™
44
© 2010 Bio-Rad
Page 45

1 - INTRODUZIONE
Brettanomyces bruxellensis è un lievito responsabile della presenza di
4-etilfenolo e 4 etilguaiacolo nelle bevande quali vini, succhi di frutta e birre
che comporta importanti perdite a livello economico. Nei vini, la sua
presenza è difficile da rilevare attraverso i metodi di coltura classici, che
sono lunghi e non specifici.
La rilevazione rapida, specifica e precoce di questo lievito nel procedimento di
elaborazione dei vini consentirebbe tuttavia all'enologo e al produttore di
adottare delle misure preventive e di eliminarlo prima della comparsa del
carattere fenolato, caratteristico dell'alterazione causata da questo lievito. In
confronto al metodo microbiologico tradizionale, la biologia molecolare apporta
soluzioni di rilevazione ad alti livelli di rapidità, di sensibilità e di specificità.
La diagnosi della presenza di questo lievito deve permettere di
determinare a che livello di rischio si trova il vino e come tale rischio evolve
nel tempo. Questo rischio varia a seconda del livello di contaminazione del
vino. Pertanto uno strumento di quantificazione adatto permette di
determinare se (i) la popolazione è in quantità bassa e quindi il rischio è
basso, cioè controllato, (ii) in quantità intermedia o critica, in questo caso il
vino deve essere sorvegliato, si impone un controllo regolare, il vino può
essere trattato (iii) e, infine, se la popolazione è molto importante, e in
questo caso lo è anche il rischio di produzione dei fenoli volatili, si impone
un'azione immediata e urgente sul vino.
In tutti i casi si consiglia un follow-up regolare. È importante seguire
l'evoluzione dei Brettanomyces bruxellensis nel vino.
VINEO™ Brettanomytest PCR Kit è un test quantitativo che permette la
rilevazione specifica di Brettanomyces bruxellensis nei vini e nei mosti in
fermentazione mediante la tecnica di polimerizzazione a catena in tempo reale
(RT-PCR). Una sequenza di DNA specifica di Brettanomyces bruxellensis viene
amplificata e rilevata simultaneamente grazie ad una sonda fluorescente. La
realizzazione di questo test permette di ottenere un risultato quantitativo in
meno di 3 ore dopo l'estrazione di DNA (VINEO™ Extract DNA Kit, 354-8100).
Il software di analisi adatto a questo test, Opticon Monitor™ oppure CFX
Manager™ Industrial Diagnostic Edition, consente, attraverso un'analisi
automatica e quantitativa, di misurare il rischio Brettanomyces bruxellensis nel
campione di vino analizzato. All'utilizzatore viene proposta anche
un'interpretazione del livello di rischio. Quest'ultima è correlata al numero di
UFC.ml-1 (Unità Formanti Colonia) rilevate nel vino:
45
© 2010 Bio-Rad
Page 46

• Negativo
• Popolazione bassa, rischio controllato
• Popolazione critica, da tenere sotto controllo
• Popolazione molto elevata, rischio di produzione di Fenoli volatili
VINEO™ Brettanomytest PCR Kit può essere utilizzato anche in modalità
qualitativa per la conferma di colonie isolate a partire dal mezzo di coltura.
2 - PRINCIPIO DEL VINEO™ Brettanomytest PCR Kit
Questo test si basa sull'amplificazione, la rilevazione e la quantificazione di
sequenze di DNA mediante la tecnica di PCR in tempo reale. Utilizza dei primer
e una sonda da DNA specifici per Brettanomyces bruxellensis. La rilevazione e
l'analisi dei risultati sono ottimizzate per l'utilizzazione con un termociclatore per
la PCR in tempo reale Bio-Rad, quale il MiniOpticon™, il Chromo4™ o il
CFX96™.
Durante la reazione di PCR, i primer si ibrideranno alla regione bersaglio poi,
catalizzati mediante polimerasi, si allungheranno nelle direzioni 5’-3’ utilizzando i
desossinucleotidi trifosfati (dNTP) presenti nella miscela di reazione, creando in
questo modo una sequenza complementare di DNA, detta amplicon.
Durante la PCR, sonde oligonucleotidiche, specifiche della sequenza
bersaglio, si ibrideranno agli amplicon. Queste sonde, marcate con
fluorofori, emettono fluorescenza solo quando ha luogo l'ibridazione. La
sonda che si lega alla sequenza bersaglio di Brettanomyces bruxellensis è
marcata con un fluoroforo specifico. L'intensità di fluorescenza aumenta
proporzionalmente all'aumento della quantità dei prodotti di amplificazione
nella provetta di PCR. La fluorescenza così generata viene misurata
direttamente attraverso il modulo ottico del termociclatore.
Ad ogni reazione viene aggiunto un DNA sintetico chiamato “Controllo
interno”. Viene amplificato simultaneamente alle sequenze bersaglio di
Brettanomyces bruxellensis, ma rilevato mediante una sonda marcata con
un secondo fluoroforo. Consente di evidenziare un eventuale fenomeno di
inibizione della reazione.
Il software associato all'apparecchio calcola automaticamente la relazione tra
l'intensità della fluorescenza e il ciclo di amplificazione. Questa relazione indica
la presenza, o l'assenza, di Brettanomyces bruxellensis nel campione.
I risultati ottenuti per l'amplificazione di questi DNA sono analizzati dal
software Opticon Monitor™ oppure CFX Manager™ programmati per
eseguire un'analisi automatica e quantitativa. Viene allora proposta
all'utilizzatore un'interpretazione relativa al rischio correlato alla presenza di
Brettanomyces bruxellensis.
46
© 2010 Bio-Rad
Page 47

