Page 1

Brilliant II QRT-PCR Master Mix Kit, 1-Step
Instruction Manual
Catalog #600809 (single kit)
#600818 (10-pack kit)
Revision C
Research Use Only. Not for Use in Diagnostic Procedures.
600809-12
Page 2

LIMITED PRODUCT WARRANTY
This warranty limits our liability to replacement of this product. No other warranties of any kind,
express or implied, including without limitation, implied warranties of merchantability or fitness for
a particular purpose, are provided by Agilent. Agilent shall have no liability for any direct, indirect,
consequential, or incidental damages arising out of the use, the results of use, or the inability to use
this product.
ORDERING INFORMATION AND TECHNICAL SERVICES
United States and Canada
Agilent Technologies
Stratagene Products Division
11011 North Torrey Pines Road
La Jolla, CA 92037
Telephone (858) 373-6300
Order Toll Free (800) 424-5444
Technical Services
email
techservices@agilent.com
World Wide Web
Europe
Location Telephone
Austria 01 25125 6800
Benelux 02 404 92 22
Denmark 45 70 13 00 30
Finland 010 802 220
France 0810 446 446
Germany 0800 603 1000
Italy 800 012575
Netherlands 020 547 2600
Spain 901 11 68 90
Sweden 08 506 4 8960
Switzerland 0848 8035 60
UK/Ireland 0845 712 5292
(800) 894-1304
www.genomics.agilent.com
All Other Countries
Please contact your local distributor. A complete list of distributors is available at www.genomics.agilent.com.
Page 3

Brilliant II QRT-PCR Master Mix Kit, 1-Step
CONTENTS
Materials Provided.............................................................................................................................. 1
Storage Conditions.............................................................................................................................. 1
Additional Materials Required .......................................................................................................... 1
Notices to Purchaser ........................................................................................................................... 1
Introduction......................................................................................................................................... 2
Features of Kit Components.................................................................................................. 2
Molecular Beacons Probes .................................................................................................... 3
TaqMan® Probes (Hydrolysis Probes)................................................................................... 3
Fluorescence Monitoring in Real-Time................................................................................. 5
Preprotocol Considerations................................................................................................................ 7
RNA Isolation........................................................................................................................ 7
Quantitative PCR Human Reference Total RNA .................................................................. 7
Probe Design ......................................................................................................................... 8
Optimal Concentrations for Experimental Probes and PCR Primers .................................... 8
Reference Dye ....................................................................................................................... 9
Magnesium Chloride Concentration...................................................................................... 9
Preparing a Single Mixture for Multiple Samples................................................................. 9
Mixing and Pipetting Enzymes ........................................................................................... 10
Temperature and Duration of cDNA Synthesis Reaction.................................................... 10
Preventing Sample Contamination ...................................................................................... 10
Recommended Control Reactions ....................................................................................... 10
Endpoint vs. Real-Time Measurements............................................................................... 11
Data Acquisition with a Spectrofluorometric Thermal Cycler............................................ 11
Multiplex RT-PCR .............................................................................................................. 11
Protocol .............................................................................................................................................. 13
Preparing the Reactions....................................................................................................... 13
RT-PCR Cycling Programs ................................................................................................. 15
Troubleshooting: TaqMan® Probes................................................................................................. 16
Troubleshooting: Molecular Beacons..............................................................................................17
References .......................................................................................................................................... 18
Endnotes............................................................................................................................................. 18
MSDS Information............................................................................................................................ 18
Quick-Reference Protocol ................................................................................................................ 19
Page 4

