Page 1

SureSelect XT HS2 RNA
System
Strand-Specific RNA Library
Preparation and Target Enrichment for
the Illumina Platform
Protocol
Version A1, September 2020
SureSelect platform manufactured with Agilent
SurePrint Technology
For Research Use Only. Not for use in diagnostic
procedures.
Agilent Technologies
Page 2

Notices
© Agilent Technologies, Inc. 2020
No part of this manual may be reproduced in
any form or by any means (including electronic storage and retrieval or translation
into a foreign language) without prior agreement and written consent from Agilent
Technologies, Inc. as governed by United
States and international copyright laws.
Manual Part Number
G9989-90000
Edition
Version A1, September 2020
Printed in USA
Agilent Technologies, Inc.
5301 Stevens Creek Blvd
Santa Clara, CA 95051 USA
Acknowledgment
Oligonucleotide sequences © 2006, 2008,
and 2011 Illumina, Inc. All rights reserved.
Only for use with the Illumina sequencer
systems and associated assays.
Technical Support
For US and Canada
Call (800) 227-9770 (option 3,4,4)
Or send an e-mail to:
ngs.support@agilent.com
For all other regions
Agilent’s world-wide Sales and Support
Center contact details for your location can
be obtained at
www.agilent.com/en/contact-us/page.
Warranty
The material contained in this
document is provided “as is,” and
is subject to being changed, without notice, in future editions. Further, to the maximum extent
permitted by applicable law, Agilent disclaims all warranties,
either express or implied, with
regard to this manual and any
information contained herein,
including but not limited to the
implied warranties of merchantability and fitness for a particular
purpose. Agilent shall not be liable for errors or for incidental or
consequential damages in connection with the furnishing, use,
or performance of this document
or of any information contained
herein. Should Agilent and the
user have a separate written
agreement with warranty terms
covering the material in this document that conflict with these
terms, the warranty terms in the
separate agreement shall control.
Technology Licenses
The hardware and/or software described in
this document are furnished under a license
and may be used or copied only in accordance with the terms of such license.
Restricted Rights Legend
U.S. Government Restricted Rights. Software and technical data rights granted to
the federal government include only those
rights customarily provided to end user customers. Agilent provides this customary
commercial license in Software and technical data pursuant to FAR 12.211 (Technical
Data) and 12.212 (Computer Software) and,
for the Department of Defense, DFARS
252.227-7015 (Technical Data - Commercial
Items) and DFARS 227.7202-3 (Rights in
Commercial Computer Software or Computer Software Documentation).
Notice to Purchaser
This product is provided under an agreement between Bio-Rad Laboratories and
Agilent Technologies Inc., and the manufacture, use, sale or import of this product is
subject to U.S. Pat. No. 6,627,424 and EP
Pat. No.1 283 875 81, owned by Bio-Rad
Laboratories, Inc. Purchase of this product
conveys to the buyer the non-transferable
right to use the purchased amount of the
product and components of the product in
PCR (but not including real-time PCR) in the
Research Field (including all Applied
Research Fields, including but not limited to
forensics, animal testing, and food testing)
and in real-time PCR in the Diagnostics and
Prognostics Fields. No rights are granted for
use of this product for real-time PCR in the
Research Field, including all Applied
Research Fields (including but not limited to
forensics, animal testing and food testing).
Limited Use Label License: This product and
its use are the subject of one or more issued
and/or pending U.S. and foreign patent
applications owned by Max Planck
Gesellschaft exclusively licensed to New
England Biolabs, Inc. and sublicensed to
Agilent Technologies. The purchase of this
product from Agilent Technologies, Inc., its
affiliates, or its authorized resellers and distributors conveys to the buyer the
non-transferable right to use the purchased
amount of the product and components of
the product in research conducted by the
buyer (whether the buyer is an academic or
for profit entity). The purchase of this product does not convey a license under any
claims in the foregoing patents or patent
applications directed to producing the product. The buyer cannot sell or otherwise
transfer this product or its components to a
third party or otherwise use this product for
the following COMMERCIAL PURPOSES: (1)
use of the product or its components in
manufacturing; or (2) use of the product or
its components for therapeutic or prophylactic purposes in humans or animals.
2 SureSelect XT HS2 RNA Library Preparation and Target Enrichment
Page 3
Safety Notices
CAUTION
A CAUTION notice denotes a hazard. It calls attention to an operating procedure, practice, or the like that, if not
correctly performed or adhered to, could result in damage to the product or loss of important data. Do not proceed
beyond a CAUTION notice until the indicated conditions are fully understood and met.
WARNING
A WARNING notice denotes a hazard. It calls attention to an operating procedure, practice, or the like that, if
not correctly performed or adhered to, could result in personal injury or death. Do not proceed beyond a
WARNING notice until the indicated conditions are fully understood and met.
SureSelect XT HS2 RNA Library Preparation and Target Enrichment 3
Page 4

In this Guide...
This guide provides an optimized protocol for preparation of
target- enriched Illumina paired- end multiplexed sequencing
libraries using the SureSelect XT HS2 RNA system.
1 Before You Begin
This chapter contains information that you should read and
understand before you start an experiment.
2 Preparation of Input RNA and Conversion to cDNA
This chapter describes the steps to prepare, qualify and
fragment the RNA samples, then convert RNA to cDNA
fragments.
3 Library Preparation
This chapter describes the steps to prepare dual- indexed,
molecular- barcoded cDNA sequencing libraries for target
enrichment.
4 Hybridization and Capture
This chapter describes the steps to hybridize and capture
the prepared cDNA library using a SureSelect or ClearSeq
probe capture library.
5 Post-Capture Sample Processing for Multiplexed Sequencing
This chapter describes the steps for post- capture
amplification and guidelines for sequencing sample
preparation.
6 Reference
This chapter contains reference information, including
component kit contents and index sequences.
4 SureSelect XT HS2 RNA Library Preparation and Target Enrichment
Page 5

What’s New in Version A1
• Updates to index pair sequence tables (page 80 through
• Updates to downstream sequencing support information
• Updates to thawing conditions in Table 13 on page 30
page 87) including updates to P5 index platform
descriptions and correction of well position typographical
errors
(see Table 38 on page 67 and Note on page 79)
SureSelect XT HS2 RNA Library Preparation and Target Enrichment 5
Page 6

6 SureSelect XT HS2 RNA Library Preparation and Target Enrichment
Page 7

Content
1 Before You Begin 7
Overview of the Workflow 8
Procedural Notes 9
Safety Notes 9
Materials Required 10
Optional Materials 15
2 Preparation of Input RNA and Conversion to cDNA 17
Step 1A. Prepare and qualify FFPE RNA samples 19
Step 1B. Prepare and fragment intact RNA samples 22
Step 2. Synthesize first-strand cDNA 24
Step 3. Synthesize second-strand cDNA 25
Step 4. Purify cDNA using AMPure XP beads 26
3 Library Preparation 29
Step 1. Prepare the Ligation master mix 31
Step 2. Repair and dA-Tail the cDNA 3' ends 32
Step 3. Ligate the molecular-barcoded adaptor 34
Step 4. Purify the sample using AMPure XP beads 35
Step 5. Amplify the adaptor-ligated cDNA library 37
Step 6. Purify the amplified library with AMPure XP beads 40
Step 7. Assess quality and quantity 42
4 Hybridization and Capture 45
Step 1. Hybridize cDNA libraries to the probe 46
Step 2. Prepare streptavidin-coated magnetic beads 51
Step 3. Capture the hybridized DNA using streptavidin-coated beads 52
SureSelect XT HS2 RNA Library Preparation and Target Enrichment 5
Page 8

Contents
5 Post-Capture Sample Processing for Multiplexed Sequencing 55
Step 1. Amplify the captured libraries 56
Step 2. Purify the amplified captured libraries using AMPure XP beads 59
Step 3. Assess sequencing library DNA quantity and quality 61
Step 4. Pool samples for multiplexed sequencing 64
Step 5. Prepare sequencing samples 66
Step 6. Do the sequencing run and analyze the data 68
Sequence analysis resources 73
6 Reference 75
Kit Contents 76
SureSelect XT HS2 Index Primer Pair Information 79
Troubleshooting Guide 91
Quick Reference Protocol 94
6 SureSelect XT HS2 RNA Library Preparation and Target Enrichment
Page 9
SureSelect XT HS2 RNA System Protocol
1
Before You Begin
Overview of the Workflow 8
Procedural Notes 9
Safety Notes 9
Materials Required 10
Optional Materials 15
Make sure you read and understand the information in this chapter and
have the necessary equipment and reagents listed before you start an
experiment.
NOTE
Agilent guarantees performance and provides technical support for the SureSelect
reagents required for this workflow only when used as directed in this Protocol.
Agilent Technologies
7
Page 10
1 Before You Begin
Overview of the Workflow
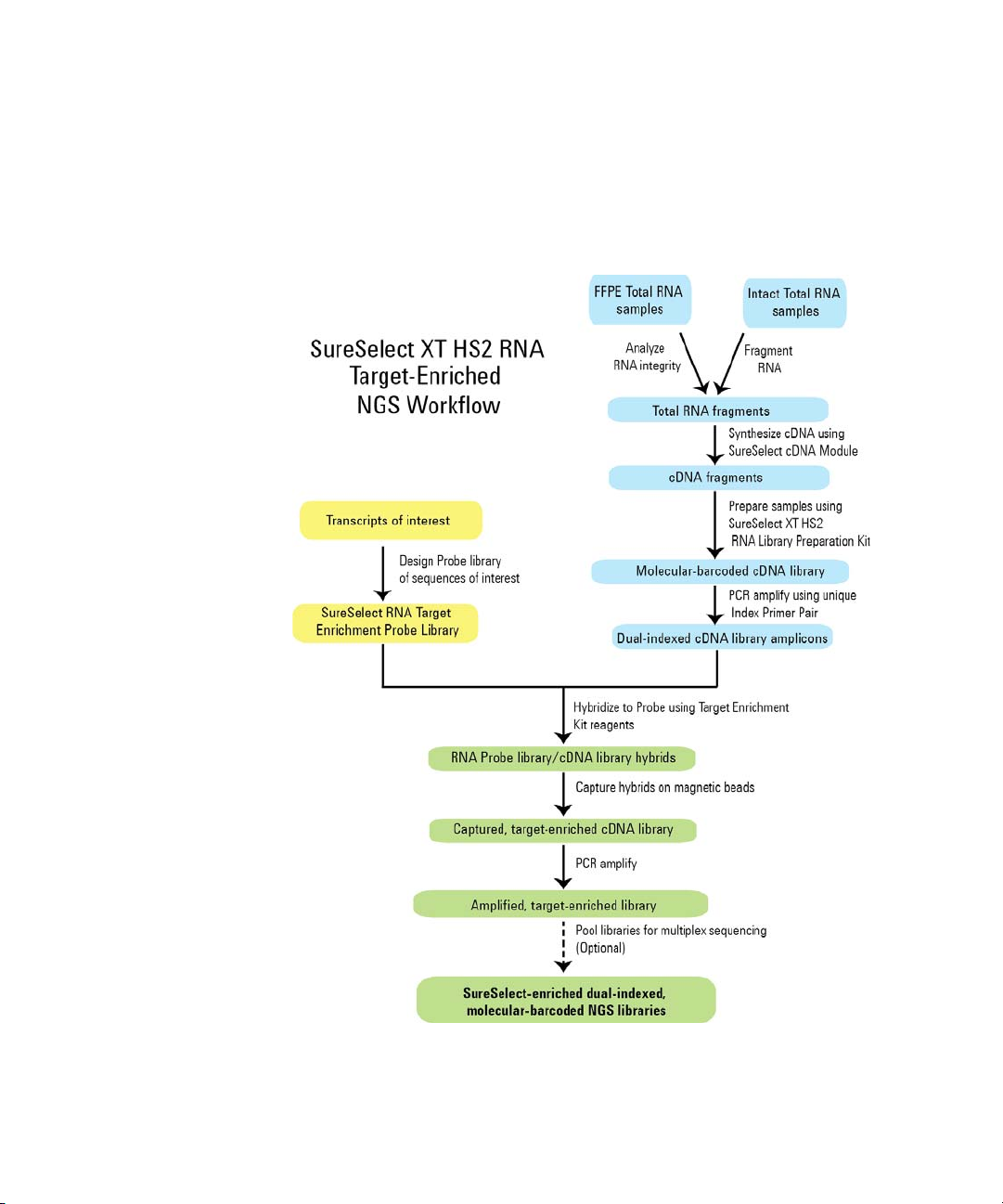
Overview of the Workflow
The SureSelect XT HS2 RNA workflow for the preparation of NGS- ready
libraries is summarized in Figure 1.
Figure 1 Overall target-enriched RNA sequencing sample preparation workflow.
8 SureSelect XT HS2 RNA Library Preparation and Target Enrichment
Page 11

Procedural Notes
• To prevent contamination of reagents by nucleases, always wear
powder- free laboratory gloves and use dedicated solutions and pipettors
with nuclease- free aerosol- resistant tips.
• Use best- practices to prevent PCR product and ribonuclease
contamination of samples throughout the workflow:
1 Assign separate pre- PCR and post- PCR work areas and use
dedicated equipment, supplies, and reagents in each area. In
particular, never use materials designated to post- PCR work areas for
pre- PCR segments of the workflow.
2 Maintain clean work areas. Clean the surfaces that pose the highest
risk of contamination daily using a 10% bleach solution, or
equivalent.
3 Always use dedicated pre- PCR pipettors with nuclease- free
aerosol- resistant tips to pipette dedicated pre- PCR solutions.
4 Wear powder- free gloves. Use good laboratory hygiene, including
changing gloves after contact with any potentially- contaminated
surfaces.
• For each protocol step that requires removal of tube cap strips, reseal
the tubes with a fresh strip of domed caps. Cap deformation may result
from exposure of the cap strips to the heated lid of the thermal cycler
and from other procedural steps. Reuse of strip caps can cause sample
loss, sample contamination, or imprecision in sample temperatures
during thermal cycler incubation steps.
• In general, follow Biosafety Level 1 (BSL1) safety rules.
• Possible stopping points, where samples may be stored at 4°C or –20°C,
are marked in the protocol. Do not subject the samples to multiple
freeze/thaw cycles.
Before You Begin 1
Procedural Notes
Safety Notes
CAUTION
SureSelect XT HS2 RNA Library Preparation and Target Enrichment 9
• Wear appropriate personal protective equipment (PPE) when working in the
laboratory.
Page 12

1 Before You Begin
Materials Required
Materials Required
Materials required to complete the SureSelect XT HS2 RNA protocol are
listed in the tables in this section. Select the preferred SureSelect XT HS2
RNA Reagent Kit format from Table 1, and a target enrichment probe from
Table 2. Then refer to Table 3 through Table 5 for additional materials
needed to complete the protocols using the selected kit format/RNA
sample type.
Table 1 SureSelect XT HS2 RNA Reagent Kit Varieties
Description Kit Part Number
16 Reaction Kit
SureSelect XT HS2 RNA Reagent Kit G9989A (with Index Pairs 1–16) G9991A (with Index Pairs 1–96)
SureSelect XT HS2 RNA Reagent Kit with
AMPure
®
XP/Streptavidin Beads
‡
G9990A (with Index Pairs 1–16) G9992A (with Index Pairs 1–96)
*
96 Reaction Kit
G9991B (with Index Pairs 97–192)
G9991C (with Index Pairs 193–288)
G9991D (with Index Pairs 289–384)
G9992B (with Index Pairs 97–192)
G9992C (with Index Pairs 193–288)
G9992D (with Index Pairs 289–384)
†
* 16-reaction kits contain enough reagents for 2 runs containing 8 samples per run.
† 96-reaction kits contain enough reagents for 4 runs containing 24 samples per run.
‡ AMPure, Beckman, and Beckman Coulter are trademarks or registered trademarks of Beckman Coulter, Inc.
10 SureSelect XT HS2 RNA Library Preparation and Target Enrichment
Page 13

Before You Begin 1
Materials Required
Table 2 Compatible Probes
Probe Capture Library Design Target 16 Reactions 96 Reactions
Pre-designed Probes
SSel XT HS and XT Low Input Human All Exon V7 Genome 5191-4028 5191-4029
SureSelect XT Clinical Research Exome V2 Genome 5190-9491 5190-9492
SureSelect XT Mouse All Exon Genome 5190-4641 5190-4642
ClearSeq Comprehensive Cancer XT Genome 5190-8011 5190-8012
ClearSeq Inherited Disease XT Genome 5190-7518 5190-7519
ClearSeq RNA Kinome Transcriptome 5190-4801 5190-4802
Custom Probes
SureSelect Custom Tier1 1–499 kb
SureSelect Custom Tier2 0.5 –2.9 Mb
SureSelect Custom Tier3 3 –5.9 Mb
SureSelect Custom Tier4 6 –11.9 Mb
SureSelect Custom Tier5 12–24 Mb
Pre-designed Probes customized with additional Plus custom content
SSel XT HS and XT Low Input Human All Exon V7 Plus 1 Genome
SSel XT HS and XT Low Input Human All Exon V7 Plus 2 Genome
SureSelect XT Clinical Research Exome V2 Plus 1 Genome
SureSelect XT Clinical Research Exome V2 Plus 2 Genome
ClearSeq Comprehensive Cancer Plus XT Genome
ClearSeq Inherited Disease Plus XT Genome
*
Custom probes may designed for either genomic or
transcriptomic targets. Please contact the SureSelect
support team (see page 2) or your local representative
for assistance with custom probe design and ordering
for RNA library target enrichment.
Please visit the SureDesign
website to design the customized
Plus content and obtain ordering
information. Contact the
SureSelect support team (see
page 2) or your local
representative if you need
assistance.
* Custom Probes designed August 2020 or later are produced using an updated manufacturing process; design-size Tier is
shown on labeling for these products. Custom Probes designed and ordered prior to August 2020 may be reordered, with
these probes produced using the legacy manufacturing process; design-size Tier is not shown on labeling for the legacy-process products. Custom Probes of both categories use the same optimized target enrichment protocols detailed in this publication.
SureSelect XT HS2 RNA Library Preparation and Target Enrichment 11
Page 14

1 Before You Begin
Materials Required
Table 3 Required Reagents
Description Vendor and Part Number Notes
1X Low TE Buffer Thermo Fisher Scientific p/n
12090-015, or equivalent
100% Ethanol (Ethyl Alcohol, 200 proof) Millipore p/n EX0276 —
Nuclease-free Water Thermo Fisher Scientific p/n
AM9930
AMPure® XP Kit
5 ml
60 ml
450 ml
Dynabeads MyOne Streptavidin T1
2 ml
10 ml
50 ml
QPCR Human Reference Total RNA Agilent p/n 750500 Control input RNA (optional)
Beckman Coulter Genomics
p/n A63880
p/n A63881
p/n A63882
Thermo Fisher Scientific
p/n 65601
p/n 65602
p/n 65604D
10 mM Tris-HCl, pH 7.5-8.0, 0.1 mM EDTA
Water should not be DEPC-treated
Separate purchase not required for use
with SureSelect XT HS2 RNA Reagent
Kits that include SureSelect DNA
®
AMPure
Streptavidin Beads (Agilent p/n G9990A,
G9992A, G9992B, G9992C, or G9992D)
XP Beads and SureSelect
12 SureSelect XT HS2 RNA Library Preparation and Target Enrichment
Page 15