Questo test consente la rilevazione e la quantificazione di Brettanomyces
bruxellensis nei DNA estratti a partire da bevande fermentate ottenute da
uva e dai mosti in fermentazione.
3 - COMPOSIZIONE DEL KIT
Questo kit contiene la quantità di reagente necessaria per 96 reazioni.
4 - PERIODO DI VALIDITÀ E CONSERVAZIONE
Dal momento della ricezione, il kit deve essere conservato ad una
temperatura tra +2°C e +8°C. Ogni reagente conservato tra +2°C e +8°C
può essere utilizzato fino alla data di scadenza indicata sulla provetta.
NB: Non congelare i reagenti.
5 - ATTREZZATURA E MATERIALE NECESSARI NON FORNITI
Attrezzature:
• Industrial Diagnostic CFX96™ per la PCR in tempo reale, blocco di
reazione da 96 pozzetti, rif. Bio-Rad: 359-3990
• CFX Manager™ Software, Industrial Diagnostic Edition, rif. Bio-Rad :
359-3893
• Sistema MiniOpticon™ per la PCR in tempo reale, blocco di reazione
da 48 pozzetti, rif. Bio-Rad: 359-3995
• Vortex
• Opzionale: centrifuga con rotore per piastra o barretta (max. 2000 x G)
• Micropipette da 20 µl, 200 µl e 1000 µl
• Multidispensatore, quale pipette combitip
Nota: Consigliamo di utilizzare un ondulatore elettrico con il
termociclatore.
47
© 2010 Bio-Rad
PRELIEVO DEL
CAMPIONE
ESTRAZIONE DNA
VINEO™ Extract
DNA Kit
(Rif.: 354-8100)
AMPLIFICAZIONE
DNA mediante PCR IN
TEMPO REALE
VINEO™
Brettanomytest PCR
Kit (Rif.: 354-8101)
ANALISI e
INTERPRETAZIONE
Opticon Monitor™
o
CFX Manager™ IDE
Tappi Designazione Reagente Volume
Rosa Fluorescent probes Sonde fluorescenti 1 provetta (0,55 ml)
Bianco Amplification mix Soluzione di amplificazione 2 provette (2 x 2,2 ml)
Verde Negative PCR control Controllo negativo PCR 1 provetta (0,5 ml)
Rosso Positive Qs PCR control Controllo positivo Qs PCR 1 provetta (0,5 ml)
Page 48