Page 5

Brilliant II QRT-PCR Master Mix Kit, 1-Step
ATERIALS PROVIDED
M
Catalog #600809 (single kit), #600818 (10-pack kit)
Materials provided Quantity
2× Brilliant II QRT-PCR Master Mix 2 × 2.5 ml
RT/RNase Block Enzyme Mixture 400 μl
Reference dye c, 1 mM 100 μl
a
Sufficient PCR reagents are provided for four hundred, 25-μl QRT-PCR reactions
b
Quantities listed are for a single kit. For 10-pack kits, each item is provided at 10 times the listed quantity.
c
The reference dye is light sensitive and should be kept away from light whenever possible.
a,b
STORAGE CONDITIONS
All Components: Upon receipt, store all components at –20°C. Store the 2× master mix at 4°C after
thawing. Once thawed, full activity is guaranteed for 6 months.
Note The reference dye is light sensitive and should be kept away from light whenever possible.
ADDITIONAL MATERIALS REQUIRED
Spectrofluorometric thermal cycler
Nuclease-free PCR-grade water
NOTICES TO PURCHASER
Notice to Purchaser: Limited License
Practice of the patented 5’ Nuclease Process requires a license from Applied Biosystems. The
purchase of this product includes an immunity from suit under patents specified in the product insert
to use only the amount purchased for the purchaser's own internal research when used with the
separate purchase of Licensed Probe. No other patent rights are conveyed expressly, by implication,
or by estoppel. Further information on purchasing licenses may be obtained from the Director of
Licensing, Applied Biosystems, 850 Lincoln Centre Drive, Foster City, California 94404, USA.
Revision C © Agilent Technologies, Inc. 2010.
Brilliant II QRT-PCR Master Mix Kit, 1-Step 1
Page 6

INTRODUCTION
Quantitative PCR is a powerful tool for gene expression analysis. Many
fluorescent chemistries are used to detect and quantitate gene transcripts.
The use of fluorescent probe technologies reduces the risk of sample
contamination while maintaining convenience, speed, and high-throughput
screening capabilities. The Brilliant II QRT-PCR Master Mix Kit, 1-Step
can be used with both hairpin and linear fluorescent probe technologies to
perform absolute or relative quantitation of gene expression. The single-step
master mix format is ideal for most high-throughput QPCR applications
where it is not necessary to archive cDNA.
The Brilliant II QRT-PCR master mix kit includes the components
necessary to carry out cDNA synthesis and PCR amplification in one tube
and one buffer.* Brilliant kits support quantitative amplification and
detection with multiplex capability and show consistent high performance
with various fluorescent detection systems, including molecular beacons and
®
TaqMan
probes. The Brilliant II QRT-PCR master mix kit has been
successfully used to amplify and detect a variety of high- and lowabundance RNA targets from experimental samples including total RNA,
+
poly(A)
RNA, and synthetic RNA.
The Brilliant II QRT-PCR master mix has been optimized for maximum
performance on the Stratagene Mx3000P and Mx3005P real-time PCR
systems and the Stratagene Mx4000 multiplex quantitative PCR system, as
well as on the ABI 7900HT real-time PCR instrument.
Features of Kit Components
RT/RNase Block Enzyme Mixture
The reverse transcriptase (RT) provided in the kit is a Moloney-based RT
specifically formulated for Stratagene Brilliant II kits. This RT performs
optimally at a reaction temperature of 50°C when used in 1-step QRT-PCR
with the Brilliant II master mix. It is stringently quality-controlled to verify
the absence of nuclease contaminants that adversely affect cDNA synthesis
and to ensure sensitive and reproducible performance in QRT-PCR
experiments with a broad range of RNA template amounts and a variety of
RNA targets that vary in size, abundance, and GC-content. The RNase
block, provided in the same tube, serves as a safeguard against
contaminating RNases.
* Primers and template are not included.
2 Brilliant II QRT-PCR Master Mix Kit, 1-Step
Page 7