Before You Begin 1
Materials Required
CAUTION
Sample volumes exceed 0.2 ml in certain steps of this protocol. Make sure that the
plasticware
used with the selected thermal cycler holds 0.25 ml per well.
Table 4 Required Equipment
Description Vendor and Part Number
Thermal Cycler with 96-well, 0.2 ml block Various suppliers
Plasticware compatible with the selected thermal cycler:
96-well plates or 8-well strip tubes
Tube cap strips, domed
Low-adhesion tubes (RNase, DNase, and DNA-free)
1.5 mL
0.5 mL
Microcentrifuge Eppendorf microcentrifuge, model 5417C or equivalent
Plate or strip tube centrifuge Labnet International MPS1000 Mini Plate Spinner, p/n
96-well plate mixer Eppendorf ThermoMixer C, p/n 5382000023 and Eppendorf
Small-volume spectrophotometer NanoDrop 2000, Thermo Fisher Scientific p/n ND-2000 or
Multichannel pipette Rainin Pipet-Lite Multi Pipette or equivalent
Single channel pipettes (10-, 20-, 200-, and 1000-µl capacity) Rainin Pipet-Lite Pipettes or equivalent
Sterile, nuclease-free aerosol barrier pipette tips general laboratory supplier
Vortex mixer general laboratory supplier
Ice bucket general laboratory supplier
Powder-free gloves general laboratory supplier
Magnetic separator Thermo Fisher Scientific p/n 12331D or equivalent
Consult the thermal cycler manufacturer’s
recommendations
USA Scientific
p/n 1415-2600
p/n 1405-2600
C1000 (requires adapter, p/n C1000-ADAPT, for use with
strip tubes) or equivalent
SmartBlock 96 PCR, p/n 5306000006, or equivalent
equivalent
*
* Select a magnetic separator configured to collect magnetic particles on one side of each well. Do not use a magnetic sep-
arator configured to collect the particles in a ring formation.
SureSelect XT HS2 RNA Library Preparation and Target Enrichment 13
Page 16

1 Before You Begin
Materials Required
Table 5 Nucleic Acid Analysis Platform Options--Select One
Description Vendor and Part Number
Agilent 4200/4150 TapeStation
Consumables:
96-well sample plates
96-well plate foil seals
8-well tube strips
8-well tube strip caps
RNA ScreenTape
RNA ScreenTape Sample Buffer
RNA ScreenTape Ladder
High Sensitivity RNA ScreenTape
High Sensitivity RNA ScreenTape Sample Buffer
High Sensitivity RNA ScreenTape Ladder
D1000 ScreenTape
D1000 Reagents
High Sensitivity D1000 ScreenTape
High Sensitivity D1000 Reagents
Agilent 2100 Bioanalyzer Instrument
Agilent 2100 Expert SW Laptop Bundle (optional)
Consumables:
RNA 6000 Pico Kit
RNA 6000 Nano Kit
DNA 1000 Kit
High Sensitivity DNA Kit
Agilent 5200/5300/5400 Fragment Analyzer Instrument
Consumables:
RNA Kit (15NT)
HS RNA Kit (15NT)
NGS Fragment Kit (1-6000 bp)
HS NGS Fragment Kit (1-6000 bp)
Agilent p/n G2991AA/G2992AA
Agilent p/n 5042-8502
Agilent p/n 5067-5154
Agilent p/n 401428
Agilent p/n 401425
Agilent p/n 5067-5576
Agilent p/n 5067-5577
Agilent p/n 5067-5578
Agilent p/n 5067-5579
Agilent p/n 5067-5580
Agilent p/n 5067-5581
Agilent p/n 5067-5582
Agilent p/n 5067-5583
Agilent p/n 5067-5584
Agilent p/n 5067-5585
Agilent p/n G2939BA
Agilent p/n G2953CA
Agilent p/n 5067-1513
Agilent p/n 5067-1511
Agilent p/n 5067-1504
Agilent p/n 5067-4626
Agilent p/n M5310AA/M5311AA/M5312AA
p/n DNF-471-0500
p/n DNF-472-0500
p/n DNF-473-0500
p/n DNF-474-0500
14 SureSelect XT HS2 RNA Library Preparation and Target Enrichment
Page 17

Before You Begin 1
Optional Materials
Optional Materials
Table 6 Supplier Information for Optional Materials
Description Vendor and Part Number Purpose
Tween 20 Sigma-Aldrich p/n P9416-50ML Sequencing library
storage (see page 69)
Optical Caps, 8× strip (flat) Consult the thermal cycler
manufacturer’s recommendations
MicroAmp Clear Adhesive Film Thermo Fisher Scientific p/n 4311971 Improved sealing for
PlateLoc Thermal Microplate Sealer with Small Hotplate
and Peelable Aluminum Seal for PlateLoc Sealer
* Flat strip caps may be used instead of domed strip caps for protocol steps performed outside of the thermal cycler. Adhesive
film may be applied over the flat strip caps for improved sealing properties.
Please contact the SureSelect support
team (see page 2) or your local
representative for ordering information
Sealing wells for
protocol steps
performed outside of
the thermal cycler
flat strip caps*
Sealing wells for
protocol steps
performed inside or
outside of the thermal
cycler
*
SureSelect XT HS2 RNA Library Preparation and Target Enrichment 15
Page 18

1 Before You Begin
Optional Materials
16 SureSelect XT HS2 RNA Library Preparation and Target Enrichment
Page 19

SureSelect XT HS2 RNA System Protocol
2
Preparation of Input RNA and
Conversion to cDNA
Step 1A. Prepare and qualify FFPE RNA samples 19
Step 1B. Prepare and fragment intact RNA samples 22
Step 2. Synthesize first-strand cDNA 24
Step 3. Synthesize second-strand cDNA 25
Step 4. Purify cDNA using AMPure XP beads 26
This chapter describes the steps to prepare input RNA samples, including
RNA fragmentation when required, and the steps to convert the RNA
fragments to strand- specific cDNA prior to sequencing library preparation
and target enrichment.
The protocol is compatible with both intact RNA prepared from fresh or
fresh frozen samples and lower- quality RNA prepared from FFPE samples.
For FFPE- derived RNA samples, begin the protocol using “Step 1A.
Prepare and qualify FFPE RNA samples” on page 19. For intact RNA
samples, begin the protocol using “Step 1B. Prepare and fragment intact
RNA samples” on page 22.
RNA sequencing library preparation requires RNA fragments sized
appropriately for the NGS workflow. In this section of the protocol, intact
total RNA samples are chemically- fragmented by treatment with metal
ions present in the 2X Priming Buffer at elevated temperature.
FFPE- derived RNA samples are already sufficiently fragmented. The FFPE
samples must be combined with the same 2X Priming Buffer, but the
mixtures are held on ice, preventing further fragmentation of the
FFPE- derived RNA.
Protocols in this section for both intact RNA and FFPE sample types are
applicable to either 2 x 100 bp or 2 x 150 bp read- length sequencing.
Agilent Technologies
17
Page 20

2 Preparation of Input RNA and Conversion to cDNA
The protocol steps in this section use the components listed in Table 7.
Thaw and mix each component as directed in Table 7 before use (refer to
the Where Used column). Remove the AMPure XP beads from cold storage
and equilibrate to room temperature for at least 30 minutes in preparation
for use on page 26. Do not freeze the beads at any time.
Table 7 Reagents thawed before use in protocol
Kit Component Storage Location Thawing
Conditions
2X Priming Buffer (tube with
purple cap)
First Strand Master Mix (amber
tube with amber cap)
Second Strand Enzyme Mix
(tube with blue cap or bottle)
Second Strand Oligo Mix (tube
with yellow cap)
* The First Strand Master Mix contains actinomycin-D and is provided ready-to-use. Keep the reagent in the supplied amber
vial to protect the contents from exposure to light.
*
SureSelect cDNA Module
(Pre PCR), –20°C
SureSelect cDNA Module
(Pre PCR), –20°C
SureSelect cDNA Module
(Pre PCR), –20°C
SureSelect cDNA Module
(Pre PCR), –20°C
Thaw on ice then
keep on ice
Thaw on ice for
30 minutes then
keep on ice
Thaw on ice then
keep on ice
Thaw on ice then
keep on ice
Mixing
Method
Vortexing page 21 (FFPE RNA) OR
Vortexing page 24
Vortexing page 25
Vortexing page 25
Where Used
page 23 (intact RNA)
18 SureSelect XT HS2 RNA Library Preparation and Target Enrichment
Page 21

Preparation of Input RNA and Conversion to cDNA 2
Step 1A. Prepare and qualify FFPE RNA samples
Step 1A. Prepare and qualify FFPE RNA samples
The instructions in this section are for FFPE- derived RNA samples. For
intact (non- FFPE) RNA samples, instead follow the instructions in “Step
1B. Prepare and fragment intact RNA samples” on page 22.
Prepare total RNA from each FFPE sample in the run. The library
preparation protocol requires 10–200 ng of FFPE total RNA in a 10 µl
volume of nuclease- free water.
Consider preparing an additional sequencing library in parallel, using a
high- quality control RNA sample, such as Agilent’s QPCR Human
Reference Total RNA (p/n 750500). Use of this control is especially
recommended during the first run of the protocol, to verify that all
protocol steps are being successfully performed. Routine use of this
control is helpful for any required troubleshooting, in order to differentiate
any performance issues related to RNA input from other factors.
Before you begin the library preparation protocol, assess the initial quality
of each sample in order to determine the appropriate reaction conditions
at several steps in the workflow. Use the steps below to qualify each FFPE
total RNA sample.
1 Use a small- volume spectrophotometer to determine sample absorbance
at 260 nm, 280 nm, and 230 nm. Determine the RNA concentration and
the 260/280 and 260/230 absorbance ratio values for the sample.
High- quality RNA samples are indicated by values of approximately 1.8
to 2.0 for both ratios. Ratios with significant deviation from 2.0 indicate
the presence of organic or inorganic contaminants, which may require
further purification or may indicate that the sample is not suitable for
use in RNA target enrichment applications.
SureSelect XT HS2 RNA Library Preparation and Target Enrichment 19
Page 22

2 Preparation of Input RNA and Conversion to cDNA
Step 1A. Prepare and qualify FFPE RNA samples
2 Examine the starting size distribution of RNA in the sample using one
of the RNA qualification systems described in Table 8. Select the
specific assay appropriate for your sample based on the RNA
concentration determined in step 1 on page 19.
Determine the DV200 (percentage of RNA in the sample that is >200 nt)
using the analysis mode described in Table 8. RNA molecules must be
>200 nt for efficient conversion to cDNA library.
Table 8 RNA qualification platforms
Analysis Instrument RNA Qualification Assay Analysis to Perform
4200/4150 TapeStation RNA ScreenTape or High Sensitivity
RNA ScreenTape
2100 Bioanalyzer RNA 6000 Pico Chip or NanoChip Smear/Region analysis using 2100 Expert Software
5200 Fragment Analyzer RNA Kit (15NT) or HS RNA Kit
(15NT)
Region analysis using TapeStation Analysis Software
Analysis using ProSize Data Analysis Software
NOTE
Grading of FFPE RNA quality by RNA Integrity Number (RIN) is not recommended for this
application.
3 Grade each RNA sample based on the percentage of RNA in the sample
>200 nucleotides, according to Table 9.
Table 9 Classification of FFPE RNA samples based on starting RNA size
Grade DV200 Recommended input
amount
Good FFPE RNA >50% 200 ng 10 ng
Poor FFPE RNA 20% to 50% 200 ng 50 ng
Inapplicable FFPE RNA <20% Not recommended for further processing
* For optimal results, prepare libraries from poor-grade FFPE RNA samples using a minimum of 50 ng
input RNA. Libraries may be prepared from 10–50 ng poor-grade FFPE RNA with potential negative
impacts on yield or NGS performance.
Minimum input
amount
*
4 Place 10 µl of each sample, containing 10–200 ng of FFPE total RNA in
nuclease- free water, into wells of a thermal cycler- compatible strip tube
or PCR plate.
Poor- quality FFPE samples should contain at least 50 ng RNA.
20 SureSelect XT HS2 RNA Library Preparation and Target Enrichment
Page 23

Preparation of Input RNA and Conversion to cDNA 2
Step 1A. Prepare and qualify FFPE RNA samples
5 Add 10 µl of 2X Priming Buffer to each sample well.
6 Mix well by pipetting up and down 15–20 times or seal the wells and
vortex at high speed for 5–10 seconds. Spin briefly to collect the liquid
then place the RNA samples on ice.
NOTE
All samples, including highly degraded FFPE samples, must be combined with 2X Priming
Buffer, which supplies the random primers for cDNA synthesis. FFPE RNA samples are not
subjected to the high-temperature incubation step used for fragmentation in this buffer.
7 Proceed immediately to “Step 2. Synthesize first- strand cDNA” on
page 24.
SureSelect XT HS2 RNA Library Preparation and Target Enrichment 21
Page 24

2 Preparation of Input RNA and Conversion to cDNA
Step 1B. Prepare and fragment intact RNA samples
Step 1B. Prepare and fragment intact RNA samples
The instructions in this section are for intact RNA prepared from fresh or
fresh frozen samples. For FFPE- derived RNA samples, instead follow the
instructions in “Step 1A. Prepare and qualify FFPE RNA samples” on
page 19.
Consider preparing an additional sequencing library in parallel, using a
high- quality control RNA sample, such as Agilent’s QPCR Human
Reference Total RNA (p/n 750500). Use of this control is especially
recommended during the first run of the protocol, to verify that all
protocol steps are being successfully performed. Routine use of this
control is helpful for any required troubleshooting, in order to differentiate
any performance issues related to RNA input from other factors.
The 2X Priming Buffer used in this step includes both fragmentation
agents and primers used for cDNA synthesis in the following steps. The
fragmentation conditions shown in this section are appropriate for both
2 x 100 bp and 2 x 150 bp NGS read- length workflows.
1 Prepare total RNA from each sample in the run. The library preparation
protocol requires 10–200 ng of intact total RNA in a 10 µl volume of
nuclease- free water.
Verify the RNA concentration and quality using a small volume
spectrophotometer and one of the RNA qualification platforms listed in
Table 5 on page 14.
2 Preprogram a thermal cycler with the program in Table 10. Immediately
pause the program, and keep paused until samples are loaded in step 6.
Table 10 Thermal cycler program for fragmentation of intact RNA samples
Step Temperature Time
Step 1 94°C 4 minutes
Step 2 4°C 1 minute
Step 3 4°C Hold
* Use a reaction volume setting of 20 l, if required for thermal cycler set up.
NOTE
22 SureSelect XT HS2 RNA Library Preparation and Target Enrichment
When using the SureCycler 8800 thermal cycler, the heated lid may be left on (default
setting) throughout the RNA library preparation incubation steps. The heated lid must be on
during the amplification and hybridization steps on page 38, page 47 and page 57.
*
Page 25

Preparation of Input RNA and Conversion to cDNA 2
Step 1B. Prepare and fragment intact RNA samples
3 Place 10 µl of each sample, containing 10–200 ng total RNA in
nuclease- free water, into wells of a thermal cycler- compatible strip tube
or PCR plate.
4 Add 10 µl of 2X Priming Buffer to each sample well.
5 Mix well by pipetting up and down 15–20 times or seal the wells and
vortex at high speed for 5–10 seconds. Spin briefly to collect the liquid.
6 Place the samples in the thermal cycler, and resume the thermal cycling
program in Table 10 for RNA fragmentation.
7 Once the thermal cycler program in Table 10 reaches the 4°C Hold
step, transfer the fragmented RNA sample plate or strip tube from the
thermal cycler to ice or a cold block. Proceed immediately to “Step 2.
Synthesize first- strand cDNA” on page 24.
SureSelect XT HS2 RNA Library Preparation and Target Enrichment 23
Page 26

2 Preparation of Input RNA and Conversion to cDNA
Step 2. Synthesize first-strand cDNA
Step 2. Synthesize first-strand cDNA
CAUTION
The First Strand Master Mix used in this step is viscous. Mix thoroughly by vortexing at
high speed for 5 seconds before removing an aliquot for use and after combining with
other solutions. Pipetting up and down is not sufficient to mix this reagent.
The First Strand Master Mix is provided with actinomycin-D already supplied in the
mixture. Do not supplement with additional actinomycin-D.
1 Preprogram a thermal cycler with the program in Table 11. Immediately
pause the program, and keep paused until samples are loaded in step 5.
Table 11 Thermal cycler program for first-strand cDNA synthesis
Step Temperature Time
Step 1 25°C 10 minutes
Step 2 37°C 40 minutes
Step 3 4°C Hold
* Use a reaction volume setting of 28 l, if required for thermal cycler set up.
2 Vortex the thawed vial of First Strand Master Mix for 5 seconds at high
speed to ensure homogeneity.
3 Add 8.5 µl of First Strand Master Mix to each RNA sample well.
4 Mix well by pipetting up and down 15–20 times or seal the wells and
vortex at high speed for 5–10 seconds. Spin briefly to collect the liquid.
5 Place the samples in the thermal cycler, and resume the program in
Table 11.
*
24 SureSelect XT HS2 RNA Library Preparation and Target Enrichment
Page 27

Preparation of Input RNA and Conversion to cDNA 2
Step 3. Synthesize second-strand cDNA
Step 3. Synthesize second-strand cDNA
CAUTION
The Second Strand Enzyme Mix used in this step is viscous. Mix thoroughly by
vortexing at high speed for 5 seconds before removing an aliquot for use and after
combining with other solutions. Pipetting up and down is not sufficient to mix this
reagent.
1 Once the thermal cycler program in Table 11 begins the 4°C hold step,
transfer the samples to ice.
2 Preprogram the thermal cycler with the program in Table 12.
Immediately pause the program, and keep paused until samples are
loaded in step 7
Table 12 Thermal cycler program for second-strand synthesis
Step Temperature Time
Step 1 16°C 60 minutes
Step 2 4°C Hold
* Use a reaction volume setting of 58 l, if required for thermal cycler set up.
3 Vortex the thawed vials of Second Strand Enzyme Mix and of Second
Strand Oligo Mix at high speed for 5 seconds to ensure homogeneity.
4 Add 25 µl of Second Strand Enzyme Mix to each sample well. Keep on
ice.
5 Add 5 µl of Second Strand Oligo Mix to each sample well, for a total
reaction volume of 58.5 µl. Keep on ice.
6 Mix well by pipetting up and down 15–20 times or seal the wells and
vortex at high speed for 5–10 seconds. Spin briefly to collect the liquid.
7 Place the plate or strip tubes in the thermal cycler, and resume the
program in Table 12.
.
*
SureSelect XT HS2 RNA Library Preparation and Target Enrichment 25
Page 28

