Page 1

Application Note
Cell Analysis
Dynamic Monitoring of Receptor
Tyrosine Kinase Activation in
LivingCells
xCELLigence real-time cell analysis
Author
Brandon Lamarche,
JoyceVelez, and Leyna Zhao
Agilent Technologies, Inc.
Introduction
Over 500 different protein kinases have been identified, constituting ~1.7% of the
human genome. Of these, 11% are known to be receptor tyrosine kinases (RTKs).1
RTKs and their growth factor ligands mediate important cellular processes including
proliferation, survival, differentiation, metabolism, motility, and gene expression.
Loss of regulation of RTK expression or activity has been implicated in initiation
and progression of cancer, inflammation, diabetes, and cardiovascular disease.
Their central role in these cellular processes and disease states has made RTKs
an attractive and important target for the development of inhibitors that could
be therapeutic for these diseases. Several antibody- and small molecule-based
inhibitors specific for various RTKs have been approved by the FDA for the treatment
of different cancers.
Page 2

RTKs are membrane receptors that
contain an intracellular kinase domain,
which transfers a phosphate group from
an ATP molecule to the hydroxyl group
on tyrosine residues. When binding to
ligands, RTKs dimerize or oligomerize,
causing autophosphorylation
and increased activation of their
intrinsic kinase activity. This leads to
phosphorylation of several downstream
effector proteins, resulting in activation
of multiple signaling pathways.
These pathways include activation of
Ras/MAPK, phosphoinositide-3 kinase,
and PLC pathways. Another pathway
activated is the phosphorylation of
effector proteins such as Src, Paxillin,
and FAK. Activation or phosphorylation
of these proteins leads to cytoskeletal
changes including membrane ruffling,
lamellipodia, and filopodia formation.2
These cellular changes are a result of
actin remodeling and are mediated by
the activities of small GTPases Rac, Rho,
and Cdc42.3
Numerous screening platforms have
been developed for the identification of
inhibitors for RTK. They are generally
subdivided into:
– Antibody-dependent technologies,
including AlphaScreen, TR-FRET, FP,
TRF, SPA, Luminex, and ELISA
– Antibody-independent methods, such
as incorporation of radioactivity,
ATP consumption, and technologies
based on change of substrate size
and charge
Although these technologies offer
some advantages, they are limited by
one or more of the following factors:
complicated and tedious optimization
steps, limited substrate capacity, assay
component interference, and expensive
assay components. All of these issues
can affect the signal, throughput, time,
and utility of the assay.
The xCELLigence system offers a unique
cell sensor arrangement, with electrodes
integrated into the wells of a microplate
(E-Plate). These sensors are arrayed in a
design covering 80% of the well surface
area, allowing for sensitive, quantitative
detection of cellular changes. Signals
from these sensors are relayed in real
time to the xCELLigence to monitor
and analyze the kinetic aspects of
cellularbehavior.
The signals relayed to the system are
impedance changes resulting from an
ionic environment created by application
of an electric field. Disruption of this
ionic environment on the sensor
surface, due to the presence of cells or
changes in cell morphology, can cause
changes in measured impedance.
This is then converted to a Cell Index
value. The extent of the cell-electrode
impedance response depends on the
quality of the cell attachment and the
sensor area covered by the cell. When
cell number or degree of attachment
increases, it causes a corresponding
increase in measured impedance value,
and, therefore, in observed Cell Index.
This system has been successfully
used in monitoring cell proliferation
and cytotoxicity, cell adhesion, and
G-protein-coupled receptor function.
This application note highlights
the development and utility of an
alternative RTK assay that uses the
impedance-based system. This assay
addresses several of the limitations
in previous methods and provides a
simple and user-friendly platform for
identification and further characterization
of RTK inhibitors.
It is known that growth factor binding to
RTK results in immediate morphological
changes. The impedance-based system
makes it possible to quantitatively
assay these cellular changes and,
hence, measure receptor tyrosine kinase
activity and function. Experiments
described here show that these cell
assays are specific, robust, reproducible,
and in concurrence with other RTK
cell-based assays, such as ELISA. The
impedance-based system was used
to screen a small, diverse library of
inhibitors and a collection of kinase
inhibitors. This screen identified a
specific and potent EGFR inhibitor.
The assay was also used to generate
dose-response curves, further
characterizing the inhibitor.
Materials and methods
Cell culture and reagents
COS7 cells were acquired from ATCC.