Materiali monouso:
• Per le piastre, provette per PCR e cappucci:
- Piastre bianche da 96 pozzetti “Low-profile”, rif. Bio-Rad: MLL-9651
- Piastre bianche da 48 pozzetti “Low-profile”, rif. Bio-Rad: MLL-4851
- Barrette da 8 provette da 200 l “Low-profile”, rif. Bio-Rad: TLS-0851
- Barrette da 8 cappucci ottici piatti, per provette o piastre per pozzetti da
0,2 ml, 120 barrette Rif. Bio-Rad: TCS-0803
• Coni per Combitip o multidispensatore, sterili confezionati
individualmente
• Coni con filtro sterili, adattabili a micropipette da 10 µl, 20 µl, 100 µl,
200 µl e 1000 µl
• Provette sterili da 2 ml e da 5 ml
• Guanti non talcati in latice o nitrile
• Acqua distillata sterile
• Candeggina al 5%
• Agente decontaminante quale DNA AWAY
®
o RNAse AWAY
®
6 - PRECAUZIONI E RACCOMANDAZIONI
• Questo test deve essere realizzato da persone che hanno ricevuto
una formazione adeguata.
• La qualità dei risultati dipende dal rispetto scrupoloso delle Buone
Prassi di Laboratorio, in particolare in materia di PCR:
- Il materiale (pipette, provette, ecc....) non deve circolare da una
postazione di lavoro all'altra.
- È indispensabile utilizzare un controllo positivo e un controllo
negativo per ogni serie di reazioni di amplificazione.
- Non utilizzare i reagenti oltre la data di scadenza.
- Agitare con il vortex i reagenti del kit prima di utilizzarli in modo da
lavorare con soluzioni omogenee.
- Verificare l'accuratezza e la precisione delle pipette e il corretto
funzionamento degli strumenti.
- Sostituire i guanti regolarmente e ogniqualvolta si ritiene che
possano essere contaminati.
- Pulire regolarmente le superfici di lavoro con candeggina al 5% e
altri agenti quali DNA AWAY
®
o RNase AWAY®.
- Indossare guanti non talcati per non lasciare impronte sulla
pellicola ottica utilizzata per chiudere le micropiastre. Non scrivere
sui tappi delle provette PCR. In entrambi i casi, la registrazione dei
dati da parte dell'apparecchio può risultare falsata.
48
© 2010 Bio-Rad
Page 49

Vi raccomandiamo vivamente di leggere l'intero protocollo prima di
iniziare il test.
Rispettare scrupolosamente il protocollo proposto.
7 - PRELIEVO E TRASPORTO DEI CAMPIONI
I campioni di vini vengono prelevati, in condizioni di asepsi, in recipienti sterili di
vetro, di polietilene o di materiale simile.
I campioni devono essere consegnati al laboratorio il più rapidamente
possibile, preferibilmente entro le 24 ore e non oltre le 48 ore successive al
prelievo. Se i campioni sono trasportati e analizzati entro 24 ore, il trasporto e
la conservazione hanno luogo a temperatura ambiente (da +18°C a +30°C).
Se i campioni vengono trasportati e analizzati entro le 48 ore, il trasporto e la
conservazione hanno luogo ad una temperatura tra +2°C e +8°C.
8 - ESTRAZIONE DEL DNA
L'estrazione del DNA va effettuata utilizzando il kit messo a punto da Bio-Rad
per l'estrazione del DNA a partire da campioni di vino, VINEO™ Extract DNA
kit (rif. Bio-Rad 354-8100).
9 - PCR IN TEMPO REALE
1. Accensione dell'apparecchio PCR
Accendere nell'ordine il termociclatore, il computer poi aprire il software
Opticon Monitor. Per maggiori informazioni e per la definizione dei parametri
del software consultare il manuale d'uso del termociclatore Bio-Rad per il kit
VINEO™ Brettanomytest PCR Kit.
2. Preparazione delle reazioni PCR
2.1 Preparare la miscela di reazione VINEO™ Brettanomytest PCR Kit
mescolando (40 µl/campione) della soluzione di amplificazione
(provetta con il tappo bianco) e (5 µl/campione) delle sonde fluorescenti
(provetta con il tappo rosa). Realizzare la miscela di reazione in funzione
del numero di campioni e dei controlli da analizzare (devono essere
utilizzati minimo 1 duplicato (2) del controllo positivo Qs PCR e un
controllo negativo per piastra). La tabella di preparazione della miscela di
reazione di amplificazione PCR presente nell'Allegato A indica le quantità
necessarie di ogni reagente da miscelare in funzione del numero di
campioni da analizzare. Non dimenticare di registrare i 3 punti di controlli.
Nota: non mischiare i lotti di reagenti.
49
© 2010 Bio-Rad
Page 50