2 Preparation of Input RNA and Conversion to cDNA
Step 4. Purify cDNA using AMPure XP beads
Step 4. Purify cDNA using AMPure XP beads
1 Verify that the AMPure XP beads have been held at room temperature
for at least 30 minutes before use.
2 Prepare 400 µl of 70% ethanol per sample, plus excess, for use in
step 9.
NOTE
The freshly-prepared 70% ethanol may be used for subsequent purification steps run on the
same day. The complete RNA Library Preparation protocol requires 1.2 mL of fresh 70%
ethanol per sample and the Target Enrichment protocol requires an additional 0.4 mL of
fresh 70% ethanol per sample.
3 Mix the bead suspension well so that the suspension appears
homogeneous and consistent in color.
4 Transfer the samples in the PCR plate or strip tube to room
temperature, then add 105 µl of the homogeneous bead suspension to
each cDNA sample well.
5 Pipette up and down 15–20 times or cap the wells and vortex at high
speed for 5–10 seconds to mix. If the beads have splashed into the well
caps, spin briefly to collect the samples, being careful not to pellet the
beads.
6 Incubate samples for 5 minutes at room temperature.
7 Put the plate or strip tube into a magnetic separation device. Wait for
the solution to clear (approximately 2 to 5 minutes).
8 Keep the plate or strip tube in the magnetic stand. Carefully remove
and discard the cleared solution from each well. Do not touch the beads
while removing the solution.
9 Continue to keep the plate or strip tube in the magnetic stand while
you dispense 200 µl of fresh 70% ethanol in each sample well.
10 Wait for 1 minute to allow any disturbed beads to settle, then remove
the ethanol.
11 Repeat step 9 and step 10 once for a total of two washes.
12 Seal the wells with strip caps, then briefly spin the samples to collect
the residual ethanol. Return the plate or strip tube to the magnetic
stand for 30 seconds. Remove the residual ethanol with a P20 pipette.
26 SureSelect XT HS2 RNA Library Preparation and Target Enrichment
Page 29
Preparation of Input RNA and Conversion to cDNA 2
Step 4. Purify cDNA using AMPure XP beads
13 Dry the samples by placing the unsealed plate or strip tube on the
thermal cycler, set to hold samples at 37°C, until the residual ethanol
has just evaporated (up to 2 minutes).
NOTE
Stopping Point If you do not continue to the next step, seal the wells and store at 4°C
Do not dry the bead pellet to the point that the pellet appears cracked during any of the
bead drying steps in the protocol. Elution efficiency is significantly decreased when the
bead pellet is excessively dried.
14 Add 52 µl nuclease- free water to each sample well.
15 Seal the wells with strip caps, then vortex the plate or strip tube for
5 seconds. Verify that all beads have been resuspended, with no visible
clumps in the suspension or bead pellets retained on the sides of the
wells. Briefly spin to collect the liquid, being careful not to pellet the
beads.
16 Incubate for 2 minutes at room temperature.
17 Put the plate or strip tube in the magnetic stand and leave until the
solution is clear (up to 5 minutes).
18 Remove 50 µl of cleared supernatant to a fresh PCR plate or strip tube
sample well and keep on ice. You can discard the beads at this time.
overnight or at –20°C for prolonged storage.
SureSelect XT HS2 RNA Library Preparation and Target Enrichment 27
Page 30

2 Preparation of Input RNA and Conversion to cDNA
Step 4. Purify cDNA using AMPure XP beads
28 SureSelect XT HS2 RNA Library Preparation and Target Enrichment
Page 31

SureSelect XT HS2 RNA System Protocol
3
Library Preparation
Step 1. Prepare the Ligation master mix 31
Step 2. Repair and dA-Tail the cDNA 3' ends 32
Step 3. Ligate the molecular-barcoded adaptor 34
Step 4. Purify the sample using AMPure XP beads 35
Step 5. Amplify the adaptor-ligated cDNA library 37
Step 6. Purify the amplified library with AMPure XP beads 40
Step 7. Assess quality and quantity 42
This chapter describes the steps to prepare cDNA NGS libraries for
sequencing using the Illumina paired- read platform. For each sample to be
sequenced, an individual dual- indexed and molecular- barcoded library is
prepared.
Protocol steps in this section use the components listed in Table 13. Thaw
and mix each component as directed in Table 13 before use (refer to the
Where Used column). Remove the AMPure XP beads from cold storage and
equilibrate to room temperature for at least 30 minutes in preparation for
use on page 35. Do not freeze the beads at any time.
To process multiple samples, prepare reagent mixtures with overage at
each step, without the cDNA library sample. Mixtures for preparation of
8 or 24 samples (including excess) are shown in tables as examples.
Agilent Technologies
29
Page 32

3 Library Preparation
Table 13 Reagents thawed before use in protocol
Kit Component Storage Location Thawing Conditions Mixing Method Where Used
Ligation Buffer (purple cap
or bottle)
T4 DNA Ligase (blue cap) SureSelect XT HS2 RNA Library
End Repair-A Tailing Buffer
(yellow cap or bottle)
End Repair-A Tailing
Enzyme Mix (orange cap)
XT HS2 RNA Adaptor Oligo
Mix (green cap)
SureSelect XT HS2 RNA Library
Preparation Kit for ILM (Pre PCR),
–20°C
Preparation Kit for ILM (Pre PCR),
–20°C
SureSelect XT HS2 RNA Library
Preparation Kit for ILM (Pre PCR),
–20°C
SureSelect XT HS2 RNA Library
Preparation Kit for ILM (Pre PCR),
–20°C
SureSelect XT HS2 RNA Library
Preparation Kit for ILM (Pre PCR),
–20°C
Thaw on ice (may
require >20 minutes)
then keep on ice
Place on ice just before
use
Thaw on ice (may
require >20 minutes)
then keep on ice
Place on ice just before
use
Thaw on ice then keep
on ice
Vortexing page 31
Inversion page 31
Vortexing page 33
Inversion page 33
Vortexing page 34
30 SureSelect XT HS2 RNA Library Preparation and Target Enrichment
Page 33

Library Preparation 3
Step 1. Prepare the Ligation master mix
Step 1. Prepare the Ligation master mix
Prepare the Ligation master mix to allow equilibration to room
temperature before use on page 34. Initiate this step before starting the
End Repair/dA- tailing protocol; leave samples on ice while completing this
step.
1 Vortex the thawed vial of Ligation Buffer for 15 seconds at high speed
to ensure homogeneity.
CAUTION
The Ligation Buffer used in this step is viscous. Mix thoroughly by vortexing at high
speed for 15 seconds before removing an aliquot for use. When combining with other
reagents, mix well by pipetting up and down 15–20 times using a pipette set to at least
80% of the mixture volume or by vortexing at high speed for 10–20 seconds.
Use flat top vortex mixers when vortexing strip tubes or plates throughout the protocol.
If reagents are mixed by vortexing, visually verify that adequate mixing is occurring.
2 Prepare the appropriate volume of Ligation master mix by combining
the reagents in Table 14.
Slowly pipette the Ligation Buffer into a 1.5- ml tube, ensuring that the
full volume is dispensed. Slowly add the T4 DNA Ligase, rinsing the
enzyme tip with buffer solution after addition. Mix well by slowly
pipetting up and down 15–20 times or seal the tube and vortex at high
speed for 10–20 seconds. Spin briefly to collect the liquid.
Keep at room temperature for 30–45 minutes before use on page 34.
Table 14 Preparation of Ligation master mix
Reagent Volume for 1 reaction Volume for 8 reactions
(includes excess)
Ligation Buffer (purple cap or bottle) 23 µl 207 µl 575 µl
T4 DNA Ligase (blue cap) 2 µl 18 µl 50 µl
*
Volume for 24 reactions
(includes excess)
†
Total 25 µl 225 µl 625 µl
* The minimum supported run size for 16-reaction kits is 8 samples per run, with kits containing enough reagents for 2 runs of
8 samples each.
† The minimum supported run size for 96-reaction kits is 24 samples per run, with kits containing enough reagents for 4 runs
of 24 samples each.
SureSelect XT HS2 RNA Library Preparation and Target Enrichment 31
Page 34

3 Library Preparation
Step 2. Repair and dA-Tail the cDNA 3' ends
Step 2. Repair and dA-Tail the cDNA 3' ends
1 Preprogram a thermal cycler with the program in Table 15. Immediately
pause the program, and keep paused until samples are loaded in step 5.
CAUTION
Table 15 Thermal cycler program for End Repair/dA-Tailing
Step Temperature Time
Step 1 20°C 15 minutes
Step 2 72°C 15 minutes
Step 3 4°C Hold
* Use a reaction volume setting of 70 l, if required for thermal cycler set up.
2 Vortex the thawed vial of End Repair- A Tailing Buffer for 15 seconds at
high speed to ensure homogeneity. Visually inspect the solution; if any
solids are observed, continue vortexing until all solids are dissolved.
*
The End Repair-A Tailing Buffer used in this step must be mixed thoroughly by
vortexing at high speed for 15 seconds before removing an aliquot for use. When
combining with other reagents, mix well either by pipetting up and down 15–20 times
using a pipette set to at least 80% of the mixture volume or by vortexing at high speed
for 5–10 seconds.
3 Prepare the appropriate volume of dA- Tailing master mix, by combining
the reagents in Table 16.
Slowly pipette the End Repair- A Tailing Buffer into a 1.5- ml tube,
ensuring that the full volume is dispensed. Slowly add the End
Repair- A Tailing Enzyme Mix, rinsing the enzyme tip with buffer
solution after addition. Mix well by pipetting up and down 15–20 times
or seal the tube and vortex at high speed for 5–10 seconds. Spin briefly
to collect the liquid and keep on ice.
32 SureSelect XT HS2 RNA Library Preparation and Target Enrichment
Page 35

Table 16 Preparation of End Repair/dA-Tailing master mix
Library Preparation 3
Step 2. Repair and dA-Tail the cDNA 3' ends
Reagent Volume for 1 reaction Volume for 8 reactions
(includes excess)
End Repair-A Tailing Buffer (yellow cap or bottle) 16 µl 144 µl 400 µl
End Repair-A Tailing Enzyme Mix (orange cap) 4 µl 36 µl 100 µl
Total 20 µl 180 µl 500 µl
Volume for 24 reactions
(includes excess)
4 Add 20 µl of the End Repair/dA- Tailing master mix to each sample well
containing approximately 50 µl of purified cDNA sample. Mix by
pipetting up and down 15–20 times using a pipette set to 50 µl or cap
the wells and vortex at high speed for 5–10 seconds.
5 Briefly spin the samples, then immediately place the plate or strip tube
in the thermal cycler and resume the thermal cycling program in
Table 15.
SureSelect XT HS2 RNA Library Preparation and Target Enrichment 33
Page 36

3 Library Preparation
Step 3. Ligate the molecular-barcoded adaptor
Step 3. Ligate the molecular-barcoded adaptor
1 Once the thermal cycler reaches the 4°C Hold step, transfer the samples
to ice while setting up this step.
2 Preprogram a thermal cycler with the program in Table 17. Immediately
pause the program, and keep paused until samples are loaded in step 5.
NOTE
Table 17 Thermal cycler program for Ligation
Step Temperature Time
Step 1 20°C 30 minutes
Step 2 4°C Hold
* Use a reaction volume setting of 100 l, if required for thermal cycler set up.
3 To each end- repaired/dA- tailed DNA sample (approximately 70 µl), add
25 µl of the Ligation master mix that was prepared on page 31 and
kept at room temperature. Mix by pipetting up and down at least
10 times using a pipette set to 70 µl or cap the wells and vortex at
high speed for 5–10 seconds. Briefly spin the samples.
4 Add 5 µl of XT HS2 RNA Adaptor Oligo Mix (green- capped tube) to
each sample. Mix by pipetting up and down 15–20 times using a pipette
set to 70 µl or cap the wells and vortex at high speed for 5–10 seconds.
Make sure to add the Ligation master mix and the XT HS2 RNA Adaptor Oligo Mix to the
samples in separate addition steps as directed above, mixing after each addition.
5 Briefly spin the samples, then immediately place the plate or strip tube
in the thermal cycler and resume the thermal cycling program in
Table 17.
*
NOTE
34 SureSelect XT HS2 RNA Library Preparation and Target Enrichment
Unique molecular barcode sequences are incorporated into both ends of each library DNA
fragment at this step.
Page 37

Library Preparation 3
Step 4. Purify the sample using AMPure XP beads
Step 4. Purify the sample using AMPure XP beads
1 Verify that the AMPure XP beads were held at room temperature for at
least 30 minutes before use.
2 Prepare 400 µl of 70% ethanol per sample, plus excess, for use in
step 8.
3 Mix the AMPure XP bead suspension well so that the reagent appears
homogeneous and consistent in color.
4 Add 80 µl of homogeneous AMPure XP beads to each cDNA library
sample (approximately 100 µl) in the PCR plate or strip tube. Pipette
up and down 15–20 times or cap the wells and vortex at high speed for
5–10 seconds to mix.
5 Incubate samples for 5 minutes at room temperature.
6 Put the plate or strip tube into a magnetic separation device. Wait for
the solution to clear (approximately 5 to 10 minutes).
7 Keep the plate or strip tube in the magnetic stand. Carefully remove
and discard the cleared solution from each well. Do not touch the beads
while removing the solution.
8 Continue to keep the plate or strip tube in the magnetic stand while
you dispense 200 µl of freshly- prepared 70% ethanol in each sample
well.
9 Wait for 1 minute to allow any disturbed beads to settle, then remove
the ethanol.
10 Repeat step 8 to step 9 once.
11 Seal the wells with strip caps, then briefly spin the samples to collect
the residual ethanol. Return the plate or strip tube to the magnetic
stand for 30 seconds. Remove the residual ethanol with a P20 pipette.
12 Dry the samples by placing the unsealed plate or strip tube on the
thermal cycler, set to hold samples at 37°C, until the residual ethanol
has just evaporated (typically 1–2 minutes).
13 Add 35 µl nuclease- free water to each sample well.
14 Seal the wells with strip caps, then mix well on a vortex mixer and
briefly spin the plate or strip tube to collect the liquid.
15 Incubate for 2 minutes at room temperature.
16 Put the plate or strip tube in the magnetic stand and leave for
approximately 5 minutes, until the solution is clear.
SureSelect XT HS2 RNA Library Preparation and Target Enrichment 35
Page 38

3 Library Preparation
Step 4. Purify the sample using AMPure XP beads
17 Remove the cleared supernatant (approximately 34 µl) to a fresh PCR
plate or strip tube sample well and keep on ice. You can discard the
beads at this time.
NOTE
It may not be possible to recover the entire 34-µl supernatant volume at this step; transfer
the maximum possible amount of supernatant for further processing. To maximize recovery,
transfer the cleared supernatant to a fresh well in two rounds of pipetting, using a P20
pipette set at 17 µl.
36 SureSelect XT HS2 RNA Library Preparation and Target Enrichment
Page 39

Library Preparation 3
Step 5. Amplify the adaptor-ligated cDNA library
Step 5. Amplify the adaptor-ligated cDNA library
This step uses the components listed in Table 18. Before you begin, thaw
the reagents listed below and keep on ice.
Table 18 Reagents for pre-capture PCR amplification
Component Storage Location Mixing Method Where Used
Herculase II Fusion DNA
Polymerase (red cap)
5× Herculase II Buffer with
dNTPs (clear cap)
SureSelect XT HS2 Index Primer
Pairs
* Indexing primer pairs are provided in individual wells of strip tubes (16 reaction kits) or plates (96 reaction kits).
SureSelect XT HS2 RNA Library
Preparation Kit for ILM (Pre PCR), –20°C
SureSelect XT HS2 RNA Library
Preparation Kit for ILM (Pre PCR), –20°C
SureSelect XT HS2 Index Primer Pairs
for ILM (Pre PCR),
*
–20°C
Pipette up and down
15–20 times
Vortexing page 39
Vortexing page 39
page 39
1 Determine the appropriate index pair assignment for each sample. See
Table 46 on page 80 through Table 53 on page 87 for nucleotide
sequences of the 8 bp index portion of the primers used to amplify the
cDNA libraries in this step.
Use a different indexing primer pair for each sample to be sequenced in
the same lane.
CAUTION
The SureSelect XT HS2 Index Primer Pairs are provided in single-use aliquots. To avoid
cross-contamination of libraries, do not retain and re-use any residual volume for
subsequent experiments.
SureSelect XT HS2 RNA Library Preparation and Target Enrichment 37
Page 40

3 Library Preparation
Step 5. Amplify the adaptor-ligated cDNA library
2 Preprogram a thermal cycler (with heated lid ON) with the program in
Table 19. Immediately pause the program, and keep paused until
samples are loaded in step 6.
Table 19 Pre-Capture PCR Thermal Cycler Program
Segment Number of Cycles Temperature Time
1 1 98°C 2 minutes
2 10–14
(See Table 20 for RNA input-based
cycle number recommendations)
3 1 72°C 5 minutes
4 1 4°C Hold
* Use a reaction volume setting of 50 l, if required for thermal cycler set up.
*
98°C 30 seconds
60°C 30 seconds
72°C 1 minute
Table 20 Pre-capture PCR cycle number recommendations
Quality of Input RNA Quantity of Input RNA Cycle Number
Intact RNA 100 to 200 ng 10 cycles
50 ng 11 cycles
10 ng 12 cycles
Good quality FFPE RNA
(DV200 >50%)
Poor quality FFPE RNA
(DV200 20% to 50%)
100 to 200 ng 12 cycles
50 ng 13 cycles
10 ng 14 cycles
100 to 200 ng 13 cycles
50 ng 14 cycles
CAUTION
To avoid cross-contaminating libraries, set up PCR reactions (all components except
the library DNA) in a dedicated clean area or PCR hood with UV sterilization and
positive air flow.
38 SureSelect XT HS2 RNA Library Preparation and Target Enrichment
Page 41

3 Prepare the appropriate volume of pre- capture PCR reaction mix, as
described in Table 21, on ice. Mix well on a vortex mixer.
Table 21 Preparation of Pre-Capture PCR Reaction Mix
Library Preparation 3
Step 5. Amplify the adaptor-ligated cDNA library
Reagent Volume for 1 reaction Volume for 8 reactions
(includes excess)
5× Herculase II Buffer with dNTPs (clear cap) 10 µl 90 µl 250 µl
Herculase II Fusion DNA Polymerase (red cap) 1 µl 9 µl 25 µl
Total 11 µl 99 µl 275 µl
Volume for 24 reactions
(includes excess)
4 Add 11 µl of the PCR reaction mixture prepared in Table 21 to each
purified DNA library sample (34 µl) in the PCR plate wells.
5 Add 5 µl of the appropriate SureSelect XT HS2 Index Primer Pair to
each reaction.
Cap the wells then vortex at high speed for 5 seconds. Spin the plate
or strip tube briefly to collect the liquid and release any bubbles.
6 Before adding the samples to the thermal cycler, resume the thermal
cycling program in Table 19 to bring the temperature of the thermal
block to 98°C. Once the cycler has reached 98°C, immediately place the
sample plate or strip tube in the thermal block and close the lid.
CAUTION
The lid of the thermal cycler is hot and can cause burns. Use caution when working
near the lid.
SureSelect XT HS2 RNA Library Preparation and Target Enrichment 39
Page 42

3 Library Preparation
Step 6. Purify the amplified library with AMPure XP beads
Step 6. Purify the amplified library with AMPure XP beads
1 Verify that the AMPure XP beads were held at room temperature for at
least 30 minutes before use.
2 Prepare 400 µl of 70% ethanol per sample, plus excess, for use in
step 8.
3 Mix the AMPure XP bead suspension well so that the reagent appears
homogeneous and consistent in color.
4 Add 50 µl of homogeneous AMPure XP beads to each 50- µl
amplification reaction in the PCR plate or strip tube. Pipette up and
down 15–20 times or cap the wells and vortex at high speed for
5–10 seconds to mix.
5 Incubate samples for 5 minutes at room temperature.
6 Put the plate or strip tube into a magnetic separation device. Wait for
the solution to clear (approximately 5 minutes).
7 Keep the plate or strip tube in the magnetic stand. Carefully remove
and discard the cleared solution from each well. Do not touch the beads
while removing the solution.
8 Continue to keep the plate or strip tube in the magnetic stand while
you dispense 200 µl of freshly- prepared 70% ethanol into each sample
well.
9 Wait for 1 minute to allow any disturbed beads to settle, then remove
the ethanol.
10 Repeat step 8 and step 9 step once.
11 Seal the wells with strip caps, then briefly spin the samples to collect
the residual ethanol. Return the plate or strip tube to the magnetic
stand for 30 seconds. Remove the residual ethanol with a P20 pipette.
12 Dry the samples by placing the unsealed plate or strip tube on the
thermal cycler, set to hold samples at 37°C, until the residual ethanol
has just evaporated (typically 1–2 minutes).
13 Add 15 µl nuclease- free water to each sample well.
14 Seal the wells with strip caps, then mix well on a vortex mixer and
briefly spin the plate or strip tube to collect the liquid.
15 Incubate for 2 minutes at room temperature.
16 Put the plate or strip tube in the magnetic stand and leave for 2 to
3 minutes, until the solution is clear.
40 SureSelect XT HS2 RNA Library Preparation and Target Enrichment
Page 43