They were maintained in DMEM
supplemented with 10% fetal bovine
serum and incubated at 37 °C with
5% CO2. Cells were plated in E-Plates
at 1×104 cells per well and incubated
overnight. On the day of the assay,
cells were serum-starved in DMEM
supplemented with 0.25% BSA for a total
of 4 hours. If pretreated with inhibitors,
cells were incubated with the inhibitors
during the last hour of serum starvation
and then stimulated with growth factors.
Inhibitors (Calbiochem) and LOPAC
enzyme inhibitor ligand set (Sigma) were
resuspended and stored according to
manufacturers’ instructions.
RTK assays using impedance
technology
Cells were continuously monitored with
the xCELLigence system. RTK-induced
effects were detected as changes in
impedance and expressed in CellIndex
units.
ELISA
Cells were plated on E-Plates at
1×104cells per well and incubated
overnight. On the day of assay, cells were
serum-starved in DMEM supplemented
with 0.25% BSA for a total of 4 hours.
If pretreated with inhibitors, cells were
incubated with the inhibitors during
the last hour of serum starvation and
2
Page 3
then stimulated with growth factor
Normalized Cell Index
2.0
Normalized Cell Index
r
for 15minutes. After growth factor
stimulation, cells were washed twice
with cold PBS and lysed. EGFR and
phospho-EGFR (1068) were detected by
ELISA at 450 nm.
Statistical and data analysis
All dose-response curves were generated
by plotting the average %control
(±standard deviation) versus ligand or
inhibitor concentrations. The average
%control was calculated relative to
samples treated with growth factor
alone without inhibitor. Samples were
measured in quadruplicate. The EC50
for ligands and IC50 for inhibitors were
determined from a fitted curve generated
by XLfit 4.0.
Results and discussion
Specificity of cellular response to EGF
and insulin treatments
Cells plated in the E-Plates were
monitored from the time of plating to
the end of the experiment. This allowed
the cells and assay conditions to be
monitored constantly before and during
the experiment. 1×104 COS7 cells in
E-Plates were serum-starved for a total
of 4 hours and stimulated with 25 ng/mL
EGF or insulin, then monitored every
minute from the time of ligand addition.
Ligand addition resulted in a rapid and
transient increase in CellIndex for both
EGF- and insulin-treated cells (Figure1A).
This increase was immediately
followed by a decrease in Cell Index,
with EGF-treated cells showing a faster
decrease than insulin-treated cells. The
transient increase in Cell Index was a
result of cytoskeletal rearrangements
due to growth factor treatment,
which is a well-documented effect of
RTKactivation.2
To characterize the specificity of these
responses to ligand treatment, cells
were pretreated for 1 hour with 10 µM
of the EGFR inhibitor (EGFRI), 4557W,
before addition of EGF or insulin. Since
the inhibitor was specific to EGFR,
application of the EGFRI should only
affect cellular changes induced by EGF
The absence of cell response in
EGF-treated cells was a result of the
specific inhibition of EGFR and its
signaling pathways by the EGFRI. The
specificity of this inhibitor and ligand
response was demonstrated by the
lack of effect on the transient Cell Index
increase in insulin-treated cells.
treatment. Indeed, after ligand addition,
insulin-treated cells showed the transient
increase in Cell Index, but EGF-treated
cells did not (Figure 1B).
A
1.8
1.6
1.4
1.2
1.0
0.8
0 2 4 6 8 10 12
Time (hours)
2.0
B
1.8
1.6
1.4
1.2
1.0
0.8
0 2 4 6 8 10 12
Time (hours)
Figure 1. Assessment of specificity of cellular response to EGF and insulin treatments.
COS7 cells were pretreated for 1 hour with either a specific EGFR inhibitor or vehicle. Cells
were then stimulated with insulin or EGF. (A) Cells treated with insulin or EGF showed a
characteristic rise in Cell Index. (B) When pretreated with 10 µM EGFR inhibitor, 4557W,
the EGF response is inhibited while the insulin response remains intact.