Brilliant II QRT-PCR 2× Master Mix
The 2× master mix contains an optimized RT-PCR buffer, MgCl2,
nucleotides (GAUC), stabilizers, and SureStart Taq DNA polymerase.
SureStart Taq DNA polymerase is a modified version of Taq2000 DNA
polymerase with hot start capability. SureStart Taq DNA polymerase
improves PCR performance by decreasing background and increasing
amplification of desired products. Using SureStart Taq, hot start is easily
incorporated into PCR protocols already optimized with Taq DNA
polymerase, with little or no modification of cycling parameters or reaction
conditions.
Reference dye
A passive reference dye (an optional reaction component) is provided in a
third tube. The passive reference dye (with excitation and emission
wavelengths of 584 nm and 612 nm, respectively) is provided as an optional
reagent that may be added to compensate for non-PCR related variations in
fluorescence. Providing the reference dye in a separate tube makes the
Brilliant II QRT-PCR master mix kit adaptable for many real-time QPCR
platforms (see Reference Dye in Preprotocol Considerations for more
information).
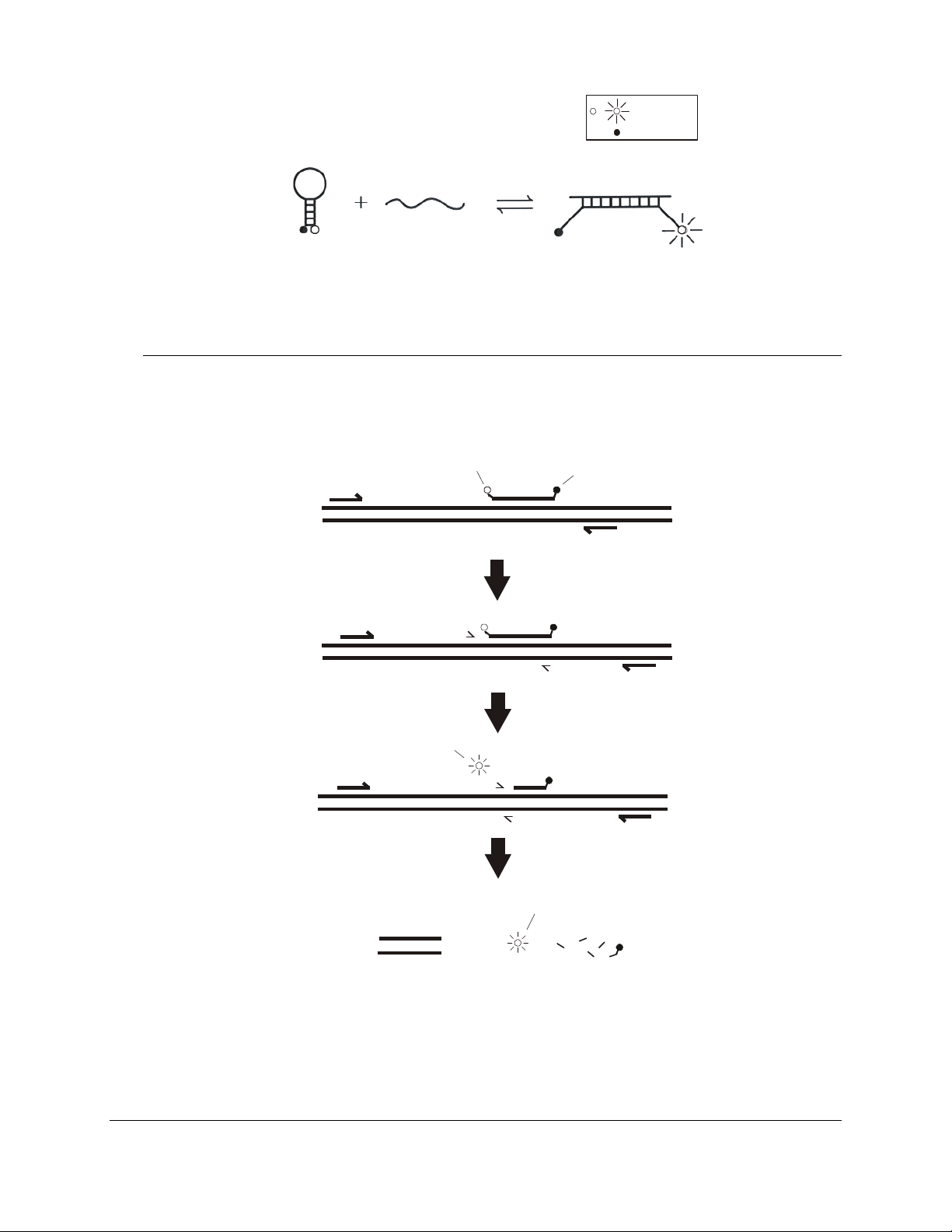
Molecular Beacons Probes
Molecular beacons are hairpin-shaped fluorescent hybridization probes that
can be used to monitor the accumulation of specific product during or after
1–5
PCR.
opposite ends of an oligonucleotide (see Figure 1). The ends of the
oligonucleotide are designed to be complementary to each other. When the
unhybridized probe is in solution, it adopts a hairpin structure that brings the
fluorophore and quencher sufficiently close to each other to allow efficient
quenching of the fluorophore. If, however, the molecular beacon is bound to
its complementary target, the fluorophore and quencher are far enough apart
that the fluorophore cannot be quenched and the molecular beacon
fluoresces. As PCR proceeds, product accumulates and the molecular
beacon fluoresces at a wavelength characteristic of the particular
fluorophore used. The amount of fluorescence at any given cycle depends
on the amount of specific product present at that time.
Molecular beacons have a fluorophore and a quencher molecule at
TaqMan® Probes (Hydrolysis Probes)
TaqMan probes are linear.
probe, and the quencher is either internal or is at the 3´ end (see Figure 2).
As long as the probe is intact, regardless of whether it is hybridized with the
target or free in solution, no fluorescence is observed from the fluorophore.
During the combined annealing-extension step of PCR, the primers and the
TaqMan probe hybridize with the target. The DNA polymerase displaces the
TaqMan probe by 3 or 4 nucleotides, and the 5´-nuclease activity of the
DNA polymerase separates the fluorophore from the quencher. Because of
this mechanism of action, these probes are also referred to as hydrolysis
probes. Fluorescence can be detected during each PCR cycle, and
fluorescence accumulates during the course of PCR.
Brilliant II QRT-PCR Master Mix Kit, 1-Step 3
6, 7
The fluorophore is usually at the 5´ end of the
Page 8
A
e
,
Fluorophore
Quencher
Molecular
beacon
FIGURE 1 The molecular beacon binds to a complementary target and fluoresces.
Ta rg et
Hybrid
PCR Primer
PCR Primer
Fluorophor
Polymerization
Ta q Ma n
probe
mplification assay
Quencher
PCR Primer
- - - - - - - -
- - - - - -
Probe displacement
and cleavage
Fluorescence
- - - - - - - - - - -
- - - - - - - - - -
Result
Fluorescence
PCR products
FIGURE 2 TaqMan probe fluoresces when the 5´-nuclease activity of the DNA polymerase separates the fluorophore from
quencher.
4 Brilliant II QRT-PCR Master Mix Kit, 1-Step
Cleavage products
Page 9