Library Preparation 3
Step 6. Purify the amplified library with AMPure XP beads
17 Remove the cleared supernatant (approximately 15 µl) to a fresh PCR
plate or strip tube sample well and keep on ice. You can discard the
beads at this time.
NOTE
Stopping Point If you do not continue to the next step, seal the sample wells and store at
It may not be possible to recover the entire 15-µl supernatant volume at this step; transfer
the maximum possible amount of supernatant for further processing.
4°C overnight or at –20°C for prolonged storage.
SureSelect XT HS2 RNA Library Preparation and Target Enrichment 41
Page 44

3 Library Preparation
Step 7. Assess quality and quantity
Step 7. Assess quality and quantity
Analyze each sample using one of the platforms listed in Table 22. Follow
the instructions in the linked user guide provided for each assay in
Table 22, after reviewing the SureSelect library qualification steps on
page 43. Each analysis method provides an electropherogram showing the
size distribution of fragments in the sample and tools for determining the
concentration of DNA in the sample. See Table 23 for fragment size
distribution guidelines. Representative electropherograms generated using
the TapeStation system are provided to illustrate typical results for
libraries prepared from either high- quality or FFPE RNA samples.
Table 22 Pre-capture library analysis options
Analysis platform Assay used at this step Link to assay instructions Amount of library
sample to analyze
Agilent 4200 or 4150 TapeStation
system
Agilent 2100 Bioanalyzer system DNA 1000 Kit Agilent DNA 1000 Kit Guide 1 µl
Agilent 5200, 5300, or 5400
Fragment Analyzer system
D1000 ScreenTape Agilent D1000 Assay Quick
Guide
NGS Fragment Kit (1-6000 bp) Agilent NGS Fragment Kit
(1-6000 bp) Kit Guide
1 µl
2 µl
Table 23 Pre-capture library qualification guidelines
Input RNA type Expected library DNA fragment
size peak position
High-quality RNA or FFPE
RNA
200 to 700 bp 2 ×100 reads or 2 ×150 reads
NGS read lengths supported
Observation of a low molecular weight peak, in addition to the expected
library fragment peak, indicates the presence of adaptor- dimers in the
library. It is acceptable to proceed to target enrichment with library
samples for which adaptor- dimers are observed in the electropherogram at
low abundance, similar to that seen in example electropherograms in this
section. See Troubleshooting on page 92 for additional considerations.
42 SureSelect XT HS2 RNA Library Preparation and Target Enrichment
Page 45

Library Preparation 3
Step 7. Assess quality and quantity
1 Set up the instrument as instructed in the appropriate user guide (links
provided in Table 22).
2 Prepare the samples for analysis and set up the assay as instructed in
the appropriate user guide. Load the analysis assay into the instrument
and complete the run.
3 Verify that the electropherogram shows the expected DNA fragment size
peak position (see Table 23 for guidelines). Sample TapeStation system
electropherograms are shown for libraries prepared from high- quality
RNA in Figure 2 and from FFPE RNA in Figure 3.
Electropherograms obtained using the other analysis platform options
listed in Table 22 are expected to show similar fragment size profiles.
4 Determine the concentration of the library DNA by integrating under
the peak. For accurate quantification, make sure that the concentration
falls within the linear range of the assay.
Figure 2 Pre-capture library prepared from high-quality RNA sample (Human Reference
Total RNA) analyzed using a D1000 ScreenTape.
SureSelect XT HS2 RNA Library Preparation and Target Enrichment 43
Page 46

3 Library Preparation
Step 7. Assess quality and quantity
Figure 3 Pre-capture library prepared from a typical FFPE RNA sample analyzed using a
D1000 ScreenTape.
44 SureSelect XT HS2 RNA Library Preparation and Target Enrichment
Page 47

SureSelect XT HS2 RNA System Protocol
4
Hybridization and Capture
Step 1. Hybridize cDNA libraries to the probe 46
Step 2. Prepare streptavidin-coated magnetic beads 51
Step 3. Capture the hybridized DNA using streptavidin-coated beads 52
This chapter describes the steps to hybridize the prepared cDNA libraries
with a target- specific Probe Capture Library. After hybridization, the
targeted molecules are captured on streptavidin- coated beads. Each cDNA
library sample is hybridized and captured individually.
The standard single- day protocol includes the hybridization step
(approximately 90 minutes) immediately followed by capture and
amplification steps. If required, the hybridized samples may be held
overnight with capture and amplification steps completed the following
day by using the simple protocol modifications noted on page 47.
CAUTION
The ratio of probe to cDNA library is critical for successful capture.
Agilent Technologies
45
Page 48

4 Hybridization and Capture
Step 1. Hybridize cDNA libraries to the probe
Step 1. Hybridize cDNA libraries to the probe
In this step, the prepared cDNA libraries are hybridized to a
target- specific Probe Capture Library. For each RNA sample library
prepared, do one hybridization and capture. Do not pool samples at this
stage.
The hybridization reaction requires 200 ng of prepared cDNA library in a
volume of 12 µl.
This step uses the components listed in Table 24. Thaw each component
under the conditions indicated in the table. Vortex each reagent to mix,
then spin tubes briefly to collect the liquid.
Table 24 Reagents for Hybridization
Kit Component Storage Location Thawing Conditions Where Used
SureSelect XT HS2 Blocker Mix
(blue cap)
SureSelect RNase Block (purple
cap)
SureSelect Fast Hybridization Buffer
(bottle)
Probe Capture Library –80°C Thaw on ice page 49
SureSelect XT HS2 Target Enrichment Kit,
ILM Hyb Module, Box 2 (Post PCR), –20°C
SureSelect XT HS2 Target Enrichment Kit
ILM Hyb Module, Box 2 (Post PCR), –20°C
SureSelect XT HS2 Target Enrichment Kit
ILM Hyb Module, Box 2 (Post PCR), –20°C
Thaw on ice page 47
Thaw on ice page 48
Thaw and keep at
Room Temperature
page 49
46 SureSelect XT HS2 RNA Library Preparation and Target Enrichment
Page 49

Hybridization and Capture 4
Step 1. Hybridize cDNA libraries to the probe
1 Preprogram a thermal cycler (with heated lid ON) with the program in
Table 25. Immediately pause the program, and keep paused until
samples are loaded in step 4.
NOTE
Table 25 Pre-programmed thermal cycler program for Hybridization
Segment Number Number of Cycles Temperature Time
1 1 95°C 5 minutes
2 1 65°C 10 minutes
3 1 65°C 1 minute
460
5 1 65°C Hold
* When setting up the thermal cycling program, use a reaction volume setting of 30 l (final volume
of hybridization reactions during cycling in Segment 4).
65°C 1 minute
37°C 3 seconds
*
The Hybridization reaction may be run overnight with the following protocol modifications:
• In segment 5 of the thermal cycler program (Table 25), replace the 65°C Hold step with a
21°C Hold step.
• Pause the thermal cycler as directed in step 1 and complete the hybridization setup
steps as directed on page 47 through page 50 before resuming the paused program.
• The hybridized samples may be held at 21°C for up to 16 hours. Complete the
streptavidin bead preparation steps on page 51 just before you are ready to start the
capture steps on page 52. Move the hybridized samples to room temperature just before
adding the washed streptavidin beads to each sample.
2 Place 200 ng of each prepared cDNA library sample into the well of a
fresh hybridization plate or strip tube and then bring the final volume
in each well to 12 µl using nuclease- free water.
3 To each DNA library sample well, add 5 µl of SureSelect XT HS2
Blocker Mix. Cap the wells then vortex at high speed for 5 seconds.
Spin the plate or strip tube briefly to collect the liquid and release any
bubbles.
SureSelect XT HS2 RNA Library Preparation and Target Enrichment 47
Page 50

4 Hybridization and Capture
Step 1. Hybridize cDNA libraries to the probe
CAUTION
The lid of the thermal cycler is hot and can cause burns. Use caution when working
near the lid.
4 Transfer the sealed sample plate or strip to the thermal cycler and
resume the thermal cycling program set up on page 47 and shown in
Table 26 below.
Important: Notice that the thermal cycler must be paused during
Segment 3 (see Table 26) to allow additional reagents to be added to
the Hybridization wells, as described in step 7 on page 50.
During Segments 1 and 2 of the thermal cycling program below, begin
preparing the additional reagents as described in step 5 below and
step 6 on page 49. If needed, you can finish these preparation steps
after pausing the thermal cycler in Segment 3.
Table 26 Thermal cycler program for Hybridization with required pause
Segment Number Number of Cycles Temperature Time
1 1 95°C 5 minutes
2 1 65°C 10 minutes
3 1 65°C 1 minute (PAUSE cycler here)
4 60 65°C 1 minute
37°C 3 seconds
*
5 1 65°C Hold
* Begin the capture steps on page 51 when the thermal cycler starts the 65°C Hold segment.
5 Prepare a 25% solution of SureSelect RNase Block (containing
1 part RNase Block:3 parts water), according to Table 27. Prepare the
amount required for the number of hybridization reactions in the run,
plus excess. Mix well and keep on ice.
Table 27 Preparation of RNase Block solution
Reagent Volume for 1 reaction Volume for 8 reactions
(includes excess)
SureSelect RNase Block 0.5 µl 4.5 µl 12.5 µl
Nuclease-free water 1.5 µl 13.5 µl 37.5 µl
Total 2 µl 18 µl 50 µl
Volume for 24 reactions
(includes excess)
48 SureSelect XT HS2 RNA Library Preparation and Target Enrichment
Page 51

Hybridization and Capture 4
Step 1. Hybridize cDNA libraries to the probe
NOTE
Prepare the mixture described in step 6, below, just before pausing the thermal cycler in
Segment 3 as described on page 48. Keep the mixture at room temperature briefly until the
mixture is added to the DNA samples in step 7 on page 50. Do not keep solutions
containing the Probe at room temperature for extended periods.
6 Prepare the Capture Library Hybridization Mix appropriate for your
probe design size. Use Table 28 for Probe Capture Libraries 3 Mb or
Table 29 for Probe Capture Libraries <3 Mb.
Combine the listed reagents at room temperature. Mix well by
vortexing at high speed for 5 seconds then spin down briefly. Proceed
immediately to step 7.
Table 28 Preparation of Capture Library Hybridization Mix for probes≥3 Mb
Reagent Volume for 1 reaction Volume for 8 reactions
(includes excess)
25% RNase Block solution (from step 5) 2 µl 18 µl 50 µl
Probe (with design 3 Mb) 5 µl 45 µl 125 µl
SureSelect Fast Hybridization Buffer 6 µl 54 µl 150 µl
Total 13 µl 117 µl 325 µl
Volume for 24 reactions
(includes excess)
Table 29 Preparation of Capture Library Hybridization Mix for probes<3 Mb
Reagent Volume for 1 reaction Volume for 8 reactions
(includes excess)
25% RNase Block solution (from step 5) 2 µl 18 µl 50 µl
Probe (with design<3 Mb) 2 µl 18 µl 50 µl
SureSelect Fast Hybridization Buffer 6 µl 54 µl 150 µl
Nuclease-free water 3 µl 27 µl 75 µl
Total 13 µl 117 µl 325 µl
Volume for 24 reactions
(includes excess)
SureSelect XT HS2 RNA Library Preparation and Target Enrichment
49
Page 52

4 Hybridization and Capture
Step 1. Hybridize cDNA libraries to the probe
7 Once the thermal cycler starts Segment 3 of the program in Table 26
(1 minute at 65°C), pause the program. With the cycler paused, and
while keeping the DNA + Blocker samples in the cycler, transfer 13 µl
of the room- temperature Capture Library Hybridization Mix from step 6
to each sample well.
Mix well by pipetting up and down slowly 8 to 10 times.
The hybridization reaction wells now contain approximately 30 µl.
8 Seal the wells with fresh domed strip caps. Make sure that all wells are
completely sealed. Vortex briefly, then spin the plate or strip tube
briefly to remove any bubbles from the bottom of the wells. Immediately
return the plate or strip tube to the thermal cycler.
9 Resume the thermal cycling program to allow hybridization of the
prepared DNA samples to the Probe Capture Library.
CAUTION
Wells must be adequately sealed to minimize evaporation, or your results can be
negatively impacted.
Before you do the first experiment, make sure the plasticware and capping method are
appropriate for the thermal cycler. Check that no more than 4 µl is lost to evaporation
under the conditions used for hybridization.
50 SureSelect XT HS2 RNA Library Preparation and Target Enrichment
Page 53

Hybridization and Capture 4
Step 2. Prepare streptavidin-coated magnetic beads
Step 2. Prepare streptavidin-coated magnetic beads
The remaining hybridization capture steps use the components listed in
Table 30.
Begin the bead preparation steps described below approximately one hour
after starting hybridization in step 9 on page 50.
Table 30 Reagents for Capture
Kit Component Storage Location Where Used
SureSelect Binding Buffer SureSelect XT HS Target Enrichment Kit ILM Hyb
Module, Box 1 (Post PCR), RT
SureSelect Wash Buffer 1 SureSelect XT HS Target Enrichment Kit ILM Hyb
Module, Box 1 (Post PCR), RT
SureSelect Wash Buffer 2 SureSelect XT HS Target Enrichment Kit ILM Hyb
Module, Box 1 (Post PCR), RT
SureSelect Streptavidin Beads
OR
Dynabeads MyOne Streptavidin T1 Beads
4°C page 51
page 51
page 52
page 52
1 Vigorously resuspend the vial of streptavidin beads on a vortex mixer.
The magnetic beads settle during storage.
2 For each hybridization sample, add 50 µl of the resuspended beads to
wells of a fresh PCR plate or a strip tube.
3 Wash the beads:
a Add 200 µl of SureSelect Binding Buffer.
b Mix by pipetting up and down 20 times or cap the wells and vortex
at high speed for 5–10 seconds then spin down briefly.
c Put the plate or strip tube into a magnetic separator device.
d Wait 5 minutes or until the solution is clear, then remove and
discard the supernatant.
e Repeat step a through step d two more times for a total of 3 washes.
4 Resuspend the beads in 200 µl of SureSelect Binding Buffer.
NOTE
If you are equipped for higher-volume magnetic bead captures, the streptavidin beads may
instead be batch-washed in a microcentrifuge tube or conical vial.
SureSelect XT HS2 RNA Library Preparation and Target Enrichment 51
Page 54

4 Hybridization and Capture
Step 3. Capture the hybridized DNA using streptavidin-coated beads
Step 3. Capture the hybridized DNA using streptavidin-coated
beads
1 After all streptavidin bead preparation steps are complete, and once the
hybridization thermal cycling program reaches the 65°C hold step
(Segment 5; see
temperature.
2 Immediately transfer the entire volume (approximately 30 µl) of each
hybridization mixture to wells containing 200 µl of washed streptavidin
beads using a multichannel pipette.
Pipette up and down 5–8 times to mix then seal the wells with fresh
caps.
3 Incubate the capture plate or strip tube on a 96- well plate mixer,
mixing vigorously (at 1400–1900 rpm), for 30 minutes at room
temperature.
Make sure the samples are properly mixing in the wells.
4 During the 30- minute incubation for capture, prewarm SureSelect Wash
Buffer 2 at 70°C as described below.
a Place 200- µl aliquots of Wash Buffer 2 in wells of a fresh 96- well
plate or strip tubes. Aliquot 6 wells of buffer for each sample in the
run.
b Cap the wells and then incubate in the thermal cycler, held at 70°C,
until used in step 9.
5 When the 30- minute capture incubation period initiated in step 3 is
complete, spin the samples briefly to collect the liquid.
6 Put the plate or strip tube in a magnetic separator to collect the beads.
Wait until the solution is clear, then remove and discard all of the
supernatant.
7 Resuspend the beads in 200 µl of SureSelect Wash Buffer 1. Mix by
pipetting up and down 15–20 times, until beads are fully resuspended.
8 Put the plate or strip tube in the magnetic separator. Wait for the
solution to clear (approximately 1 minute), then remove and discard all
of the supernatant.
Table 26 on page 48), transfer the samples to room
52 SureSelect XT HS2 RNA Library Preparation and Target Enrichment
Page 55

Hybridization and Capture 4
Step 3. Capture the hybridized DNA using streptavidin-coated beads
CAUTION
It is important to maintain bead suspensions at 70°C during the washing procedure
below to ensure specificity of capture.
Make sure that the SureSelect Wash Buffer 2 is pre-warmed to 70°C before use.
Do not use a tissue incubator, or other devices with significant temperature
fluctuations, for the incubation steps.
9 Remove the plate or strip tubes from the magnetic separator and
transfer to a rack at room temperature. Wash the beads with Wash
Buffer 2, using the protocol steps below.
a Resuspend the beads in 200 µl of 70°C prewarmed Wash Buffer 2.
Pipette up and down 15–20 times, until beads are fully resuspended.
b Carefully seal the wells with fresh caps and then vortex at high
speed for 8 seconds. Spin the plate or strip tube briefly to collect the
liquid without pelleting the beads.
Make sure the beads are in suspension before proceeding.
c Incubate the samples for 5 minutes at 70°C in the thermal cycler
with the heated lid on.
d Put the plate or strip tube in the magnetic separator at room
temperature.
e Wait 1 minute for the solution to clear, then remove and discard the
supernatant.
f Repeat step a through step e five more times for a total of 6 washes.
10 After verifying that all wash buffer has been removed, add 25 µl of
nuclease- free water to each sample well. Pipette up and down 8 times
to resuspend the beads.
Keep the samples on ice until they are used on page 58.
NOTE
SureSelect XT HS2 RNA Library Preparation and Target Enrichment 53
Captured DNA is retained on the streptavidin beads during the post-capture amplification
step.
Page 56

4 Hybridization and Capture
Step 3. Capture the hybridized DNA using streptavidin-coated beads
54 SureSelect XT HS2 RNA Library Preparation and Target Enrichment
Page 57