2.2 Dopo la preparazione, la miscela di reazione (Soluzioni di amplificazione e
di sonde) va utilizzata immediatamente, o può essere conservata per 2
ore ad una temperatura da +2°C a +8°C.
2.3 Ripartire 45 μl di questa miscela di reazione per pozzetto, secondo il
piano di piastra definito. Gli Allegati B e C propongono piani di piastra da
utilizzare rispettivamente sui termociclatori Chromo4™/CFX96™ e
MiniOpticon™.
Aggiungere 5 μl di campione o di controllo negativo o di controllo positivo
Qs PCR e chiudere ermeticamente i pozzetti della piastra o delle barrette.
È importante insistere sulla chiusura dei pozzetti della piastra o delle
barrette per evitare qualunque fenomeno di evaporazione nel corso della
reazione PCR.
È importante evitare il formarsi di bolle sul fondo dei pozzetti pipettando
con cautela. Per eliminare le bolle, dopo aver sigillato la piastra o aver
chiuso le barrette PCR, è possibile centrifugare brevemente la piastra o le
barrette PCR. La piastra può essere conservata per 2 ore ad una
temperatura da +2°C a +8°C.
2.4 Posizionare la piastra o le barrette PCR nel termociclatore. Accertarsi che
siano orientate correttamente (pozzetti A1 in alto a sinistra).
Chiudere il modulo di reazione.
3. Avvio della reazione di amplificazione
Per avviare la PCR, consultare il manuale d'uso del termociclatore Bio-Rad
per il kit VINEO™ Brettanomytest PCR Kit.
10 - ANALISI DEI DATI
L'analisi dei dati può essere effettuata direttamente al termine della reazione
di amplificazione o in seguito riaprendo il file dati. Per aprire dei file dati e
analizzare i risultati della PCR, consultare il manuale d'uso del termociclatore
Bio-Rad per il kit VINEO™ Brettanomytest PCR Kit.
11 - INTERPRETAZIONE DEI RISULTATI
Per ottenere i risultati dell'analisi, è sufficiente leggere i valori di Ct (Cycle
treshold): valore del ciclo di amplificazione a partire dal quale la fluorescenza
si innalza significativamente al di sopra del rumore di fondo.
L'analisi manuale consentirà solo un'analisi qualitativa (presenza o assenza).
50
© 2010 Bio-Rad
Page 51

È necessario utilizzare il software Opticon Monitor™ (versione 3.3 o
superiore) oppure il CFX Manager™ software Industrial Diagnostic Edition
per un'interpretazione automatizzata e quantitativa. Sarà possibile stampare
una relazione completa (Cfr. istruzioni del software).
1. Controlli
Prima dell'interpretazione finale dei risultati è necessario verificare i risultati dei
controlli negativo e positivo.
Perché il test sia valido, i risultati dei controlli negativo e positivo devono
essere i seguenti:
* N/A significa “Non Applicabile”. Il software indica N/A per il Ct di un campione quando la curva di
fluorescenza non interseca la soglia.
Se i risultati dei controlli positivo Qs PCR e negativo differiscono da quelli
descritti nella tabella qui sopra, è necessario ripetere la PCR.
2. Campioni
Un campione è considerato positivo per Brettanomyces bruxellensis se
viene ottenuto un valore di Ct ≥ 10 per il fluoroforo FAM.
Se non viene ottenuto alcun valore per il CtFAM (Ct
FAM
= N/A),
l'interpretazione del risultato dipende dal valore del controllo interno:
- Un campione è considerato negativo per Brettanomyces bruxellensis se
non viene ottenuto alcun valore di Ct per il fluoroforo FAM (Ct
FAM
=N/A) e
se il Ct del controllo interno è superiore o uguale a 28. (Ct
HEX
≥ 28).
- Un valore N/A per il Ct
HEX
del controllo interno indica, quando il Ct
FAM
del bersaglio è ugualmente N/A, che probabilmente ha avuto luogo un
fenomeno di inibizione della reazione di PCR. In tal caso, il campione di
DNA deve essere diluito (a 1:10 per esempio) in acqua distillata sterile,
poi sottoposto nuovamente a PCR.
- Se il Ct
HEX
del controllo interno è inferiore a 28 non è possibile
interpretare il risultato. Verificare che la soglia sia stata posta
correttamente o che la curva grezza abbia un aspetto caratteristico di
amplificazione esponenziale. Se la curva osservata non è corretta, sarà
necessario ripetere il test di PCR per questo campione.
51
© 2010 Bio-Rad
Rilevazione di Brettanomyces
bruxellensis (FAM)
Rilevazione del controllo
interno (Canale HEX)
Controllo negativo Ct = N/A* 25 ≤ Ct ≤ 40
Controllo positivo Qs PCR 27 ≤ Ct ≤ 37 Non significativo
Page 52