Fluorescence Monitoring in Real-Time
When fluorescence signal from a PCR reaction is monitored in real-time, the
results can be displayed as an amplification plot, which reflects the change
in fluorescence during cycling. This information can be used during realtime PCR experiments to quantitate initial copy number based on the
threshold cycle (Ct).
determined to be statistically significant above background. The threshold
cycle is inversely proportional to the log of the initial copy number.
more template that is initially present, the fewer the number of cycles it
takes to reach the point where the fluorescence signal is detectable above
background. Quantitative information based on threshold cycle is more
accurate than information based on endpoint determinations because
threshold cycle is based on measurements taken during the exponential
phase of PCR amplification when PCR efficiency is not yet influenced by
limiting reagents, small differences in reaction components, or cycling
conditions.
Ct values determined for a set of standard wells, containing known amounts
of the target, may be plotted to generate a standard curve that can be used to
relate Ct values to initial copy number for unknown samples. Figure 3
shows Mx3000P instrument standard curve plots for the GAPDH gene and
the cyclophilin gene from a multiplex QRT-PCR experiment using TaqMan
probes. In this experiment, serial dilutions of total RNA were reversetranscribed and amplified with fluorescence detected at each cycle. The
table shows the R
instrument from the standard curve plots. The R
and 1) is an indication of the quality of the fit of the standard curve to the
standard data points plotted, with values closer to 1 indicating a better fit of
the data to the line. The slope of the standard curve is directly related to the
average efficiency of amplification throughout the cycling program and may
be used to calculate the PCR efficiency for a given template in a given
experiment. A reaction with 100% efficiency will produce a slope of –3.322.
6
Ct is defined as the cycle at which fluorescence is
2
values and PCR efficiencies calculated by the Mx3000P
6
The
2
value (always between 0
Brilliant II QRT-PCR Master Mix Kit, 1-Step 5
Page 10

Target Symbol R2 Value Slope Efficiency (%)
Cyclophilin (closed squares) 0.999 -3.455 94.7
GAPDH (open squares) 0.999 -3.404 96.7
Figure 3 Mx3000P quantitative PCR instrument standard curve plots using TaqMan probes for GAPDH (open squares) or
cyclophilin (closed squares) in multiplex single-tube RT-PCR reactions. The table below the standard curve plot shows the
2
R
value, standard curve slope and amplification efficiency for each of the targets.
6 Brilliant II QRT-PCR Master Mix Kit, 1-Step
Page 11