SureSelect XT HS2 RNA System Protocol
5
Post-Capture Sample Processing for
Multiplexed Sequencing
Step 1. Amplify the captured libraries 56
Step 2. Purify the amplified captured libraries using AMPure XP beads 59
Step 3. Assess sequencing library DNA quantity and quality 61
Step 4. Pool samples for multiplexed sequencing 64
Step 5. Prepare sequencing samples 66
Step 6. Do the sequencing run and analyze the data 68
Sequence analysis resources 73
This chapter describes the steps to amplify, purify, and assess quality and
quantity of the captured libraries. Sample pooling instructions are
provided to prepare the indexed, molecular barcoded samples for
multiplexed sequencing.
Agilent Technologies
55
Page 58
5 Post-Capture Sample Processing for Multiplexed Sequencing
Step 1. Amplify the captured libraries
Step 1. Amplify the captured libraries
In this step, the SureSelect- enriched DNA libraries are PCR amplified.
This step uses the components listed in Table 31. Before you begin, thaw
the reagents listed below and keep on ice. Remove the AMPure XP beads
from cold storage and equilibrate to room temperature for at least
30 minutes in preparation for use on page 59. Do not freeze the beads at
any time.
Table 31 Reagents for post-capture PCR amplification
Component Storage Location Mixing Method Where Used
Herculase II Fusion DNA
Polymerase (red cap)
5× Herculase II Buffer with
dNTPs (clear cap)
SureSelect Post-Capture Primer
Mix (clear cap)
Prepare one amplification reaction for each DNA library.
CAUTION
To avoid cross-contaminating libraries, set up PCR mixes in a dedicated clean area or
PCR hood with UV sterilization and positive air flow.
SureSelect XT HS2 Target Enrichment
Kit, ILM Hyb Module, Box 2 (Post PCR),
–20°C
SureSelect XT HS2 Target Enrichment
Kit, ILM Hyb Module, Box 2 (Post PCR),
–20°C
SureSelect XT HS2 Target Enrichment
Kit, ILM Hyb Module, Box 2 (Post PCR),
–20°C
Pipette up and down
15–20 times
Vortexing page 58
Vortexing page 58
page 58
56 SureSelect XT HS2 RNA Library Preparation and Target Enrichment
Page 59

Post-Capture Sample Processing for Multiplexed Sequencing 5
Step 1. Amplify the captured libraries
1 Preprogram a thermal cycler (with heated lid ON) with the program in
Table 32. Immediately pause the program, and keep paused until
samples are loaded in step 5.
Table 32 Post-capture PCR Thermal Cycler Program
Segment Number of Cycles Temperature Time
1 1 98°C 2 minutes
2 12–16
(See Table 33 for probe design size-based
cycle number recommendations)
3 1 72°C 5 minutes
4 1 4°C Hold
98°C 30 seconds
60°C 30 seconds
72°C 1 minute
Table 33 Post-capture PCR cycle number recommendations
Probe Design Size Cycles
Probes <0.2 Mb 16 cycles
Probes 0.2–3 Mb 14 cycles
Probes 3–5 Mb 13 cycles
Probes>5 Mb 12 cycles
SureSelect XT HS2 RNA Library Preparation and Target Enrichment 57
Page 60

5 Post-Capture Sample Processing for Multiplexed Sequencing
Step 1. Amplify the captured libraries
2 Prepare the appropriate volume of PCR reaction mix, as described in
Table 34, on ice. Mix well on a vortex mixer.
Table 34 Preparation of post-capture PCR Reaction mix
Reagent Volume for 1 reaction Volume for 8 reactions
(includes excess)
Nuclease-free water 13 µl 117 µl 325 µl
5× Herculase II Buffer with dNTPs (clear cap) 10 µl 90 µl 250 µl
Herculase II Fusion DNA Polymerase (red cap) 1 µl 9 µl 25 µl
SureSelect Post-Capture Primer Mix (clear cap) 1 µl 9 µl 25 µl
Total 25 µl 225 µl 625 µl
Volume for 24 reactions
(includes excess)
3 Add 25 µl of the PCR reaction mix prepared in Table 34 to each sample
well containing 25 µl of bead- bound target- enriched DNA (prepared on
page 53 and held on ice).
4 Mix the PCR reactions well by pipetting up and down until the bead
suspension is homogeneous. Avoid splashing samples onto well walls; do
not spin the samples at this step.
5 Place the plate or strip tube in the thermal cycler, and resume the
thermal cycling program in Table 32.
6 When the PCR amplification program is complete, spin the plate or
strip tube briefly. Remove the streptavidin- coated beads by placing the
plate or strip tube on the magnetic stand at room temperature. Wait
2 minutes for the solution to clear, then remove each supernatant
(approximately 50 µl) to wells of a fresh plate or strip tube.
The beads can be discarded at this time.
58 SureSelect XT HS2 RNA Library Preparation and Target Enrichment
Page 61

Post-Capture Sample Processing for Multiplexed Sequencing 5
Step 2. Purify the amplified captured libraries using AMPure XP beads
Step 2. Purify the amplified captured libraries using
AMPure XP beads
1 Verify that the AMPure XP beads were held at room temperature for at
least 30
minutes before use.
2 Prepare 400 µl of fresh 70% ethanol per sample, plus excess, for use in
step 8.
3 Mix the AMPure XP bead suspension well so that the suspension
appears homogeneous and consistent in color.
4 Add 50 µl of the homogeneous AMPure XP bead suspension to each
amplified DNA sample (approximately 50 µl) in the PCR plate or strip
tube. Mix well by pipetting up and down 15–20 times or cap the wells
and vortex at high speed for 5–10 seconds.
Check that the beads are in a homogeneous suspension in the sample
wells. Each well should have a uniform color with no layers of beads or
clear liquid present.
5 Incubate samples for 5 minutes at room temperature.
6 Put the plate or strip tube on the magnetic stand at room temperature.
Wait for the solution to clear (approximately 3 to 5 minutes).
7 Keep the plate or strip tube in the magnetic stand. Carefully remove
and discard the cleared solution from each well. Do not disturb the
beads while removing the solution.
8 Continue to keep the plate or tubes in the magnetic stand while you
dispense 200 µl of freshly- prepared 70% ethanol in each sample well.
9 Wait for 1 minute to allow any disturbed beads to settle, then remove
the ethanol.
10 Repeat step 8 and step 9 once for a total of two washes. Make sure to
remove all of the ethanol at each wash step.
11 Seal the wells with strip caps, then briefly spin to collect the residual
ethanol. Return the plate or strip tube to the magnetic stand for
30 seconds. Remove the residual ethanol with a P20 pipette.
12 Dry the samples by placing the unsealed plate or strip tube on the
thermal cycler, set to hold samples at 37°C, until the residual ethanol
has just evaporated (typically 1–2 minutes).
13 Add 25 µl of Low TE buffer (10 mM Tris pH 7.5- 8.0, 0.1 mM EDTA) to
each sample well.
SureSelect XT HS2 RNA Library Preparation and Target Enrichment 59
Page 62

5 Post-Capture Sample Processing for Multiplexed Sequencing
Step 2. Purify the amplified captured libraries using AMPure XP beads
14 Seal the sample wells with strip caps, then mix well on a vortex mixer
and briefly spin to collect the liquid without pelleting the beads.
15 Incubate for 2 minutes at room temperature.
16 Put the plate or strip tube in the magnetic stand and leave for
2 minutes or until the solution is clear.
17 Remove the cleared supernatant (approximately 25 µl) to a fresh well.
You can discard the beads at this time.
60 SureSelect XT HS2 RNA Library Preparation and Target Enrichment
Page 63

Post-Capture Sample Processing for Multiplexed Sequencing 5
Step 3. Assess sequencing library DNA quantity and quality
Step 3. Assess sequencing library DNA quantity and quality
Analyze each library using one of the platforms listed in Table 35. Follow
the instructions in the linked user guide provided for each assay in
Table 35, after reviewing the post- capture library qualification steps
below. See Table 36 for expected fragment size distribution guidelines.
Representative electropherograms generated using the TapeStation system
are provided to illustrate typical results for post- capture libraries
prepared from either high- quality or FFPE RNA samples.
Table 35 Post-capture library analysis options
Analysis platform Assay used at this step Link to assay instructions Amount of library
sample to analyze
Agilent 4200 or 4150
TapeStation system
Agilent 2100 Bioanalyzer
system
Agilent 5200, 5300, or 5400
Fragment Analyzer system
Table 36 Post-capture library qualification guidelines
Input RNA type Expected library DNA fragment
High-quality RNA or FFPE
RNA
1 Set up the instrument as instructed in the appropriate user guide (links
2 Prepare the samples for analysis and set up the assay as instructed in
High Sensitivity D1000
ScreenTape
High Sensitivity DNA Kit Agilent High Sensitivity DNA Kit
HS NGS Fragment Kit
(1-6000 bp)
size peak position
200 to 700 bp 2 ×100 reads or 2 ×150 reads
Agilent High Sensitivity D1000
Assay Quick Guide
Guide
Agilent HS NGS Fragment Kit
(1-6000 bp) Kit Guide
NGS read lengths supported
2 µl
1 µl
2 µl
provided in Table 35).
the appropriate user guide. Load the analysis assay into the instrument
and complete the run.
SureSelect XT HS2 RNA Library Preparation and Target Enrichment 61
Page 64

5 Post-Capture Sample Processing for Multiplexed Sequencing
Step 3. Assess sequencing library DNA quantity and quality
3 Verify that the electropherogram shows the expected DNA fragment size
peak position (see Table 36 for guidelines). Sample TapeStation system
electropherograms are shown for libraries prepared from high- quality
intact RNA in Figure 4 and from FFPE RNA in Figure 5.
Electropherograms obtained using the other analysis platform options
listed in Table 35 are expected to show similar fragment size profiles.
4 Determine the concentration of the library DNA by integrating under
the entire peak. For accurate quantification, make sure that the
concentration falls within the linear range of the assay.
Figure 4 Post-capture library prepared from an intact RNA sample analyzed using a
High Sensitivity D1000 ScreenTape assay.
62 SureSelect XT HS2 RNA Library Preparation and Target Enrichment
Page 65

Post-Capture Sample Processing for Multiplexed Sequencing 5
Step 3. Assess sequencing library DNA quantity and quality
Figure 5 Post-capture library prepared from a typical FFPE RNA sample analyzed using
a High Sensitivity D1000 ScreenTape assay.
Stopping Point If you do not continue to the next step, seal the sample wells and store at
4°C overnight or at –20°C for prolonged storage.
SureSelect XT HS2 RNA Library Preparation and Target Enrichment 63
Page 66

5 Post-Capture Sample Processing for Multiplexed Sequencing
Step 4. Pool samples for multiplexed sequencing
Step 4. Pool samples for multiplexed sequencing
The number of indexed libraries that may be multiplexed in a single
sequencing lane is determined by the output specifications of the platform
used, together with the amount of sequencing data required for your
research design. Calculate the number of indexes that can be combined
per lane, according to the capacity of your platform and the amount of
sequencing data required per sample.
Combine the libraries such that each index- tagged sample is present in
equimolar amounts in the pool using one of the following methods:
Method 1: Dilute each sample to be pooled to the same final
concentration (typically 4 nM–15 nM, or the concentration of the most
dilute sample) using Low TE, then combine equal volumes of all
samples to create the final pool.
Method 2: Starting with samples at different concentrations, add the
appropriate volume of each sample to achieve equimolar
concentration in the pool, then adjust the pool to the desired final
volume using Low TE. The formula below is provided for
determination of the amount of each indexed sample to add to the
pool.
Volume of Index
where V(f) is the final desired volume of the pool,
C(f) is the desired final concentration of all the DNA in the pool
(typically 4 nM–15 nM or the concentration of the most dilute
sample)
# is the number of indexes, and
C(i) is the initial concentration of each indexed sample
Table 37 shows an example of the amount of 4 index- tagged samples
(of different concentrations) and Low TE needed for a final volume of
20 µl at 10 nM DNA.
64 SureSelect XT HS2 RNA Library Preparation and Target Enrichment
Vf Cf
---------------------------------=
# Ci
Page 67

Post-Capture Sample Processing for Multiplexed Sequencing 5
Step 4. Pool samples for multiplexed sequencing
Table 37 Example of volume calculation for total volume of 20 µl at 10 nM concentration
Component V(f) C(i) C(f) # Volume to use (µl)
Sample 1 20 µl 20 nM 10 nM 4 2.5
Sample 2 20 µl 10 nM 10 nM 4 5
Sample 3 20 µl 17 nM 10 nM 4 2.9
Sample 4 20 µl 25 nM 10 nM 4 2
Low TE 7.6
If you store the library before sequencing, add Tween 20 to 0.1% v/v and
store at - 20°C short term.
SureSelect XT HS2 RNA Library Preparation and Target Enrichment 65
Page 68

5 Post-Capture Sample Processing for Multiplexed Sequencing
Step 5. Prepare sequencing samples
Step 5. Prepare sequencing samples
The final SureSelect XT HS2 RNA library pool is ready for direct
sequencing using standard Illumina paired- end primers and chemistry.
Each fragment in the prepared library contains one target insert
surrounded by sequence motifs required for multiplexed sequencing using
the Illumina platform, as shown in Figure 6.
Figure 6 Content of SureSelect XT HS2 sequencing library. Each fragment contains one
target insert (blue) surrounded by the Illumina paired-end sequencing elements (black), the dual sample indexes (red and green), dual molecular barcodes (brown) and the library bridge PCR primers (yellow).
Libraries can be sequenced on the Illumina HiSeq, MiSeq, NextSeq, or
NovaSeq platform using the run type and chemistry combinations shown
in Table 38.
The optimal seeding concentration for SureSelect XT HS2 RNA
target- enriched libraries varies according to sequencing platform, run type,
and Illumina kit version. See Table 38 for guidelines. Seeding
concentration and cluster density may also need to be optimized based on
the library cDNA fragment size range and on the desired output and data
quality. Begin optimization using a seeding concentration in the middle of
the range listed in Table 38.
Follow Illumina’s recommendation for a PhiX control in a
low- concentration spike- in for improved sequencing quality control.
66 SureSelect XT HS2 RNA Library Preparation and Target Enrichment
Page 69

Post-Capture Sample Processing for Multiplexed Sequencing 5
Step 5. Prepare sequencing samples
Table 38 Illumina Kit Configuration Selection Guidelines
Platform Run Type Read Length SBS Kit Configuration Chemistry Seeding
Concentration
HiSeq 2500 Rapid Run 2 × 100 bp 200 Cycle Kit v2 9–10 pM
HiSeq 2500 High Output 2 × 100 bp 4×50 Cycle Kits
HiSeq 2500 High Output 2 × 100 bp 250 Cycle Kit v4 12–14 pM
MiSeq All Runs 2 × 100 bp or
2 × 150 bp
MiSeq All Runs 2 × 75 bp 150 Cycle Kit v3 12–16 pM
NextSeq 500/550 All Runs 2 × 100 bp or
2 × 150 bp
HiSeq 3000/4000 All Runs 2 × 100 bp or
2 × 150 bp
NovaSeq 6000 Standard
Workflow Runs
NovaSeq 6000 Xp Workflow
Runs
2 × 100 bp or
2 × 150 bp
2 × 100 bp or
2 × 150 bp
300 Cycle Kit v2 9–10 pM
300 Cycle Kit v2.5 1.2–1.5 pM
300 Cycle Kit v1 230–240 pM
300 Cycle Kit v1.0 or v1.5 300–600 pM
300 Cycle Kit v1.0 or v1.5 200–400 pM
*
v3 9–10 pM
* A single 200-cycle kit does not include enough reagents to complete Reads 1 and 2 in addition to the 8-bp i7 and 8-bp i5 index
reads in this format. If preferred, the additional reads may be supported by using one 200-cycle kit plus one 50-cycle kit.
SureSelect XT HS2 RNA Library Preparation and Target Enrichment 67
Page 70

5 Post-Capture Sample Processing for Multiplexed Sequencing
Step 6. Do the sequencing run and analyze the data
Step 6. Do the sequencing run and analyze the data
The guidelines below provide an overview of SureSelect XT HS2 RNA
library sequencing run setup and analysis considerations. Links are
provided for additional details for various NGS platforms and analysis
pipeline options.
• Each of the sample- level indexes requires an 8- bp index read. For
complete index sequence information, see page 79 through page 90.
• For the HiSeq, NextSeq, and NovaSeq platforms, set up the run using
the instrument’s user interface, following the guidelines on page 69.
• For the MiSeq platform, set up the run using Illumina Experiment
Manager (IEM) using the steps detailed on page 69 to page 72 to
generate a custom sample sheet.
• Demultiplex using Illumina’s bcl2fastq software to generate paired end
reads based on the dual indexes and remove sequences with incorrectly
paired P5 and P7 indexes.
• Library fragments include a degenerate molecular barcode (MBC) in
each strand (see Figure 6 on page 66). Note that unlike DNA, where
both strands are present and the MBCs in the strands can be matched
to form a duplex consensus read, single- stranded RNA stops at single
consensus generation.
• The MBC sequence and dark bases are located at the 5’ end of both
Read 1 and Read 2. Use the Agilent Genomics NextGen Toolkit (AGeNT)
for molecular barcode extraction and trimming (see page 73 for more
information). If your sequence analysis pipeline excludes MBCs and is
incompatible with AGeNT, you can trim or mask the first five bases
from each read before alignment as described in the Note on page 73.
• Before aligning reads to reference sequences, Illumina adaptor
sequences should be trimmed from the reads using Agilent’s AGeNT
trimmer module, which properly accounts for the degenerate MBCs in
the adaptor sequence. See page 73 for more information. Do not use the
adaptor trimming options in Illumina Experiment Manager (IEM). Make
sure any IEM adaptor trimming option checkboxes are cleared
(deselected) when setting up the sequencing run.
68 SureSelect XT HS2 RNA Library Preparation and Target Enrichment
Page 71

Post-Capture Sample Processing for Multiplexed Sequencing 5
Step 6. Do the sequencing run and analyze the data
HiSeq/NextSeq/NovaSeq platform sequencing run setup guidelines
Set up sequencing runs using the instrument control software interface,
using the settings shown in Table 39. For HiSeq runs, select Dual Index
on the Run Configuration screen of the instrument control software
interface and enter the Cycles settings in Table 39.
For the NextSeq or NovaSeq platform, open the Run Setup screen of the
instrument control software interface and enter the Read Length settings
in Table 39. In the Custom Primers section, clear (do not select) the
checkboxes for all primers (Read 1, Read 2, Index 1 and Index 2).
Table 39 Run settings
Run Segment Cycles/Read Length
Read 1 100
Index 1 (i7) 8
Index 2 (i5) 8
Read 2 100
MiSeq platform sequencing run setup guidelines
Use the Illumina Experiment Manager (IEM) software to generate a custom
Sample Sheet according to the guidelines below. Once a Sample Sheet has
been generated, index sequences need to be manually changed to the
SureSelect XT HS2 indexes used for each sample. See Table 46 on page 80
through Table 53 on page 87 for nucleotide sequences of the SureSelect
XT HS2 index pairs.
Set up a custom Sample Sheet:
1 In the IEM software, create a Sample Sheet for the MiSeq platform
using the following Workflow selections.
• Under Category, select Other.
• Under Application, select FASTQ Only.
SureSelect XT HS2 RNA Library Preparation and Target Enrichment 69
Page 72

5 Post-Capture Sample Processing for Multiplexed Sequencing
Step 6. Do the sequencing run and analyze the data
2 On the Workflow Parameters screen, enter the run information, making
sure to specify the key parameters highlighted below. In the Library
Prep Workflow field, select TruSeq Nano DNA. In the Index Adapters
field, select TruSeq DNA CD Indexes (96 Indexes). Make sure to clear
both adaptorSpecific Settings (circled below), since these are selected by default.
If TruSeq Nano DNA is not available in the Sample Prep Kit field,
instead select TruSeq HT.
trimming checkboxes under FASTQ Only Workflow-
3 Using the Sample Sheet Wizard, set up a New Plate, entering the
required information for each sample to be sequenced. In the
I7 Sequence column, assign each sample to any of the Illumina i7
indexes. The index will be corrected to the i7 sequence from the
SureSelect XT HS2 index pair at a later stage.
70 SureSelect XT HS2 RNA Library Preparation and Target Enrichment
Page 73