EGF
Insulin
EGF+EGF inhibitor
Insulin+EGF inhibito
3
Page 4

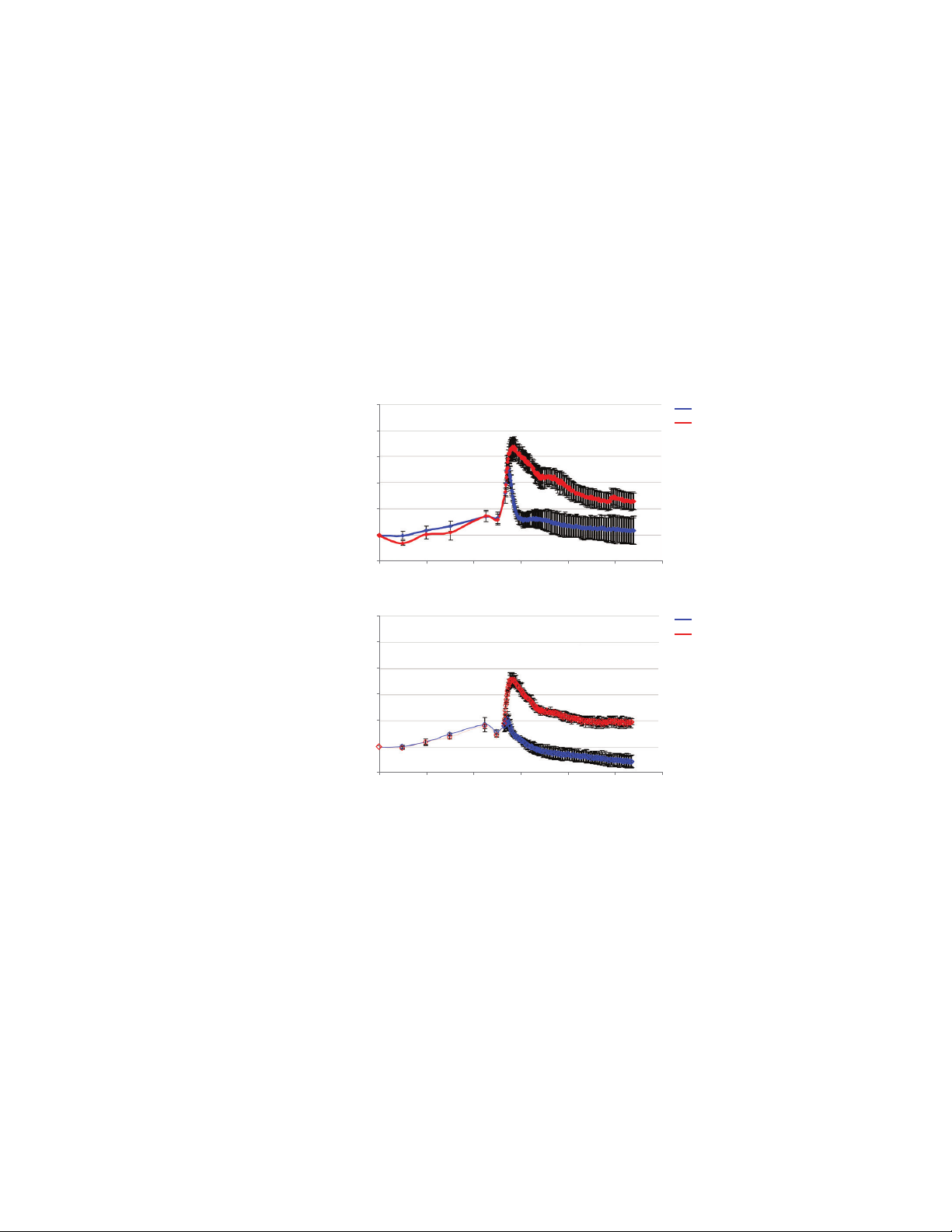
Characterization of COS7 cellular
Normalized Cell Index
100
[EGF] (ng/mL)
0 ng/mL
Normalized Cell Index
response to EGF and HGF treatments
To further characterize this cellular
response, a wide range of EGF and
HGF concentrations were used to
determine the ligand EC50 (Figure 2A
and 2B). For each concentration, the
Cell Index was measured every minute
over several hours. Cells treated with
low concentrations of the ligand showed
transient small changes in peak Cell
Index, while increasing the concentration
of ligand resulted in an increase in the
amplitude of the Cell Index peak. The
magnitude of the Cell Index was directly
related to the concentration of ligand
used and reached a saturable response.
These Cell Index traces were used to
determine the maximum Cell Index
for each ligand concentration, and to
calculate the %control (relative to the
response of the sample when it was
treated with the maximum concentration
of ligand). These values were plotted
versus ligand concentration. From the
fitted curves the EGF and HGF EC50 were
calculated to be 0.95 and 5.9ng/mL,
respectively.
