Page 1

General Recommendations
• Store the array in the original packaging and do not open the packaging until ready to use
• Prepared arrays can be used multiple times, though the peak features will degrade over time. After opening, store arrays in a dry and dark
location and their original packaging. Record the usage dates
• The ProteinChip premade array supplied with this kit has been prepared with a mixture of peptides and matrix. Avoid exposing these arrays to
vibration, moisture, or light
• ProteinChip array spots contain chemically treated surfaces. Avoid contamination of these surfaces: always handle ProteinChip arrays by
their edges, and never touch the spots
• When running single ProteinChip arrays on the ProteinChip instrument, Enterprise Edition, always fill empty locations in the cassette with
blank, expired, or used arrays.
• To ensure best results, evaluate the performance of the ProteinChip system using the ProteinChip system check kit (catalog #C70-00081) or
ProteinChip OQ kit (catalog #C70-00080), or as included in service contracts
Introduction
Mass calibration is the process by which the time-of flight data obtained by mass spectrometry are converted to molecular mass data. Data collected
using standards (known peptides or proteins) are used to derive a calibration equation, which is then applied to the spectra of experimental samples.
In the following protocol, you use the spectra obtained from the ProteinChip®peptide standard array to perform the mass calibration procedure.
The procedures for importing protocols and performing mass calibration are illustrated in a tutorial video included on the ProteinChip Peptide Mass
Calibraiton Kit CD. The tutorial video is also available online at www.bio-rad.com/proteinchip/starterkit.
Import the Pep Std Array Protocol
The ProteinChip reader uses data collection protocols to collect data from arrays. The protocol used with this kit is provided with the CD. Import the
protocol into the Protocols folder of the ProteinChip data manager software.
1. Ensure that the ProteinChip reader is turned on and connected to the computer. (This is usually done during installation.)
2. Insert the ProteinChip Peptide Mass Calibration Kit CD and launch the ProteinChip data manager software.
3. In the Explorer pane, select the Protocols folder. In the toolbar, select Protocols>Import Protocol. The Choose File(s) to Import
dialog opens.
4. Select the file named Pep Std Array.ptx and click import. Click OK at the prompt.
Read the Peptide Standard Array
1. Insert the ProteinChip peptide standard array into the instrument. If you are using an Enterprise Edition instrument, place the array
into a cassette, fill the remaining slots in the cassette with blank or used arrays, and place the cassette into the instrument.
2. In ProteinChip data manager software, click Start and open the Protocol Mode tab.
3. Select Instrument > Preferences. In the Preferences dialog, click "…". The Select Folder dialog opens.
4. Select New Folder, name the new folder Peptide Mass Calibration and click OK.
5. Optimize the laser intensity. Run protocol Pep Std Array on any partition ("n") of 10 on spot A. Adjust the laser energy until the
height of the insulin peak at 5.96 kD is 100–200 µA.
6. Run protocol Pep Std Array. Run on partition 1 of 4 on spot A of chip. Record usage on the label provided on the package. Each
spot on the array may be read up to 5 times on each of 4 partitions (1 of 4, 2 of 4, 3 of 4 and 4 of 4).
ProteinChip®Peptide Mass Calibration Kit
Catalog #C70-00070
Page 2
Perform Mass Calibration
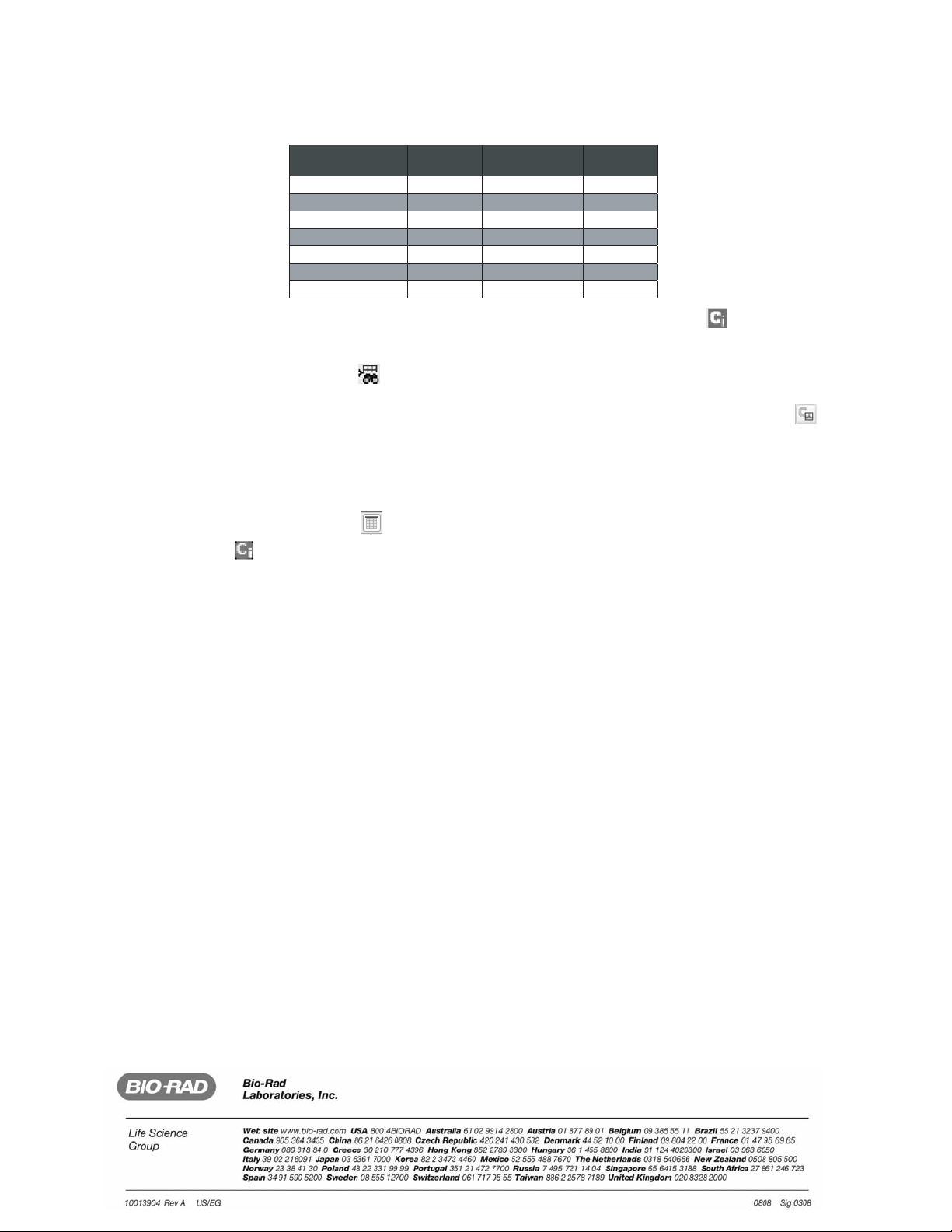
1. Open the spectrum obtained using the peptide standard array, and label the 4 peaks indicated in the table below.
Note: Label each peak at its highest point; if not, the mass will be recorded incorrectly, reducing the accuracy of the calibration.