Post-Capture Sample Processing for Multiplexed Sequencing 5
Step 6. Do the sequencing run and analyze the data
Likewise, in the I5 Sequence column, assign any of the Illumina i5
indexes, to be corrected to the i5 sequence from the SureSelect XT HS2
index pair at a later stage.
4 Finish the sample sheet setup tasks and save the sample sheet file.
SureSelect XT HS2 RNA Library Preparation and Target Enrichment 71
Page 74

5 Post-Capture Sample Processing for Multiplexed Sequencing
Step 6. Do the sequencing run and analyze the data
Editing the Sample Sheet to include SureSelect XT HS2 dual indexes
• Open the Sample Sheet file in a text editor and edit the i7 and i5 index
information for each sample in columns 5–8 (highlighted below). See
Table 46 on page 80 through Table 53 on page 87 for nucleotide
sequences of the SureSelect XT HS2 index pairs.
• In column 5 under I7_Index_ID, enter the SureSelect XT HS2 index
pair number assigned to the sample. In column 6 under index, enter
the corresponding P7 index sequence.
• In column 7 under I5_Index_ID, enter the SureSelect XT HS2 index
pair number assigned to the sample. In column 8 under index2, enter
the corresponding P5 index sequence.
Figure 7 Sample sheet for SureSelect XT HS2 library sequencing
5 Save the edited Sample Sheet in an appropriate file location for use in
the run.
72 SureSelect XT HS2 RNA Library Preparation and Target Enrichment
Page 75

NOTE
Post-Capture Sample Processing for Multiplexed Sequencing 5
Sequence analysis resources
Sequence analysis resources
Guidelines are provided below for typical NGS analysis pipeline steps
appropriate for SureSelect XT HS2 RNA library data analysis. Your NGS
analysis pipeline may vary.
Use the Illumina bcl2fastq software to generate paired end reads by
demultiplexing sequences based on the dual indexes and to remove
sequences with incorrectly paired P5 and P7 indexes.
The demultiplexed FASTQ data needs to be pre- processed to remove
sequencing adaptors and extract the molecular barcode (MBC) sequences
using the Agilent Genomics NextGen Toolkit (AGeNT). AGeNT is a set of
Java- based software modules that provide MBC pre- processing adaptor
trimming and duplicate read identification. This toolkit is designed to
enable building, integrating, maintaining, and troubleshooting internal
analysis pipelines for users with bioinformatics expertise. For additional
information and to download this toolkit, visit the AGeNT page at
www.agilent.com.
If your sequence analysis pipeline excludes MBCs, you can remove the first 5 bases from
Read 1 and Read 2 by either masking or trimming before proceeding to further analysis. To
remove during demultiplexing via masking, include the base mask N5Y*,I8,I8,N5Y* (where *
may be replaced with the actual read length, matching the read length value in the
RunInfo.xml file). Alternatively, the first 5 bases may be trimmed from the demultiplexed
fastq files using a suitable processing tool of your choice, such as seqtk. Alternatively, the
AGeNT trimmer module can be used to remove the MBCs and properly remove adaptor
sequences as well. Standard adaptor trimmers will fail to remove the MBC sequences from
the opposite adaptor (refer to Figure 6).
The trimmed reads should be aligned, and MBC tags added to the aligned
BAM file using a suitable tool such as the BWA- MEM. Once alignment and
tagging are complete, the AGeNT LocatIt module may be used to generate
consensus reads and mark or remove duplicates. The resulting BAM files
are ready for downstream analysis including variant discovery.
SureSelect XT HS2 RNA Library Preparation and Target Enrichment 73
Page 76

5 Post-Capture Sample Processing for Multiplexed Sequencing
Sequence analysis resources
Strandedness guidelines
The SureSelect XT HS2 RNA sequencing library preparation method
preserves RNA strandedness using dUTP second- strand marking. The
sequence of read 1, which starts at the P5 end, matches the reverse
complement of the poly- A RNA transcript strand. Read 2, which starts at
the P7 end, matches the poly- A RNA transcript strand. When running
analysis of this data to determine strandedness, it is important to include
this information. For example, when using the Picard tools
(https://broadinstitute.github.io/picard) to calculate RNA sequencing
metrics, it is important to include the parameter STRAND_SPECIFICITY=
SECOND_READ_TRANSCRIPTION_STRAND to correctly calculate the
strand specificity metrics.
74 SureSelect XT HS2 RNA Library Preparation and Target Enrichment
Page 77

SureSelect XT HS2 RNA System Protocol
6
Reference
Kit Contents 76
SureSelect XT HS2 Index Primer Pair Information 79
Troubleshooting Guide 91
Quick Reference Protocol 94
This chapter contains reference information, including component kit
contents, index sequences, and troubleshooting information.
Agilent Technologies
75
Page 78

6 Reference
Kit Contents
Kit Contents
SureSelect XT HS2 RNA Reagent Kits include the component kits listed in Table 40. Detailed
contents of each of the multi- part component kits listed in Table 40 are shown in Table 41
through Table 45 on the following pages.
Table 40 Component Kits
Component Kit Name Storage
Condition
Standard Component Modules
SureSelect cDNA Module (Pre PCR) –20°C 5500-0148 5500-0149
SureSelect XT HS2 RNA Library
Preparation Kit for ILM (Pre PCR)
SureSelect XT HS2 Index Primer
Pairs for ILM (Pre PCR)
SureSelect Target Enrichment Kit,
ILM Hyb Module, Box 1 (Post PCR)
SureSelect XT HS2 Target
Enrichment Kit, ILM Hyb Module,
Box 2 (Post PCR)
Optional Component Modules
SureSelect RNA AMPure® XP Beads +4°C 5191-6670*
SureSelect Streptavidin Beads +4°C 5191-5741* 5191-5742
* Provided only with 16-Reaction Reagent Kit part number G9990A.
–20°C 5500-0150 5500-0151
–20°C 5191-5687 (Index Pairs 1–16) 5191-5688 (Index Pairs 1–96),
Room
Temperature
–20°C 5191-6686 5191-6688
Component Kit Part Number
16 Reaction Kits 96 Reaction Kits
5191-5689 (Index Pairs 97–192),
5191-5690 (Index Pairs 193–288), OR
5191-5691 (Index Pairs 289–384)
5190-9685 5190-9687
5191-6671
†
†
† Provided only with 96-Reaction Reagent Kit part numbers G9992A, G9992B, G9992C, G9992D.
76 SureSelect XT HS2 RNA Library Preparation and Target Enrichment
Page 79

Reference 6
Kit Contents
The contents of each of the multi- part component kits listed in Table 40 are described in
the tables below.
Table 41 SureSelect cDNA Module (Pre PCR) Content
Kit Component 16 Reaction Kit Format 96 Reaction Kit Format
2X Priming Buffer tube with purple cap tube with purple cap
First Strand Master Mix
Second Strand Enzyme Mix tube with blue cap bottle
Second Strand Oligo Mix tube with yellow cap tube with yellow cap
* The First Strand Master Mix contains actinomycin-D. Keep the reagent in the supplied amber vial to protect the contents from
exposure to light.
*
amber tube with amber cap amber tube with amber cap
Table 42 SureSelect XT HS2 RNA Library Preparation Kit for ILM (Pre PCR) Content
Kit Component 16 Reaction Kit Format 96 Reaction Kit Format
End Repair-A Tailing Enzyme Mix tube with orange cap tube with orange cap
End Repair-A Tailing Buffer tube with yellow cap bottle
T4 DNA Ligase tube with blue cap tube with blue cap
Ligation Buffer tube with purple cap bottle
XT HS2 RNA Adaptor Oligo Mix tube with green cap tube with green cap
Herculase II Fusion DNA Polymerase tube with red cap tube with red cap
5× Herculase II Reaction Buffer with dNTPs tube with clear cap tube with clear cap
Table 43 SureSelect XT HS2 Index Primer Pairs for ILM (Pre PCR) Content
Kit Component 16 Reaction Kit Format 96 Reaction Kit Format
SureSelect XT HS2 Index
Primer Pairs for ILM (Pre PCR)
Blue 8-well strip tube (index pairs 1-8), AND
White 8-well strip tube (index pairs 9-16)
Orange 96-well plate (index pairs 1–96), OR
Blue 96-well plate (index pairs 97–192), OR
Green 96-well plate (index pairs 193–288), OR
Red 96-well plate (index pairs 289–384)
SureSelect XT HS2 RNA Library Preparation and Target Enrichment 77
Page 80

6 Reference
Kit Contents
Table 44 SureSelect Target Enrichment Kit, ILM Hyb Module Box 1 (Post PCR) Content
Kit Component 16 Reaction Kit Format 96 Reaction Kit Format
SureSelect Binding Buffer bottle bottle
SureSelect Wash Buffer 1 bottle bottle
SureSelect Wash Buffer 2 bottle bottle
Table 45 SureSelect XT HS2 Target Enrichment Kit, ILM Hyb Module Box 2 (Post PCR) Content
Kit Component 16 Reaction Kit Format 96 Reaction Kit Format
SureSelect Fast Hybridization Buffer bottle bottle
SureSelect XT HS2 Blocker Mix tube with blue cap tube with blue cap
SureSelect RNase Block tube with purple cap tube with purple cap
SureSelect Post-Capture Primer Mix tube with clear cap tube with clear cap
Herculase II Fusion DNA Polymerase tube with red cap tube with red cap
5× Herculase II Buffer with dNTPs tube with clear cap tube with clear cap
78 SureSelect XT HS2 RNA Library Preparation and Target Enrichment
Page 81

SureSelect XT HS2 Index Primer Pair Information
SureSelect XT HS2 Index Primer Pair Information
The SureSelect XT HS2 Index Primer Pairs are provided pre- combined.
Each member of the primer pair contains a unique 8- bp P5 or P7 index,
resulting in dual- indexed NGS libraries. The nucleotide sequence of the
index portion of each primer is provided in Table 46 through Table 53.
See page 68 for sequencing run setup requirements for sequencing
libraries using 8- bp indexes.
NOTE
P7 indexes are shown in a single orientation, applicable to any of the supported Illumina
platforms. P5 indexes are shown in two orientations for different platforms; check the table
column headings carefully before selecting the P5 sequences.
The first P5 index orientation is applicable to the supported platforms NovaSeq 6000 with
v1.0 chemistry, MiSeq, and HiSeq 2500. This orientation is also applicable to the HiSeq 2000
platform that is not specifically supported in this user manual.
The second P5 index orientation is applicable to the supported platforms NovaSeq 6000
with v1.5 chemistry, NextSeq 500/550, HiSeq 4000 and HiSeq 3000. This orientation is also
applicable to the iSeq 100, MiniSeq, and HiSeq X platforms that are not specifically
supported in this user manual.
Reference 6
One primer pair is provided in each well of 8- well strip tubes (16 reaction
kits; see Figure 8 for a map) or of 96- well plates (96 reaction kits; see
page 89 through page 90 for plate maps). Each well contains a single- use
aliquot of a specific pair of forward plus reverse primers.
SureSelect XT HS2 RNA Library Preparation and Target Enrichment 79
Page 82

6 Reference
SureSelect XT HS2 Index Primer Pair Information
Table 46 SureSelect XT HS2 Index Primer Pairs 1–48, provided in orange 96-well plate or in strip tubes
Primer
Pair #
Well P7 Index P5 Index
NovaSeq (v1.0
chemistry),
MiSeq,
HiSeq 2500
P5 Index
NovaSeq (v1.5
chemistry),
NextSeq,
HiSeq 4000,
HiSeq 3000
Primer
Pair #
Well P7 Index P5 Index
NovaSeq (v1.0
chemistry),
MiSeq,
HiSeq 2500
P5 Index
NovaSeq (v1.5
chemistry),
NextSeq,
HiSeq 4000,
HiSeq 3000
1 A01 CAAGGTGA ATGGTTAG CTAACCAT 25 A04 AGATGGAT TGGCACCA TGGTGCCA
2 B01 TAGACCAA CAAGGTGA TCACCTTG 26 B04 GAATTGTG AGATGGAT ATCCATCT
3 C01 AGTCGCGA TAGACCAA TTGGTCTA 27 C04 GAGCACTG GAATTGTG CACAATTC
4 D01 CGGTAGAG AGTCGCGA TCGCGACT 28 D04 GTTGCGGA GAGCACTG CAGTGCTC
5 E01 TCAGCATC AAGGAGCG CGCTCCTT 29 E04 AATGGAAC GTTGCGGA TCCGCAAC
6 F01 AGAAGCAA TCAGCATC GATGCTGA 30 F04 TCAGAGGT AATGGAAC GTTCCATT
7 G01 GCAGGTTC AGAAGCAA TTGCTTCT 31 G04 GCAACAAT TCAGAGGT ACCTCTGA
8 H01 AAGTGTCT GCAGGTTC GAACCTGC 32 H04 GTCGATCG GCAACAAT ATTGTTGC
9 A02 CTACCGAA AAGTGTCT AGACACTT 33 A05 ATGGTAGC GTCGATCG CGATCGAC
10 B02 TAGAGCTC CTACCGAA TTCGGTAG 34 B05 CGCCAATT ATGGTAGC GCTACCAT
11 C02 ATGTCAAG TAGAGCTC GAGCTCTA 35 C05 GACAATTG CGCCAATT AATTGGCG
12 D02 GCATCATA ATGTCAAG CTTGACAT 36 D05 ATATTCCG GACAATTG CAATTGTC
13 E02 GACTTGAC GCATCATA TATGATGC 37 E05 TCTACCTC ATATTCCG CGGAATAT
14 F02 CTACAATG GACTTGAC GTCAAGTC 38 F05 TCGTCGTG TCTACCTC GAGGTAGA
15 G02 TCTCAGCA CTACAATG CATTGTAG 39 G05 ATGAGAAC TCGTCGTG CACGACGA
16 H02 AGACACAC TCTCAGCA TGCTGAGA 40 H05 GTCCTATA ATGAGAAC GTTCTCAT
17 A03 CAGGTCTG AGACACAC GTGTGTCT 41 A06 AATGACCA GTCCTATA TATAGGAC
18 B03 AATACGCG CAGGTCTG CAGACCTG 42 B06 CAGACGCT AATGACCA TGGTCATT
19 C03 GCACACAT AATACGCG CGCGTATT 43 C06 TCGAACTG CAGACGCT AGCGTCTG
20 D03 CTTGCATA GCACACAT ATGTGTGC 44 D06 CGCTTCCA TCGAACTG CAGTTCGA
21 E03 ATCCTCTT CTTGCATA TATGCAAG 45 E06 TATTCCTG CGCTTCCA TGGAAGCG
22 F03 GCACCTAA ATCCTCTT AAGAGGAT 46 F06 CAAGTTAC TATTCCTG CAGGAATA
23 G03 TGCTGCTC GCACCTAA TTAGGTGC 47 G06 CAGAGCAG CAAGTTAC GTAACTTG
24 H03 TGGCACCA TGCTGCTC GAGCAGCA 48 H06 CGCGCAAT CAGAGCAG CTGCTCTG
80 SureSelect XT HS2 RNA Library Preparation and Target Enrichment
Page 83

SureSelect XT HS2 Index Primer Pair Information
Table 47 SureSelect XT HS2 Index Primer Pairs 49–96, provided in orange 96-well plate
Reference 6
Primer
Pair #
Well P7 Index P5 Index
NovaSeq (v1.0
chemistry),
MiSeq,
HiSeq 2500
P5 Index
NovaSeq (v1.5
chemistry),
NextSeq,
HiSeq 4000,
HiSeq 3000
Primer
Pair #
Well P7 Index P5 Index
NovaSeq (v1.0
chemistry),
MiSeq,
HiSeq 2500
P5 Index
NovaSeq (v1.5
chemistry),
NextSeq,
HiSeq 4000,
HiSeq 3000
49 A07 TGAGGAGT CGCGCAAT ATTGCGCG 73 A10 AACGCATT ATAGTGAC GTCACTAT
50 B07 ATGACGAA TGAGGAGT ACTCCTCA 74 B10 CAGTTGCG AACGCATT AATGCGTT
51 C07 TACGGCGA ATGACGAA TTCGTCAT 75 C10 TGCCTCGA CAGTTGCG CGCAACTG
52 D07 AGCGAGTT TACGGCGA TCGCCGTA 76 D10 AAGGCTTA TGCCTCGA TCGAGGCA
53 E07 TGTATCAC AGCGAGTT AACTCGCT 77 E10 GCAATGAA AAGGCTTA TAAGCCTT
54 F07 GATCGCCT TGTATCAC GTGATACA 78 F10 AAGAACCT GCAATGAA TTCATTGC
55 G07 GACTCAAT GATCGCCT AGGCGATC 79 G10 CTGTGCCT AAGAACCT AGGTTCTT
56 H07 CAGCTTGC GACTCAAT ATTGAGTC 80 H10 TACGTAGC CTGTGCCT AGGCACAG
57 A08 AGCTGAAG CAGCTTGC GCAAGCTG 81 A11 AAGTGGAC TACGTAGC GCTACGTA
58 B08 ATTCCGTG AGCTGAAG CTTCAGCT 82 B11 CAACCGTG AAGTGGAC GTCCACTT
59 C08 TATGCCGC ATTCCGTG CACGGAAT 83 C11 CTGTTGTT CAACCGTG CACGGTTG
60 D08 TCAGCTCA TATGCCGC GCGGCATA 84 D11 GCACGATG CTGTTGTT AACAACAG
61 E08 AACTGCAA TCAGCTCA TGAGCTGA 85 E11 GTACGGAC GCACGATG CATCGTGC
62 F08 ATTAGGAG AACTGCAA TTGCAGTT 86 F11 CTCCAAGC GTACGGAC GTCCGTAC
63 G08 CAGCAATA ATTAGGAG CTCCTAAT 87 G11 TAGTCTGA CTCCAAGC GCTTGGAG
64 H08 GCCAAGCT CAGCAATA TATTGCTG 88 H11 TTCGCCGT TAGTCTGA TCAGACTA
65 A09 TCCGTTAA GCCAAGCT AGCTTGGC 89 A12 GAACTAAG ATACGAAG CTTCGTAT
66 B09 GTGCAACG TCCGTTAA TTAACGGA 90 B12 AAGCCATC GAGATTCA TGAATCTC
67 C09 AGTAACGC GTGCAACG CGTTGCAC 91 C12 AACTCTTG AAGCCATC GATGGCTT
68 D09 CATAGCCA AGTAACGC GCGTTACT 92 D12 GTAGTCAT AACTCTTG CAAGAGTT
69 E09 CACTAGTA CATAGCCA TGGCTATG 93 E12 CTCGCTAG GTAGTCAT ATGACTAC
70 F09 TTAGTGCG CACTAGTA TACTAGTG 94 F12 AGTCTTCA CAGTATCA TGATACTG
71 G09 TCGATACA TTAGTGCG CGCACTAA 95 G12 TCAAGCTA CTTCGTAC GTACGAAG
72 H09 ATAGTGAC TCGATACA TGTATCGA 96 H12 CTTATCCT TCAAGCTA TAGCTTGA
SureSelect XT HS2 RNA Library Preparation and Target Enrichment 81
Page 84