PREPROTOCOL CONSIDERATIONS
RNA Isolation
High-quality intact RNA is essential for successful synthesis of full-length
cDNA. Total and poly(A)
Stratagene Absolutely RNA isolation kits. Oligo(dT)-selection for poly(A)
RNA is typically not necessary, although including this step may improve
the yield of specific cDNA templates. RNA samples with OD
1.8–2.0 are optimally pure.
Preventing RNase Contamination
Take precautions to minimize the potential for contamination by
ribonucleases (RNases). RNA isolation should be performed under
RNase-free conditions. Wear gloves and use sterile tubes, pipet tips, and
RNase-free water. Do not use DEPC-treated water, which can inhibit PCR.
The RNase inhibitor that is included in the RT/RNase block enzyme mixture
provides additional protection against RNase contamination.
Preventing Genomic DNA Contamination
Contaminating DNA can be removed from the RNA preparation using an
RNase-free DNase. Additionally, PCR primers may be designed to span
adjacent exons in order to prevent amplification of the intron-containing
genomic DNA.
+
RNA can be rapidly isolated and purified using
ratios of
260/280
+
Quantitative PCR Human Reference Total RNA
Stratagene QPCR Human Reference Total RNA (Catalog #750500) is a
high-quality control for quantitative PCR gene-expression analysis.
Stratagene QPCR Human Reference Total RNA is composed of total RNA
from 10 human cell lines (see the table below), with quantities of RNA from
the individual cell lines optimized to maximize representation of gene
transcripts present in low, medium, and high abundance. The reference RNA
is carefully screened for contaminating genomic DNA, the presence of
which can complicate interpretation of QRT-PCR assay data.
Quantitative PCR Human Reference Total RNA Cell Line Derivations
Adenocarcinoma, mammary gland
Hepatoblastoma, liver
Adenocarcinoma, cervix
Embryonal carcinoma, testis
Glioblastoma, brain
Melanoma, skin
Liposarcoma
Histiocytic lymphoma; macrophage; histocyte
Lymphoblastic leukemia, T lymphoblast
Plasmacytoma; myeloma; B lymphocyte
Brilliant II QRT-PCR Master Mix Kit, 1-Step 7
Page 12

The QPCR Human Reference Total RNA is ideally suited for optimizing
QRT-PCR assays. Often only small amounts of experimental RNA template
are available for setting up an expression profiling study. Using the
extensive representation of specific mRNA species in the generic template,
assays may be optimized for a variety of primer/probe systems. This
eliminates the use of precious experimental RNA samples for assay
optimization.
Probe Design
Probes should have a melting temperature that is 7–10°C higher than the
annealing temperature of the primers. For additional considerations in
®
designing TaqMan probes, refer to Primer Express
oligo design software
from Applied Biosystems.
Resuspend lyophilized custom molecular beacon or TaqMan probes in
buffer containing 5 mM Tris-HCl, pH 8.0, and 0.1 mM EDTA (low TE
buffer).
Optimal Concentrations for Experimental Probes and PCR Primers
Probes
The optimal concentration of the experimental probe should be determined
empirically. The optimal concentration is the lowest concentration that
results in the lowest Ct and an adequate fluorescence for a given target
concentration.
A) Molecular Beacons
The molecular beacon concentration can be optimized by varying the
final concentration from 200 to 500 nM in increments of 100 nM.
®
B) TaqMan
Probes
The TaqMan probe concentration can be optimized by varying the final
concentration from 100 to 500 nM in increments of 100 nM.
PCR Primers
The optimal concentration of the upstream and downstream PCR primers
should also be determined empirically. The optimal concentration is the
lowest concentration that results in the lowest Ct and an adequate
fluorescence for a given target concentration. The primer concentration for
use with molecular beacons can be optimized by varying the concentration
from 200 to 600 nM. The primer concentration for use with TaqMan probes
can be optimized by varying the concentration from 100 to 600 nM. The
best concentrations of the upstream and downstream primers are not always
of equal molarity.
8 Brilliant II QRT-PCR Master Mix Kit, 1-Step
Page 13