An important consideration when
establishing the validity of this new
1.8
A
1.6
1.4
1.2
1.0
0.8
-60 0 60 120 180 240 300
Time (hours)
B
2.0
1.8
1.6
1.4
1.2
1.0
0.8
-30 30 90 150 210 270 330
Time (hours)
C
100
80
60
40
20
%Control (Phospho-EGFR)
0
0 0.01 0.1 1 10 100
0.025 ng/mL
0.25 ng/mL
2.50 ng/mL
12.5 ng/mL
25.0 ng/mL
50.0 ng/mL
HGF 0 ng/mL
HGF 0.1 ng/mL
HGF 0.3 ng/mL
HGF 1.0 ng/mL
HGF 10.0 ng/mL
HGF 30.0 ng/mL
HGF 100.0 ng/mL
HGF 300.0 ng/mL
100
80
60
40
%Conrol
20
0
0 0.01 0.1 1 10
[EGF] (ng/mL)
100
80
60
%Conrol
40
20
0
0 0.1 1 10 1,000100
[HGF] (ng/mL)
EC50 = 0.95 ng/mL
EC50 = 5.9 ng/mL
method was to show that these results
are consistent with other RTK assays. To
compare the EC50 values derived from
this new system with a well-established
assay used to monitor RTK activity,
ELISA assays were performed to detect
Figure 2. Characterization of COS7 cellular response to EGF and HGF treatments. Cell Index traces of
COS7 cells treated with EGF (A) and HGF (B). Maximum cell indexes were determined from each trace.
Dose-response curves were generated by plotting %control versus ligand concentration. (C) ELISA
assay of phosphorylated EGFR was performed on COS7 cells treated with varying concentrations
of EGF. Dose-response curves were generated by plotting %control of absorbance readings versus
ligandconcentration.
phosphorylated EGFR on COS7 cells
that had been treated with varying
concentrations of EGF (Figure 2C). An
EC50 value of 2.6 ng/mL was calculated
from the fitted curve. This value was
comparable to EC50 values determined
using the impedance technology. This
demonstrated that the system can be
used as an alternative or complementary
assay to RTKassays.
4
Page 5

Optimization of assay conditions for
Normalized Cell Index
screening of inhibitors against RTKs
A few system parameters had to be
optimized before the new system could
be used to screen for EGFR inhibitors.
First, the optimum concentration of
cells needed to achieve the maximum
signal-to-noise ratio (S/N) was
determined (Figure3A). A range of COS7
cells were plated and tested for response
to EGF. The peak in Cell Index due to EGF
treatment increased as the number of
cells plated increased. However, above
the critical cell density, further increase
in cell number resulted in a decrease
in Cell Index. This decrease may be
due to the absence of available space
between the cells, which prevents the
lateral expansion of cell membranes
over the sensors during ligand-mediated
cytoskeletal rearrangement. Second,
the ligand concentration was titrated
to determine the maximum amount of
ligand needed to produce the highest
signal, as well as the appropriate ligand
concentration for the type of assay used.
After these parameters were optimized,
statistical parameters, including the
Z’ factor of the assay, were calculated
(Figure 3B). The Z’ factor parameter was
used for evaluating assay quality. The
value calculated for this assay (0.6) was
above the acceptable limit for a robust
and consistent assay, and the S/N value
was 38.
Validation of the impedance-based
assay system via inhibitor screening
This assay was used to screen a diverse
collection of small molecule inhibitors
from Sigma for validation of their
inhibitor capabilities (Figure 4A). The
library was supplemented with a specific
EGFRI and was arrayed in a 96-well
concentration, between 5and 10µM,
of each component in the inhibitor
ligand set from Sigma. It also contained
several wells of full activity (positive)
and zero activity (negative) reference
controls. Maximum Cell Index due to
EGF treatment was determined for each
inhibitor-treated sample.
plate. The plate contained a single
2.0
A
1.8
1.6
1.4
1.2
Normalized Cell Index
1.0
0.8
Time (min)
2.0
B
1.75
1.50
1.25
1.0
0 20 40 60 80 100
Figure 3. Optimization of assay conditions for screening of inhibitors against RTKs. (A) Increasing number
of COS7 cells were plated and treated with EGF. Cell indexes were measured every minute over several
hours. (B) Statistical evaluation of label-free EGFR inhibitor screening assay. Z’, S/N, S/B, and %CV were
determined to assess quality of assay.