6 Reference
SureSelect XT HS2 Index Primer Pair Information
Table 48 SureSelect XT HS2 Index Primer Pairs 97–144, provided in blue 96-well plate
Primer
Pair #
Well P7 Index P5 Index
NovaSeq (v1.0
chemistry),
MiSeq,
HiSeq 2500
P5 Index
NovaSeq (v1.5
chemistry),
NextSeq,
HiSeq 4000,
HiSeq 3000
Primer
Pair #
Well P7 Index P5 Index
NovaSeq (v1.0
chemistry),
MiSeq,
HiSeq 2500
P5 Index
NovaSeq (v1.5
chemistry),
NextSeq,
HiSeq 4000,
HiSeq 3000
97 A01 TCATCCTT CTTATCCT AGGATAAG 121 A04 CAGGCAGA AGACGCCT AGGCGTCT
B01 AACACTCT TCATCCTT AAGGATGA 122 B04 TCCGCGAT CAGGCAGA TCTGCCTG
98
C01 CACCTAGA AACACTCT AGAGTGTT 123 C04 CTCGTACG TCCGCGAT ATCGCGGA
99
100
101
102
103
104
105
106
107
108
109
110
111
112
113
114
115
116
117
118
119
120
D01 AGTTCATG CACCTAGA TCTAGGTG 124 D04 CACACATA CTCGTACG CGTACGAG
E01 GTTGGTGT AGTTCATG CATGAACT 125 E04 CGTCAAGA CACACATA TATGTGTG
F01 GCTACGCA GTTGGTGT ACACCAAC 126 F04 TTCGCGCA CGTCAAGA TCTTGACG
G01 TCAACTGC GCTACGCA TGCGTAGC 127 G04 CGACTACG TTCGCGCA TGCGCGAA
H01 AAGCGAAT TCAACTGC GCAGTTGA 128 H04 GAAGGTAT CGACTACG CGTAGTCG
A02 GTGTTACA AAGCGAAT ATTCGCTT 129 A05 TTGGCATG GAAGGTAT ATACCTTC
B02 CAAGCCAT GTGTTACA TGTAACAC 130 B05 CGAATTCA TTGGCATG CATGCCAA
C02 CTCTCGTG CAAGCCAT ATGGCTTG 131 C05 TTAGTTGC CGAATTCA TGAATTCG
D02 TCGACAAC CTCTCGTG CACGAGAG 132 D05 GATGCCAA TTAGTTGC GCAACTAA
E02 TCGATGTT TCGACAAC GTTGTCGA 133 E05 AGTTGCCG GATGCCAA TTGGCATC
F02 CAAGGAAG TCGATGTT AACATCGA 134 F05 GTCCACCT AGTTGCCG CGGCAACT
G02 ATTGATGC AGAGAATC GATTCTCT 135 G05 ATCAAGGT GTCCACCT AGGTGGAC
H02 TCGCAGAT TTGATGGC GCCATCAA 136 H05 GAACCAGA ATCAAGGT ACCTTGAT
A03 GCAGAGAC TCGCAGAT ATCTGCGA 137 A06 CATGTTCT GAACCAGA TCTGGTTC
B03 CTGCGAGA GCAGAGAC GTCTCTGC 138 B06 TCACTGTG CATGTTCT AGAACATG
C03 CAACCAAC CTGCGAGA TCTCGCAG 139 C06 ATTGAGCT TCACTGTG CACAGTGA
D03 ATCATGCG CAACCAAC GTTGGTTG 140 D06 GATAGAGA ATTGAGCT AGCTCAAT
E03 TCTGAGTC ATCATGCG CGCATGAT 141 E06 TCTAGAGC GATAGAGA TCTCTATC
F03 TCGCCTGT TCTGAGTC GACTCAGA 142 F06 GAATCGCA TCTAGAGC GCTCTAGA
G03 GCGCAATT TCGCCTGT ACAGGCGA 143 G06 CTTCACGT GAATCGCA TGCGATTC
H03 AGACGCCT GCGCAATT AATTGCGC 144 H06 CTCCGGTT CTTCACGT ACGTGAAG
82 SureSelect XT HS2 RNA Library Preparation and Target Enrichment
Page 85

SureSelect XT HS2 Index Primer Pair Information
Table 49 SureSelect XT HS2 Index Primer Pairs 145–192, provided in blue 96-well plate
Reference 6
Primer
Pair #
Well P7 Index P5 Index
NovaSeq (v1.0
chemistry),
MiSeq,
HiSeq 2500
P5 Index
NovaSeq (v1.5
chemistry),
NextSeq,
HiSeq 4000,
HiSeq 3000
Primer
Pair #
Well P7 Index P5 Index
NovaSeq (v1.0
chemistry),
MiSeq,
HiSeq 2500
P5 Index
NovaSeq (v1.5
chemistry),
NextSeq,
HiSeq 4000,
HiSeq 3000
145 A07 TGTGACTA CTCCGGTT AACCGGAG 169 A10 CGCTCAGA CTAACAAG CTTGTTAG
146
147
148
149
150
151
152
153
154
155
156
157
158
159
160
161
162
163
164
165
166
167
168
B07 GCTTCCAG TGTGACTA TAGTCACA 170 B10 TAACGACA CGCTCAGA TCTGAGCG
C07 CATCCTGT GCTTCCAG CTGGAAGC 171 C10 CATACTTG TAACGACA TGTCGTTA
D07 GTAATACG CATCCTGT ACAGGATG 172 D10 AGATACGA CATACTTG CAAGTATG
E07 GCCAACAA GTAATACG CGTATTAC 173 E10 AATCCGAC AGATACGA TCGTATCT
F07 CATGACAC GCCAACAA TTGTTGGC 174 F10 TGAAGTAC AATCCGAC GTCGGATT
G07 TGCAATGC CATGACAC GTGTCATG 175 G10 CGAATCAT TGAAGTAC GTACTTCA
H07 CACATTCG TGCAATGC GCATTGCA 176 H10 TGATTGGC CGAATCAT ATGATTCG
A08 CAATCCGA CACATTCG CGAATGTG 177 A11 TCGAAGGA TGATTGGC GCCAATCA
B08 CATCGACG CAATCCGA TCGGATTG 178 B11 CAGTCATT TCGAAGGA TCCTTCGA
C08 GTGCGCTT CATCGACG CGTCGATG 179 C11 CGCGAACA CAGTCATT AATGACTG
D08 ATAGCGTT GTGCGCTT AAGCGCAC 180 D11 TACGGTTG CGCGAACA TGTTCGCG
E08 GAGTAAGA ATAGCGTT AACGCTAT 181 E11 AGAACCGT TACGGTTG CAACCGTA
F08 CTGACACA GAGTAAGA TCTTACTC 182 F11 AGGTGCTT AGAACCGT ACGGTTCT
G08 ATACGTGT CTGACACA TGTGTCAG 183 G11 ATCGCAAC AGGTGCTT AAGCACCT
H08 GACCGAGT ATACGTGT ACACGTAT 184 H11 GCCTCTCA ATCGCAAC GTTGCGAT
A09 GCAGTTAG GACCGAGT ACTCGGTC 185 A12 TCGCGTCA GCCTCTCA TGAGAGGC
B09 CGTTCGTC GCAGTTAG CTAACTGC 186 B12 GAGTGCGT TCGCGTCA TGACGCGA
C09 CGTTAACG CGTTCGTC GACGAACG 187 C12 CGAACACT GCATAAGT ACTTATGC
D09 TCGAGCAT CGTTAACG CGTTAACG 188 D12 TAAGAGTG AGAAGACG CGTCTTCT
E09 GCCGTAAC TCGAGCAT ATGCTCGA 189 E12 TGGATTGA TAAGAGTG CACTCTTA
F09 GAGCTGTA GCCGTAAC GTTACGGC 190 F12 AGGACATA TGGATTGA TCAATCCA
G09 AGGAAGAT GAGCTGTA TACAGCTC 191 G12 GACATCCT AGGACATA TATGTCCT
H09 CTAACAAG AGGAAGAT ATCTTCCT 192 H12 GAAGCCTC GACATCCT AGGATGTC
SureSelect XT HS2 RNA Library Preparation and Target Enrichment 83
Page 86

6 Reference
SureSelect XT HS2 Index Primer Pair Information
Table 50 SureSelect XT HS2 Index Primer Pairs 193–240, provided in green 96-well plate
Primer
Pair #
Well P7 Index P5 Index
NovaSeq (v1.0
chemistry),
MiSeq,
HiSeq 2500
P5 Index
NovaSeq (v1.5
chemistry),
NextSeq,
HiSeq 4000,
HiSeq 3000
Primer
Pair #
Well P7 Index P5 Index
NovaSeq (v1.0
chemistry),
MiSeq,
HiSeq 2500
P5 Index
NovaSeq (v1.5
chemistry),
NextSeq,
HiSeq 4000,
HiSeq 3000
193 A01 GTCTCTTC GAAGCCTC GAGGCTTC 217 A04 GCGGTATG CACGAGCT AGCTCGTG
194 B01 AGTCACTT GTCTCTTC GAAGAGAC 218 B04 TCTATGCG GCGGTATG CATACCGC
195 C01 AGCATACA AGTCACTT AAGTGACT 219 C04 AGGTGAGA TCTATGCG CGCATAGA
196 D01 TCAGACAA AGCATACA TGTATGCT 220 D04 CACAACTT AGGTGAGA TCTCACCT
197 E01 TTGGAGAA TCAGACAA TTGTCTGA 221 E04 TTGTGTAC CACAACTT AAGTTGTG
198 F01 TTAACGTG TTGGAGAA TTCTCCAA 222 F04 TCACAAGA TTGTGTAC GTACACAA
199 G01 CGTCTGTG TTAACGTG CACGTTAA 223 G04 GAAGACCT TCACAAGA TCTTGTGA
200 H01 AACCTAAC CGTCTGTG CACAGACG 224 H04 AGTTCTGT GAAGACCT AGGTCTTC
201 A02 AGAGTGCT AACCTAAC GTTAGGTT 225 A05 GCAGTGTT AGTTCTGT ACAGAACT
202 B02 TTATCTCG AGAGTGCT AGCACTCT 226 B05 AGGCATGC GCAGTGTT AACACTGC
203 C02 CATCAGTC TTATCTCG CGAGATAA 227 C05 AAGGTACT AGGCATGC GCATGCCT
204 D02 AAGCACAA CATCAGTC GACTGATG 228 D05 CACTAAGT AAGGTACT AGTACCTT
205 E02 CAGTGAGC AAGCACAA TTGTGCTT 229 E05 GAGTCCTA CACTAAGT ACTTAGTG
206 F02 GTCGAAGT CAGTGAGC GCTCACTG 230 F05 AGTCCTTC GAGTCCTA TAGGACTC
207 G02 TCTCATGC GTCGAAGT ACTTCGAC 231 G05 TTAGGAAC AGTCCTTC GAAGGACT
208 H02 CAGAAGAA TCTCATGC GCATGAGA 232 H05 AAGTCCAT TTAGGAAC GTTCCTAA
209 A03 CGGATAGT CAGAAGAA TTCTTCTG 233 A06 GAATACGC AAGTCCAT ATGGACTT
210 B03 CACGTGAG CGGATAGT ACTATCCG 234 B06 TCCAATCA GAATACGC GCGTATTC
211 C03 TACGATAC CACGTGAG CTCACGTG 235 C06 CGACGGTA TCCAATCA TGATTGGA
212 D03 CGCATGCT TACGATAC GTATCGTA 236 D06 CATTGCAT CGACGGTA TACCGTCG
213 E03 GCTTGCTA CGCATGCT AGCATGCG 237 E06 ATCTGCGT CATTGCAT ATGCAATG
214 F03 GAACGCAA GCTTGCTA TAGCAAGC 238 F06 GTACCTTG ATCTGCGT ACGCAGAT
215 G03 ATCTACCA GAACGCAA TTGCGTTC 239 G06 GAGCATAC GTACCTTG CAAGGTAC
216 H03 CACGAGCT ATCTACCA TGGTAGAT 240 H06 TGCTTACG GAGCATAC GTATGCTC
84 SureSelect XT HS2 RNA Library Preparation and Target Enrichment
Page 87

SureSelect XT HS2 Index Primer Pair Information
Table 51 SureSelect XT HS2 Index Primer Pairs 241–288, provided in green 96-well plate
Reference 6
Primer
Pair #
Well P7 Index P5 Index
NovaSeq (v1.0
chemistry),
MiSeq,
HiSeq 2500
P5 Index
NovaSeq (v1.5
chemistry),
NextSeq,
HiSeq 4000,
HiSeq 3000
Primer
Pair #
Well P7 Index P5 Index
NovaSeq (v1.0
chemistry),
MiSeq,
HiSeq 2500
P5 Index
NovaSeq (v1.5
chemistry),
NextSeq,
HiSeq 4000,
HiSeq 3000
241 A07 AAGAGACA TGCTTACG CGTAAGCA 265 A10 CAATGCTG CATGAATG CATTCATG
242 B07 TAGCTATG AAGAGACA TGTCTCTT 266 B10 CTTGATCA CAATGCTG CAGCATTG
243 C07 TCTGCTAC TAGCTATG CATAGCTA 267 C10 GCGAATTA CTTGATCA TGATCAAG
244 D07 GTCACAGA TCTGCTAC GTAGCAGA 268 D10 GTTCGAGC GCGAATTA TAATTCGC
245 E07 CGATTGAA GTCACAGA TCTGTGAC 269 E10 GCCAGTAG GTTCGAGC GCTCGAAC
246 F07 GAGAGATT CGATTGAA TTCAATCG 270 F10 AAGGTCGA GCCAGTAG CTACTGGC
247 G07 TCATACCG GAGAGATT AATCTCTC 271 G10 AGTGAAGT CACTTATG CATAAGTG
248 H07 TCCGAACT TCATACCG CGGTATGA 272 H10 GTTGCAAG ATAACGGC GCCGTTAT
249 A08 AGAGAGAA TCCGAACT AGTTCGGA 273 A11 AGCCGGAA GTTGCAAG CTTGCAAC
250 B08 GATCGTTA AGAGAGAA TTCTCTCT 274 B11 AACAGCCG AGCCGGAA TTCCGGCT
251 C08 GCGCTAGA GATCGTTA TAACGATC 275 C11 CTAGTGTA AACAGCCG CGGCTGTT
252 D08 ATGACTCG GCGCTAGA TCTAGCGC 276 D11 GAGGCTCT CTAGTGTA TACACTAG
253 E08 CAATAGAC ATGACTCG CGAGTCAT 277 E11 CTCCGCAA GAGGCTCT AGAGCCTC
254 F08 CGATATGC CAATAGAC GTCTATTG 278 F11 CGCTATTG CTCCGCAA TTGCGGAG
255 G08 GTCAGAAT CGATATGC GCATATCG 279 G11 GTGTTGAG CGCTATTG CAATAGCG
256 H08 CATAAGGT GCACTACT AGTAGTGC 280 H11 TCACCGAC GTGTTGAG CTCAACAC
257 A09 TGTTGGTT GATTCGGC GCCGAATC 281 A12 CGGTAATC TCACCGAC GTCGGTGA
258 B09 ATACTCGC TGTTGGTT AACCAACA 282 B12 GTGACTGC CGGTAATC GATTACCG
259 C09 AATGCTAG ATACTCGC GCGAGTAT 283 C12 CGACTTGT GTGACTGC GCAGTCAC
260 D09 GCCTAGGA AATGCTAG CTAGCATT 284 D12 GATAGGAC CGACTTGT ACAAGTCG
261 E09 GCAACCGA GCCTAGGA TCCTAGGC 285 E12 AAGTACTC GATAGGAC GTCCTATC
262 F09 ATACTGCA GCAACCGA TCGGTTGC 286 F12 GCTCTCTC AAGTACTC GAGTACTT
263 G09 TCTCCTTG ATACTGCA TGCAGTAT 287 G12 CTACCAGT GCTCTCTC GAGAGAGC
264 H09 CATGAATG TCTCCTTG CAAGGAGA 288 H12 GATGAGAT CTACCAGT ACTGGTAG
SureSelect XT HS2 RNA Library Preparation and Target Enrichment 85
Page 88

6 Reference
SureSelect XT HS2 Index Primer Pair Information
Table 52 SureSelect XT HS2 Index Primer Pairs 289–336, provided in red 96-well plate
Primer
Pair #
Well P7 Index P5 Index
NovaSeq (v1.0
chemistry),
MiSeq,
HiSeq 2500
P5 Index
NovaSeq (v1.5
chemistry),
NextSeq,
HiSeq 4000,
HiSeq 3000
Primer
Pair #
Well P7 Index P5 Index
NovaSeq (v1.0
chemistry),
MiSeq,
HiSeq 2500
P5 Index
NovaSeq (v1.5
chemistry),
NextSeq,
HiSeq 4000,
HiSeq 3000
289 A01 AGATAGTG GATGAGAT ATCTCATC 313 A04 AGCTACAT GATCCATG CATGGATC
290 B01 AGAGGTTA AGATAGTG CACTATCT 314 B04 CGCTGTAA AGCTACAT ATGTAGCT
291 C01 CTGACCGT AGAGGTTA TAACCTCT 315 C04 CACTACCG CGCTGTAA TTACAGCG
292 D01 GCATGGAG CTGACCGT ACGGTCAG 316 D04 GCTCACGA CACTACCG CGGTAGTG
293 E01 CTGCCTTA GCATGGAG CTCCATGC 317 E04 TGGCTTAG GCTCACGA TCGTGAGC
294 F01 GCGTCACT CTGCCTTA TAAGGCAG 318 F04 TCCAGACG TGGCTTAG CTAAGCCA
295 G01 GCGATTAC GCGTCACT AGTGACGC 319 G04 AGTGGCAT TCCAGACG CGTCTGGA
296 H01 TCACCACG GCGATTAC GTAATCGC 320 H04 TGTACCGA AGTGGCAT ATGCCACT
297 A02 AGACCTGA TCACCACG CGTGGTGA 321 A05 AAGACTAC TGTACCGA TCGGTACA
298 B02 GCCGATAT AGACCTGA TCAGGTCT 322 B05 TGCCGTTA AAGACTAC GTAGTCTT
299 C02 CTTATTGC GCCGATAT ATATCGGC 323 C05 TTGGATCT TGCCGTTA TAACGGCA
300 D02 CGATACCT CTTATTGC GCAATAAG 324 D05 TCCTCCAA TTGGATCT AGATCCAA
301 E02 CTCGACAT CGATACCT AGGTATCG 325 E05 CGAGTCGA TCCTCCAA TTGGAGGA
302 F02 GAGATCGC CTCGACAT ATGTCGAG 326 F05 AGGCTCAT CGAGTCGA TCGACTCG
303 G02 CGGTCTCT GAGATCGC GCGATCTC 327 G05 GACGTGCA AGGCTCAT ATGAGCCT
304 H02 TAACTCAC CGGTCTCT AGAGACCG 328 H05 GAACATGT GACGTGCA TGCACGTC
305 A03 CACAATGA TAACTCAC GTGAGTTA 329 A06 AATTGGCA GAACATGT ACATGTTC
306 B03 GACTGACG CACAATGA TCATTGTG 330 B06 TGGAGACT AATTGGCA TGCCAATT
307 C03 CTTAAGAC GACTGACG CGTCAGTC 331 C06 AACTCACA TGGAGACT AGTCTCCA
308 D03 GAGTGTAG CTTAAGAC GTCTTAAG 332 D06 GTAGACTG AACTCACA TGTGAGTT
309 E03 TGCACATC GAGTGTAG CTACACTC 333 E06 CGTAGTTA GTAGACTG CAGTCTAC
310 F03 CGATGTCG TGCACATC GATGTGCA 334 F06 CGTCAGAT CGTAGTTA TAACTACG
311 G03 AACACCGA CGATGTCG CGACATCG 335 G06 AACGGTCA CGTCAGAT ATCTGACG
312 H03 GATCCATG AACACCGA TCGGTGTT 336 H06 GCCTTCAT AACGGTCA TGACCGTT
86 SureSelect XT HS2 RNA Library Preparation and Target Enrichment
Page 89