Reference Dye
A passive reference dye is included in this kit and may be added to
compensate for non-PCR related variations in fluorescence. Fluorescence
from the passive reference dye does not change during the course of the
PCR reaction but provides a stable baseline to which samples are
normalized. In this way, the reference dye compensates for changes in
fluorescence between wells caused by slight volume differences in reaction
tubes. The excitation and emission wavelengths of the reference dye are 584
nm and 612 nm, respectively. Although addition of the reference dye is
optional when using the Mx4000, Mx3000P or Mx3005P system, with other
®
instruments (including the ABI 7900HT and ABI PRISM
7700) the use of
the reference dye may be required for optimal results.
Reference Dye Dilution Recommendations
Prepare fresh* dilutions of the reference dye prior to setting up the
reactions, and keep all tubes containing the reference dye protected from
light as much as possible. Make initial dilutions of the reference dye using
nuclease-free PCR-grade H
Mx3005P, or Mx4000 instrument, use the reference dye at a final
concentration of 30 nM. If you are using the ABI 7900HT real-time PCR
instrument, use the reference dye at a final concentration of 300 nM. For
other instruments, use the following guidelines for passive reference dye
optimization. For instruments that allow excitation at ~584 nm (including
most tungsten/halogen lamp-based instruments and instruments equipped
with a ~584 nm LED), begin optimization using the reference dye at a final
concentration of 30 nM. For instruments that do not allow excitation near
584 nm, (including most laser-based instruments) begin optimization using
the reference dye at a final concentration of 300 nM.
O. If you are using a Stratagene Mx3000P,
2
Magnesium Chloride Concentration
Magnesium chloride concentration affects the specificity of the PCR primers
and probe hybridization. The Brilliant II QRT-PCR master mix contains
MgCl
at a concentration of 5.5 mM (in the 1× solution), which is suitable
2
for most targets.
Preparing a Single Mixture for Multiple Samples
If running multiple samples containing the same primers and probes, prepare
a single mixture of reaction components and then aliquoting the mixture into
individual reaction tubes using a fresh pipet tip for each addition. Preparing
a common mixture facilitates the accurate dispensing of reagents, minimizes
the loss of reagents during pipetting, and helps to minimize sample-tosample variation.
* The diluted reference dye, if stored in a light-protected tube at 4°C, can be used within the
day for setting up additional assays.
Brilliant II QRT-PCR Master Mix Kit, 1-Step 9
Page 14

Mixing and Pipetting Enzymes
Solutions that contain enzymes (including reverse transcriptase and
SureStart Taq DNA polymerase) should be mixed gently by inversion or
gentle vortexing without generating bubbles. Pipet the enzymes carefully
and slowly; otherwise, the viscosity of the buffer, which contains 50%
glycerol, can lead to pipetting errors.
Temperature and Duration of cDNA Synthesis Reaction
For cDNA synthesis, we recommend a 50°C incubation for most targets
using the Brilliant II QRT-PCR master mix kit. However, incubation up to
55°C can be employed to reduce secondary structures or to improve
specificity. A 30-minute incubation for the first-strand synthesis reaction is
sufficient for most targets. Rare RNA sequences or long amplicons may
benefit from an extended incubation time (up to 60 minutes) at a lower
temperature (42°C).
Preventing Sample Contamination
Take precautions to minimize the potential for carryover of nucleic acids
from one experiment to the next. Use separate work areas and pipettors for
pre- and post-amplification steps. Use positive displacement pipets or
aerosol-resistant pipet tips.
Treatment with Uracil-N-glycosylase (UNG) is NOT recommended for
decontamination of single tube RT-PCR reactions since UNG would be
active during the 50°C incubation necessary for reverse transcription.
Recommended Control Reactions
No Template Control (NTC)
We recommend performing no-template control reactions for each
experimental sample to screen for contamination of reagents or false
amplification.
No-RT Control
We recommend performing no-RT control reactions for each experimental
sample by omitting the RT/RNase block enzyme mixture from the reaction.
The no-RT control is expected to generate no signal if there is no
amplification of genomic DNA. No signal indicates that the RNA
preparation is free of contaminating genomic DNA or that the primers are
specific for the cDNA. See Preventing Genomic DNA Contamination in
RNA Isolation.
Endogenous Control
Consider performing an endogenous control reaction to normalize variation
in the amount of RNA template across samples. See Reference 8 for
guidelines on the use of endogenous controls for QPCR.
10 Brilliant II QRT-PCR Master Mix Kit, 1-Step
Page 15