Interpretazione dei risultati ottenuti sui campioni:
**: In caso di valore nullo per un campione e il suo controllo interno (Ct = N/A), l'analisi deve essere
ripetuta con il campione di DNA diluito ( a 1/5° o a 1/10° per esempio).
***: Può capitare di ottenere un valore inferiore a 10, verificare che la curva come dato grezzo abbia un
aspetto caratteristico di amplificazione esponenziale (linea di base piatta, poi aumento regolare della
fluorescenza seguito da un plateau). Se la curva osservata è corretta, il campione può essere
considerato positivo per la presenza di Brettanomyces bruxellensis. in caso contrario, l'interpretazione
del risultato dipende dal valore del controllo interno, come spiegato nei paragrafi precedenti.
12 - PROTOCOLLO DI CONFERMA A PARTIRE DA COLONIE ISOLATE
È possibile utilizzare il test VINEO™ Brettanomytest PCR Kit per la conferma
di colonie isolate su agar-agar.
1. Pungere una colonia isolata, a partire da un mezzo selettivo o non, con
uno stuzzicadenti o un'ansa da 1 µl, o qualsiasi altro materiale corrente di
laboratorio adatto (cono di pipettaggio per esempio).
2. Risospendere la colonia in 100 µl di R2 in una provetta Eppendorf (è
possibile risospendere la colonia in soluzione di acqua fisiologica sterile,
l'efficienza della PCR può quindi essere influenzato). Omogeneizzare bene
la sospensione utilizzando il vortex.
3. Porre 5 µl della sospensione in 45 µl della miscela di reazione PCR (Cf.
parte 9.2 PCR in tempo reale) e seguire il resto del metodo VINEO™
Brettanomytest PCR Kit per ottenere e interpretare i risultati.
Nota: una sola analisi qualitativa viene realizzata a partire dalla PCR
effettuata su questa sospensione. Il segnale ottenuto in PCR è
spesso molto elevato poiché la quantità di cellule prelevate e
analizzate mediante PCR con questo metodo è molto considerevole.
13 - PRESTAZIONI DEL TEST E VALIDAZIONI
Il test VINEO™ Brettanomytest PCR Kit è specifico per la rilevazione di
Brettanomyces bruxellensis. È possibile rilevare il lievito a partire da:
• 500 UFC/ml di vino con il protocollo di estrazione del DNA descritto nel
VINEO™ Extract DNA Kit: su 1,8 ml per un mosto in fermentazione o
un vino carico in corso di vinificazione o di affinamento e
• 10 UFC/ml di vino secondo il protocollo che parte da 45 ml per un vino
poco carico, prossimo all'imbottigliamento o già imbottigliato la cui
analisi richiede un livello di sensibilità analitica elevato.
52
© 2010 Bio-Rad
Rilevazione di Brettanomyces
bruxellensis (FAM)
Rilevazione del controllo
interno
Interpretazione
Ct ≥ 10*** Non significativo Positivo
Ct = N/A
Ct
HEX
≥ 28
Negativo
Ct = N/A Ct = N/A Inibizione**
Page 53

ALLEGATO A
TABELLA DI PREPARAZIONE DELLA MISCELA DI REAZIONE DI
AMPLIFICAZIONE PCR
Utilizzare questa tabella per determinare le quantità necessarie di sonde fluorescenti e
soluzione di amplificazione per preparare la miscela di reazione di PCR. Il calcolo del
numero di campioni deve includere 2 controlli Qs PCR e il controllo negativo.
53
© 2010 Bio-Rad
Numero
di campioni
Sonde
fluorescenti (μl)
(Tappo rosa)
Soluzione di
amplificazione (μl)
(Tappo bianco)
Numero
di campioni
Sonde
fluorescenti (μL)
(Tappo rosa)
Soluzione di
amplificazione (μl)
(Tappo bianco)
1 5 40 49 265 2117
2 11 86 50 270 2160
3 16 130 51 275 2203
4 22 173 52 281 2246
5 27 216 53 286 2290
6 32 259 54 292 2333
7 38 302 55 297 2376
8 43 346 56 302 2419
9 49 389 57 308 2462
10 54 432 58 313 2506
11 59 475 59 319 2549
12 65 518 60 324 2592
13 70 562 61 329 2635
14 76 605 62 335 2678
15 81 648 63 340 2722
16 86 691 64 346 2765
17 92 734 65 351 2808
18 97 778 66 356 2851
19 103 821 67 362 2894
20 108 864 68 367 2938
21 113 907 69 373 2981
22 119 950 70 378 3024
23 124 994 71 383 3067
24 130 1037 72 389 3110
25 135 1080 73 394 3154
26 140 1123 74 400 3197
27 146 1166 75 405 3240
28 151 1210 76 410 3283
29 157 1253 77 416 3326
30 162 1296 78 421 3370
31 167 1339 79 427 3413
32 173 1382 80 432 3456
33 178 1426 81 437 3499
34 184 1469 82 443 3542
35 189 1512 83 448 3586
36 194 1555 84 454 3629
37 200 1598 85 459 3672
38 205 1642 86 464 3715
39 211 1685 87 470 3758
40 216 1728 88 475 3802
41 221 1771 89 481 3845
42 227 1814 90 486 3888
43 232 1858 91 491 3931
44 238 1901 92 497 3974
45 243 1944 93 502 4018
46 248 1987 94 508 4061
47 254 2030 95 513 4104
48 259 2074 96 518 4147
Page 54