SureSelect XT HS2 Index Primer Pair Information
Table 53 SureSelect XT HS2 Index Primer Pairs 337–384, provided in red 96-well plate
Reference 6
Primer
Pair #
Well P7 Index P5 Index
NovaSeq (v1.0
chemistry),
MiSeq,
HiSeq 2500
P5 Index
NovaSeq (v1.5
chemistry),
NextSeq,
HiSeq 4000,
HiSeq 3000
Primer
Pair #
Well P7 Index P5 Index
NovaSeq (v1.0
chemistry),
MiSeq,
HiSeq 2500
P5 Index
NovaSeq (v1.5
chemistry),
NextSeq,
HiSeq 4000,
HiSeq 3000
337 A07 TGAGACGC GCCTTCAT ATGAAGGC 361 A10 CTGAGCTA GCACAGTA TACTGTGC
338 B07 CATCGGAA TGAGACGC GCGTCTCA 362 B10 CTTGCGAT CTGAGCTA TAGCTCAG
339 C07 TAGGACAT CATCGGAA TTCCGATG 363 C10 GAAGTAGT CTTGCGAT ATCGCAAG
340 D07 AACACAAG TAGGACAT ATGTCCTA 364 D10 GTTATCGA GAAGTAGT ACTACTTC
341 E07 TTCGACTC AACACAAG CTTGTGTT 365 E10 TGTCGTCG GTTATCGA TCGATAAC
342 F07 GTCGGTAA TTCGACTC GAGTCGAA 366 F10 CGTAACTG TGTCGTCG CGACGACA
343 G07 GTTCATTC GTCGGTAA TTACCGAC 367 G10 GCATGCCT CGTAACTG CAGTTACG
344 H07 AAGCAGTT GTTCATTC GAATGAAC 368 H10 TCGTACAC GCATGCCT AGGCATGC
345 A08 ATAAGCTG AAGCAGTT AACTGCTT 369 A11 CACAGGTG TCGTACAC GTGTACGA
346 B08 GCTTAGCG ATAAGCTG CAGCTTAT 370 B11 AGCAGTGA CACAGGTG CACCTGTG
347 C08 TTCCAACA GCTTAGCG CGCTAAGC 371 C11 ATTCCAGA AGCAGTGA TCACTGCT
348 D08 TACCGCAT TTCCAACA TGTTGGAA 372 D11 TCCTTGAG ATTCCAGA TCTGGAAT
349 E08 AGGCAATG TACCGCAT ATGCGGTA 373 E11 ATACCTAC TCCTTGAG CTCAAGGA
350 F08 GCCTCGTT AGGCAATG CATTGCCT 374 F11 AGACCATT ATACCTAC GTAGGTAT
351 G08 CACGGATC GCCTCGTT AACGAGGC 375 G11 CGTAAGCA AGACCATT AATGGTCT
352 H08 GAGACACG CACGGATC GATCCGTG 376 H11 TCTGTCAG CGTAAGCA TGCTTACG
353 A09 AGAGTAAG GAGACACG CGTGTCTC 377 A12 CACAGACT TCTGTCAG CTGACAGA
354 B09 AGTACGTT AGAGTAAG CTTACTCT 378 B12 GTCGCCTA CACAGACT AGTCTGTG
355 C09 AACGCTGC AGTACGTT AACGTACT 379 C12 TGCGCTCT GTCGCCTA TAGGCGAC
356 D09 GTAGAGCA AACGCTGC GCAGCGTT 380 D12 GCTATAAG TGCGCTCT AGAGCGCA
357 E09 TCCTGAGA GTAGAGCA TGCTCTAC 381 E12 CAACAACT GCTATAAG CTTATAGC
358 F09 CTGAATAG TCCTGAGA TCTCAGGA 382 F12 AGAGAATC CTCTCACT AGTGAGAG
359 G09 CAAGACTA CTGAATAG CTATTCAG 383 G12 TAATGGTC AGACGAGC GCTCGTCT
360 H09 GCACAGTA CAAGACTA TAGTCTTG 384 H12 GTTGTATC TAATGGTC GACCATTA
SureSelect XT HS2 RNA Library Preparation and Target Enrichment 87
Page 90

6 Reference
Index Primer Pair Strip Tube and Plate Maps
Index Primer Pair Strip Tube and Plate Maps
SureSelect XT HS2 Index Primer Pairs 1- 16 (provided with 16 reaction
kits) are supplied in a set of two 8- well strip tubes as detailed below.
Figure 8 Map of the SureSelect XT HS2 Index Primer Pairs for ILM (Pre PCR) strip tubes
provided with 16 reaction kits
The blue strip contains Index Primer Pairs 1- 8, with pair #1 supplied in
the well proximal to the numeral 1 etched on the strip’s plastic end tab.
The white strip contains Index Primer Pairs 9- 16, with pair #9 supplied in
the well proximal to the numeral 9 etched on the strip’s plastic end tab.
When using the strip tube- supplied index primer pairs in the library
preparation protocol, re- seal any unused wells using the fresh foil seal
strips provided with the index strip tubes.
See Table 54 on page 89 through Table 57 on page 90 for plate maps
showing positions of the SureSelect XT HS2 Index Primer Pairs provided
with 96 reaction kits.
CAUTION
The SureSelect XT HS2 Index Primer Pairs are provided in single-use aliquots. To avoid
cross-contamination of libraries, use each well in only one library preparation reaction.
Do not retain and re-use any residual volume for subsequent experiments.
88 SureSelect XT HS2 RNA Library Preparation and Target Enrichment
Page 91

Reference 6
Index Primer Pair Strip Tube and Plate Maps
Table 54 Plate map for SureSelect XT HS2 Index Primer Pairs 1-96, provided in orange plate
1 2 3 4 5 6 7 8 9 10 11 12
A 1 9 17 25 33 41 49 57 65 73 81 89
B 2 1018263442 50 58 6674 82 90
C 3 1119273543 51 59 67 75 83 91
D 4 12202836 44 52 60 68 76 84 92
E 5 13212937 45 53 61 69 77 85 93
F 6 14223038 46 54 6270 78 86 94
G 7 15233139 47 55 63 71 79 87 95
H 8 16243240 48 56 64 72 80 88 96
Table 55 Plate map for SureSelect XT HS2 Index Primer Pairs 97-192, provided in blue plate
1 2 3 4 5 6 7 8 9 10 11 12
A 97 105 113 121 129 137 145 153 161 169 177 185
B 98 106 114 122 130 138 146 154 162 170 178 186
C 99 107 115 123 131 139 147 155 163 171 179 187
D 100 108 116 124 132 140 148 156 164 172 180 188
E 101 109 117 125 133 141 149 157 165 173 181 189
F 102 1110 118 126 134 142 150 158 166 174 182 190
G 103 111 119 127 135 143 151 159 167 175 183 191
H 104 112 120 128 136 144 152 160 168 176 184 192
SureSelect XT HS2 RNA Library Preparation and Target Enrichment 89
Page 92

6 Reference
Index Primer Pair Strip Tube and Plate Maps
Table 56 Plate map for SureSelect XT HS2 Index Primer Pairs 193-288, provided in green plate
1 2 3 4 5 6 7 8 9 10 11 12
A 193 201 209 217 225 233 241 249 257 265 273 281
B 194 202 210 218 226 234 242 250 258 266 274 282
C 195 203 211 219 227 235 243 251 259 267 275 283
D 196 204 212 220 228 236 244 252 260 268 276 284
E 197 205 213 221 229 237 245 253 261 269 277 285
F 198 206 214 222 230 238 246 254 262 270 278 286
G 199 207 215 223 231 239 247 255 263 271 279 287
H 200 208 216 224 232 240 248 256 264 272 280 288
Table 57 Plate map for SureSelect XT HS2 Index Primer Pairs 289-384, provided in red plate
1 2 3 4 5 6 7 8 9 10 11 12
A 289 297 305 313 321 329 337 345 353 361 369 377
B 290 298 306 314 322 330 338 346 354 362 370 378
C 291 299 307 315 323 331 339 347 355 363 371 379
D 292 300 308 316 324 332 340 348 356 364 372 380
E 293 301 309 317 325 333 341 349 357 365 373 381
F 294 302 310 318 326 334 342 350 358 366 374 382
G 295 303 311 319 327 335 343 351 359 367 375 383
H 296 304 312 320 328 336 344 352 360 368 376 384
90 SureSelect XT HS2 RNA Library Preparation and Target Enrichment
Page 93

Troubleshooting Guide
If yield of pre-capture libraries is low
✔ The library preparation protocol includes specific thawing, temperature
control, pipetting, and mixing instructions which are required for
optimal performance of the highly viscous buffer and enzyme solutions
used in the protocol. Be sure to adhere to all instructions when setting
up the reactions.
✔ Ensure that the ligation master mix (see page 31) is kept at room
temperature for 30–45 minutes before use.
✔ PCR cycle number may require optimization. Repeat library preparation
for the sample, increasing the pre- capture PCR cycle number by 1 to 2
cycles.
✔ Performance of the solid- phase reversible immobilization (SPRI)
purification step may be poor. Verify the expiration date for the vial of
AMPure XP beads used for purification. Adhere to all bead storage and
handling conditions recommended by the manufacturer. Ensure that the
beads are kept at room temperature for at least 30 minutes before use.
Use freshly- prepared 70% ethanol for each SPRI procedure.
✔ DNA elution during SPRI purification steps may be incomplete. Ensure
that the AMPure XP beads are not overdried just prior to sample
elution.
Reference 6
Troubleshooting Guide
If solids observed in the End Repair-A Tailing Buffer
✔ Vortex the solution at high speed until the solids are dissolved. The
observation of solids when first thawed does not impact performance,
but it is important to mix the buffer until all solutes are dissolved.
If pre-capture library fragment size is different than expected in
electropherograms
✔ FFPE RNA pre- capture libraries may have a smaller fragment size
distribution due to the presence of fragments in the input RNA that are
smaller than the target RNA fragment size.
SureSelect XT HS2 RNA Library Preparation and Target Enrichment 91
Page 94

6 Reference
Troubleshooting Guide
✔ DNA fragment size selection during SPRI purification depends upon
using the correct ratio of sample to AMPure XP beads. Before removing
an aliquot of beads for the purification step, mix the beads until the
suspension appears homogeneous and consistent in color and verify
that you are using the bead volume recommended for pre- capture
purification on page 40.
If low molecular weight adaptor-dimer peak is present in pre-capture library
electropherograms
✔ The presence of a low molecular weight peak, in addition to the
expected peak, indicates the presence of adaptor- dimers in the library.
It is acceptable to proceed to target enrichment with library samples for
which adaptor- dimers are observed in the electropherogram at low
abundance, similar to the samples analyzed on page 42. The presence of
excessive adaptor- dimers in the samples may be associated with
reduced yield of pre- capture libraries. If excessive adaptor- dimers are
observed, verify that the adaptor ligation protocol is being performed as
directed on page 34. In particular, ensure that the Ligation master mix
is mixed with the sample prior to adding the XT HS2 RNA Adaptor
Oligo Mix to the mixture. Do not add the Ligation master mix and the
Adaptor Oligo Mix to the sample in a single step.
If yield of post-capture libraries is low
✔ PCR cycle number may require optimization. Repeat library preparation
and target enrichment for the sample, increasing the post- capture PCR
cycle number by 1 to 2 cycles.
✔ The RNA probe capture library used for hybridization may have been
compromised. Verify the expiration date on the probe vial or Certificate
of Analysis. Adhere to the recommended storage and handling
conditions. Ensure that the Capture Library Hybridization Mix is
prepared immediately before use, as directed on page 49, and that
solutions containing the probe are not held at room temperature for
extended periods.
92 SureSelect XT HS2 RNA Library Preparation and Target Enrichment
Page 95

Reference 6
Troubleshooting Guide
If post-capture library fragment size is different than expected in
electropherograms
✔ DNA fragment size selection during SPRI purification depends upon
using the correct ratio of sample to AMPure XP beads. Before removing
an aliquot of beads for the purification step, mix the beads until the
suspension appears homogeneous and consistent in color and verify
that you are using the bead volume recommended for post- capture
purification on page 59.
If low % on-target is observed in library sequencing results
✔ Stringency of post- hybridization washes may have been lower than
required. Complete the wash steps as directed, paying special attention
to the details of SureSelect Wash Buffer 2 washes listed below:
• SureSelect Wash Buffer 2 is pre- warmed to 70°C (see page 52)
• Samples are maintained at 70°C during washes (see page 53)
• Bead suspensions are mixed thoroughly during washes by pipetting
up and down and vortexing (see page 53)
✔ Minimize the amount of time that hybridization reactions are exposed
to RT conditions during hybridization setup. Locate a vortex and plate
spinner or centrifuge in close proximity to thermal cycler to retain the
65°C sample temperature during mixing and transfer steps (step 8 to
step 9 on page 50).
SureSelect XT HS2 RNA Library Preparation and Target Enrichment 93
Page 96

6 Reference
Quick Reference Protocol
Quick Reference Protocol
An abbreviated summary of the protocol steps is provided below for
experienced users. Use the complete protocol on page 17 to page 64 until
you are familiar with all of the protocol details such as reagent mixing
instructions and instrument settings.
H
Step Summary of Conditions
RNA Fragmentation and cDNA Preparation
Prepare and qualify RNA
samples
Fragment RNA and prime
cDNA synthesis
Synthesize first-strand
cDNA
Synthesize second-strand
cDNA
Purify cDNA 58.5 µl cDNA sample + 105 µl AMPure XP bead suspension
Prepare Ligation master mix Per reaction: 23 µl Ligation Buffer + 2 µl T4 DNA Ligase
Prepare
End-Repair/dA-Tailing
master mix
End-Repair and dA-Tail the
DNA fragments
Prepare 10–200 ng total RNA in 10 µl nuclease-free water (high-quality or FFPE samples)
For FFPE DNA, qualify integrity as directed on page 20.
Add 10 µl 2× Priming Buffer to each sample. Place FFPE samples on ice (further fragmentation
not required). For high-quality samples only, fragment by incubating in thermal cycler: 4 min @
94°C, 1min @ 4°C, Hold @ 4°C.
20 µl primed RNA fragments + 8.5 µl First Strand Master Mix
Incubate in thermal cycler: 10 min @ 25°C, 40 min @ 37°C, Hold @ 4°C
28.5 µl first-strand cDNA+ 25 µl Second Strand Enzyme Mix + 5 µl Second Strand Oligo Mix
Incubate in thermal cycler: 60 min @ 16°C, Hold @ 4°C
Elute cDNA in 52 µl nuclease-free H
Keep on ice
Library Prep
Keep at room temperature 30–45 min before use
Per reaction: 16 µl End Repair-A Tailing Buffer + 4 µl End Repair-A Tailing Enzyme Mix
Keep on ice
50 µl cDNA fragments + 20 µl End Repair/dA-Tailing master mix
Incubate in thermal cycler: 15 min @ 20°C, 15 min @ 72°C, Hold @ 4°C
O, removing 50 µl to fresh well
2
Ligate adaptor 70 µl DNA sample + 25 µl Ligation master mix +5 µl SureSelect XT HS2 RNA Adaptor Oligo Mix
Incubate in thermal cycler: 30 min @ 20°C, Hold @ 4°C
94 SureSelect XT HS2 RNA Library Preparation and Target Enrichment
Page 97

Reference 6
Quick Reference Protocol
Step Summary of Conditions
Purify DNA 100 µl DNA sample + 80 µl AMPure XP bead suspension
Elute DNA in 35 µl nuclease-free H
Keep on ice
Prepare PCR master mix Per reaction: 10 µl 5× Herculase II Reaction Buffer with dNTPs + 1 µl Herculase II Fusion DNA
Polymerase
Keep on ice
Amplify the purified DNA 34 µl purified DNA + 11 µl PCR master mix + 5 µl assigned SureSelect XT HS2 Index Primer Pair
Amplify in thermal cycler using program on page 38
Purify amplified DNA 50 µl amplified DNA + 50 µl AMPure XP bead suspension
Elute DNA in 15 µl nuclease-free H
Quantify and qualify DNA Analyze quantity and quality using TapeStation, Bioanalyzer, or Fragment Analyzer System
Hybridization/Capture
Program thermal cycler Input thermal cycler program on page 47 and pause program
Prep DNA in hyb plate Adjust 200 ng purified prepared library to 12 µl volume with nuclease-free H
Run pre-hybridization
blocking protocol
12 µl library DNA + 5 µl SureSelect XT HS2 Blocker Mix
Run paused thermal cycler program segments 1 through 3; start new pause during segment 3
(1 min @ 65°C)
O, removing 34 µl to fresh well
2
O
2
O
2
Prepare Hyb Mix Prepare 25% RNAse Block dilution, then prepare appropriate Capture Library Hyb Mix below:
Probes ≥3 Mb:
Probes <3 Mb: 2 µl 25% RNase Block + 2 µl Probe + 3 µl nuclease-free H
2 µl 25% RNase Block + 5 µl Probe+ 6 µl SureSelect Fast Hybridization Buffer
O + 6 µl SureSelect
2
Fast Hybridization Buffer
Run the hybridization With cycler paused and samples retained in cycler, add 13 µl Capture Library Hyb Mix to wells
Resume the thermal cycler program, completing segments 4 (hybridization) and 5 (65°C hold)
Prepare streptavidin beads Wash 50 µl Streptavidin T1 beads 3× in 200 µl SureSelect Binding Buffer
Capture hybridized libraries Add hybridized samples (~30 µl) to washed streptavidin beads (200 µl)
Incubate 30 min at RT with vigorous shaking (1400-1900 rpm)
During incubation, pre-warm 6 × 200 µl aliquots per sample of SureSelect Wash Buffer 2 to 70°C
Wash captured libraries Collect streptavidin beads with magnetic stand, discard supernatant
Wash beads 1× with 200 µl SureSelect Wash Buffer 1 at RT
Wash beads 6× with 200 µl pre-warmed SureSelect Wash Buffer 2 (5 minutes at 70°C per wash)
Resuspend washed beads in 25 µl nuclease-free H
O
2
SureSelect XT HS2 RNA Library Preparation and Target Enrichment 95
Page 98

6 Reference
Quick Reference Protocol
Step Summary of Conditions
Post-capture amplification
Prepare PCR master mix Per reaction: 13 µl nuclease-free H
+ 1 µl SureSelect Post-Capture Primer Mix + 1 µl Herculase II Fusion DNA Polymerase
Keep on ice
O+ 10 µl 5× Herculase II Reaction Buffer with dNTPs
2
Amplify the bead-bound
captured libraries
Purify amplified DNA Remove streptavidin beads using magnetic stand; retain supernatant
Quantify and qualify DNA Analyze quantity and quality using TapeStation, Bioanalyzer, or Fragment Analyzer System
25 µl DNA bead suspension+ 25 µl PCR master mix
Amplify in thermal cycler using conditions on page 57
50 µl amplified DNA + 50 µl AMPure XP bead suspension
Elute DNA in 25 µl Low TE
96 SureSelect XT HS2 RNA Library Preparation and Target Enrichment
Page 99

www.agilent.com
In This Book
This guide contains
instructions for using the
SureSelect XT HS2 RNA
Reagent Kits to prepare
target- enriched NGS
libraries for the Illumina
platform.
Agilent Technologies, Inc. 2020
Version A1, September 2020
*G9989-90000*
p/n G9989-90000
Agilent Technologies
 Loading...
Loading...