Mass standards included in the ProteinChip peptide standard array. Use the four peaks marked "yes" in the table below.
2. From the toolbar, select Spectra > Calibrate > Internal Calibration (or click the Internal Calibration icon ). The Internal
Calibration dialog opens. The spectrum appears in the bottom half of the window with existing peak labels appearing in red. A table of
calibrants and peaks appears in the top half of the window; the calibrant names are blank.
3. Click the Match Calibrants to Peak icon . Confirm that the peaks are labeled correctly by comparing the peak names to those in the
table.
4. Select 3 parameters from the Calibration type drop-down list and select the Weighted check box. Click the Calibrate icon . The
labels now appear in the spectra.
5. Open the Calibration equation tab. The residual plot displays the error for each calibrant in parts per million (ppm).
6. Select 300 from the Scale (ppm) drop-down list. If the error of a calibrant is greater than ±300 ppm, double-check the peak label or name
to ensure proper labeling. Close the window.
7. Click the Toggle Spectrum Table button to view all the spectrum files within your folder. Note that the Calibration type icon has
changed to green , indicating that the spectrum has been calibrated.
Save the Calibration Equation
1. Select a calibrated spectrum (green icon) from within your Pep Std Array project folder.
2. Save the equation by selecting Spectra > Calibrate > Save Calibration Equation. The Save Calibration Equation dialog opens.
3. Enter a name for the calibration equation. Include the date of creation in the name. Click OK.
Note: A calibration equation can be used to externally calibrate other spectra. For best results, use the calibration equation within 1 day
for mass ranges between 1 kD and 25 kD. For more information on external calibration, refer to the ProteinChip Data Manager Software
Manual.
Ordering Information
C70-00080 ProteinChip OQ Kit, includes 2 detector calibration arrays, 6 detector qualification arrays, 2 peptide standard arrays, CD with
protocols, instructions
C70-00081 ProteinChip System Check Kit, includes 1 detector calibration array, 1 detector qualification array, 1 peptide standard array,
CD with protocols, instruction manual
C50-30013 ProteinChip Cassettes, empty, hold 12 ProteinChip arrays, 5
C10-00001 ProteinChip Protein Calibrant Kit, includes protein MW standards (2 sets of 10 standards)
C10-00002 ProteinChip Peptide Calibrant Kit, includes peptide MW standards (2 sets of 7 standards)
C10-0006 ProteinChip Human Serum Control, 500 µl
C10-0005 ProteinChip All-In-One Peptide Standard, lyophilized, 100 spots
C10-0007 ProteinChip All-In-One Protein Standard II, lyophilized, 100 spots
Legal Notice
No part of this publication may be reproduced or transmitted in any form or by any means, electronic or mechanical, including photocopy, recording, or any information storage
or retrieval system, without permission in writing from Bio-Rad.
Bio-Rad reserves the right to modify its products and services at any time. This user guide is subject to change without notice.
Although prepared to ensure accuracy, Bio-Rad assumes no liability for errors, or for any damages resulting from the application or use of this information.
The SELDI process is covered by US patents 5,719,060, 6,225,047, 6,579,719, and 6,818,411 and other issued patents and pending applications in the US and other jurisdictions.
Copyright
©
2008 by Bio-Rad Laboratories, Inc. All rights reserved.
Label Peak for
Calibration
Peak
Name
Molecular
Mass (Da)
Yes
1
Arg8-vasopressin
1,084.25
No
2
Somatostatin
1,637.90
Yes
3
Dynorphin A
2,147.50
No
4
ACTH
2,933.50
Yes
5
Beta endorphin
3,465.00
No
6
Arg-insulin
5,963.80
Yes
7
Cytochrome C
12,230.92
 Loading...
Loading...