Well number
12600-60-120-180-240
COS7 2.5K
COS7 5K
COS7 10K
COS7 20K
COS7 30K
Z' S/N S/B CV%
0.6381.5 3.8
5
Page 6

Additionally, the %control (relative to the
A
B
180
Log[4557W] (µM)
100
positive-reference, so EGF-treated cell
without inhibitor) was calculated. Using
60% (or 40% inhibition) as the cutoff
criteria, the screening study identified
four potent inhibitors or "hits". The
most potent of these was the EGFRI
4557W (Figure 4B). The assay was also
tested against a collection of kinase
inhibitors, which similarly identified the
EGFRI as the one that produced the
most significant inhibition (Figure 4B). A
dose-response curve was generated for
this inhibitor (Figure 4C). From the fitted
curve, an IC50 of 161nM was calculated.
These experiments demonstrated
that the system is able to identify a
potent and selective inhibitor from a
diverse inhibitor library and also from
a chemically focused kinase inhibitor
library. The system can also be used to
further characterize identifiedhits.
160
140
120
100
80
%Control%Control
60
40
20
0
0 10 20 30 40 50 60 70 80 90
200
150
Well number
EGF
Insulin N = 4
100
50
0
4557W
Genistein
C
Figure 4. Validation of the impedance-based assay system using inhibitor screening. (A) Graphical representation of a screen
of 81 compounds, mostly from the enzyme inhibitor ligand set of sigma. Compounds were screened in singlets at 5 to 10 µM
concentrations. The red circle represents the negative control; the blue circle represents the positive control. (B) A collection of
kinase inhibitors were screened for inhibition of EGFR activity. (C) EGFRI, 4557W, was identified as potent inhibitor of EGFR signaling
from both screens. Cellular response to EGF was measured after the cells were pretreated with varying concentrations of inhibitor.
Dose-response curves were generated by plotting %control of maximum Cell Index versus ligand concentration.
EGFR
1.6
1.4
1.2
1.0
Normalized Cell Index
0.8
0 35 70 105 140 175 210 245 280 315 350-35
EGFR
PD98059
Mek
Time (min)
SP600125
Jnk
PP2
Piceatannol
Src
Kinase inhibitors
Pyk
BisIM
PKC
ROCK
Rho
0.0 µM
0.001 µM
0.01 µM
0.1 µM
0.3 µM
1.0 µM
10.0 µM
30.0 µM
Wortmannin
PI3K
120
100
80
60
% Control
40
20
TCN
Akt
0
0 0.001 0.1 10
6
Page 7

Reaction of selected immortalized
Insulin
cell lines to treatment with various
ligands for RTK
In another set of experiments, several
human cell lines were treated with
various growth factors (Figure 5).
A431 PC3 COS7
EGF
FGF
Theseresults demonstrate that
responses are cell-specific and
factor-specific. For example, the cell line
A431 showed a robust response to EGF
and HGF, but only responded minimally
to other growth factors.
These data demonstrate a simple and
novel cell-based assay for RTK activity
and function. The assay quantifies
morphological changes in response
to growth factor treatment and
therefore mimics proximal events in
kinaseactivation.
HGF
PDGF
Figure 5. Comparison of unique signaling patterns of selected immortalized cell lines after treatment with
various ligands for RTK. Cells were plated, serum-starved, and treated with ligands. Green traces represent cells
treated with ligand, and red traces represent cells treated with vehicle. Response was measured every minute
and data normalized to time of ligand addition. Error bars represent a standard deviation of n = 4.
7
Page 8

Unlike other RTK assays, this assay is:
– Cell-based
– Label-free
– Capable of monitoring cellular
changes in real time
– Noninvasive
The assay also provides valuable
information about the state of the
cell and the signaling pathways being
activated. In addition, the RTK assay
described here does not require
expensive reagents or suffer from
assay component interference. Since
the readout is noninvasive, multiple
treatments can be performed in the
same well. The assay can also be used
with other existing cell-based assays for
RTK. Finally, it requires little optimization
and user training, making it amenable for
use in primary and secondary screens.
References
1. Manning, G. et al., Science 2002, 298,
1912
2. Hall, A. Science 1998, 279, 509.
3. Etienne-Manneville, S.; Hall, A. Nature
2002, 420, 629.
www.agilent.com/chem
For Research Use Only. Not for use in diagnostic procedures.
This information is subject to change without notice.
© Agilent Technologies, Inc. 2020, 2021
Printed in the USA, February 2, 2021
5994-1693EN
 Loading...
Loading...