54
© 2010 Bio-Rad
ALLEGATO B
PIANO DI PIASTRA DA UTILIZZARE SUI TERMOCICLATORI Chromo4™ o
CFX96™
I primi 3 pozzetti della colonna 1 possono essere utilizzati per analizzare i 3 punti di
controlli. Gli altri pozzetti della piastra saranno utilizzati per deporre i campioni da
analizzare.
NC: Controllo negativo PCR
QS: Controllo positivo Qs PCR
ALLEGATO C
PIANO DI PIASTRA DA UTILIZZARE SUL TERMOCICLATORE MiniOpticon™
I primi 3 pozzetti della colonna 1 possono essere utilizzati per analizzare i 3 punti di
controlli. Gli altri pozzetti della piastra saranno utilizzati per deporre i campioni da
analizzare.
NC: Controllo negativo PCR
QS: Controllo positivo Qs PCR
1 2 3 4 5 6 7 8 9 10 11 12
A QS
B QS
C NC
D Campione
E Campione
F …
G
H
1 2 3 4 5 6
A QS
B QS
C NC
D Campione
E Campione
F …
G
H
Page 55

55
© 2010 Bio-Rad
AVVISO PER L'ACQUIRENTE: LICENZA LIMITATA
L'utilizzazione di questo prodotto è tutelata da uno o molti dei seguenti brevetti statunitensi e dalle
rivendicazioni di brevetto corrispondenti fuori degli Stati Uniti: 5 079 352, 5 789 224, 5 618 711, 6 127
155, 5 677 152 (unicamente rivendicazioni da 1 a 23) e 5 773 258 (unicamente rivendicazioni da 1 a 6) e
dalle rivendicazioni fuori degli Stati Uniti corrispondenti al brevetto USA n. 4 889 818. L'acquisto di questo
prodotto comprende un'immunità da azioni giudiziarie limitata e non trasferibile conformemente alle
rivendicazioni di brevetti precedenti quando questa quantità di prodotto viene utilizzata unicamente a scopi
di analisi alimentare, di analisi ambientale e in microbiologia industriale, compresa la pubblicazione dei
risultati delle attività dell'acquirente mediante pagamento o qualsiasi altra contropartita commerciale, e
quando viene egualmente utilizzata per le ricerche proprie dell'acquirente. Non viene espressamente
ceduto alcun diritto sulla base di una qualunque rivendicazione di brevetto (quali le rivendicazioni del
metodo 5'-nucleasi nei brevetti USA n. 5 210 015 e 5 487 972) che sia per implicazione o per
preclusione. Informazioni più complete riguardanti l'acquisto di licenze possono essere ottenute
contattando il Direttore delle licenze, Applied Biosystems, 850 Lincoln Centre Drive, Foster City, California,
94404, Stati Uniti.
Page 56

Purchaser's information: VINEO is a trademark of Bio-Rad
Information à l’acheteur : VINEO is a trademark of Bio-Rad
Información al comprador: VINEO es una marca registrada de Bio-Rad
Informazione per l'acquirente: VINEO è un marchio registrato di Bio-Rad
Bio-Rad
3, bd Raymond Poincaré
92430 Marnes-la-Coquette - France
Tel.: +33 1 47 95 60 00
Fax.: +33 1 47 41 91 33 Rev. D - 12/2010
 Loading...
Loading...