Endpoint vs. Real-Time Measurements
Fluorescence may be detected either at the endpoint of cycling or in realtime using a real-time spectrofluorometric thermal cycler. Real-time
experiments are typically performed on an instrument capable of detecting
fluorescence from samples during each cycle of a PCR protocol. For
endpoint analysis, PCR reactions can be run on any thermal cycler and can
then be analyzed with a fluorescence plate reader that has been designed to
accommodate PCR tubes and that is optimized for the detection of PCR
reactions that include fluorescent probes. If using a fluorescence plate
reader, it is recommended that readings be taken both before and after PCR
for comparison.
Data Acquisition with a Spectrofluorometric Thermal Cycler
Acquisition of real-time data generated by fluorogenic probes should be
performed as recommended by the instrument's manufacturer.
When developing an assay, it is necessary to decide whether to use a 2-step
or a 3-step PCR protocol. We recommend a 2-step protocol for the Brilliant
II QRT-PCR master mix kit. In a 2-step cycling protocol, fluorescence data
are collected during the combined annealing/extension step. When using a
3-step protocol, it is prudent to collect fluorescence data at both the
annealing step and the extension step of the PCR reaction.
Multiplex RT-PCR
Multiplex RT-PCR is the amplification of more than one target in a single
polymerase chain reaction.
been successfully used to amplify two targets in a multiplex reaction without
reoptimizing the concentrations of DNA polymerase, reverse transcriptase
or dNTPs.
In a typical multiplex RT-PCR reaction, one PCR primer pair primes the
amplification of the target of interest and another PCR primer pair primes
the amplification of an endogenous control. For accurate analysis, it is
important to minimize competition between concurrent amplifications for
common reagents. To minimize competition, the limiting primer
concentrations need to be determined.
to optimization of the other reaction components. The number of
fluorophores in each tube can influence the analysis. The use of a dark
quencher, which emits heat instead of light, might enhance the quality of
multiplex RT-PCR results by reducing the background light emission. The
following PCR primer and probe design guidelines are useful for multiplex
RT-PCR.
9
The Brilliant II QRT-PCR master mix kit has
10
Consideration should also be given
Brilliant II QRT-PCR Master Mix Kit, 1-Step 11
Page 16

PCR Primer Considerations for Multiplex RT-PCR
♦ Design primer pairs with similar annealing temperatures for all targets
to be amplified.
♦ To avoid duplex formation, analyze the sequences of primers
and probes with primer analysis software.
♦ The limiting primer concentrations are the primer concentrations that
result in the lowest fluorescence intensity without affecting the Ct. If the
relative abundance of the two targets to be amplified is known,
determine the limiting primer concentrations for the most abundant
target. If the relative abundance of the two targets is unknown,
determine the limiting primer concentrations for both targets. The
limiting primer concentrations are determined by running serial
dilutions of those forward and reverse primer concentrations optimized
for one-probe detection systems, but maintaining a constant target
concentration. A range of primer concentrations of 50–200 nM is
recommended. Running duplicates or triplicates of each combination of
primer concentrations within the matrix is also recommended.
10
Probe Considerations for Multiplex RT-PCR
A) Molecular Beacons
♦ Label each molecular beacon with a spectrally distinct
fluorophore.
♦ Consider designing probes with dark quenchers.
♦ Design molecular beacons for different targets to have different
11
stem sequences.
®
B) TaqMan
♦ Label each TaqMan probe with a spectrally distinct fluorophore.
♦ Consider designing probes with dark quenchers.
Probes
12 Brilliant II QRT-PCR Master Mix Kit, 1-Step
Page 17

PROTOCOL
Preparing the Reactions
Notes Following initial thawing of the master mix, store the unused
portion at 4°C. Multiple freeze-thaw cycles should be avoided.
It is prudent to set up a no-template control reaction to screen for
contamination of reagents or false amplification. Similarly, a
no-RT control should be included to verify that the fluorescence
signal is due to the amplification of cDNA and not of
contaminating genomic DNA.
Consider performing an endogenous control reaction to normalize
variations in the amount of RNA template across samples. For
information on the use and production of endogenous controls for
QPCR, see Reference 8.
1. If the reference dye will be included in the reaction, (optional), dilute
the dye solution provided 1:500 (for the Mx3000P, Mx3005P, and
Mx4000 instruments) or 1:50 (for the ABI 7900HT real-time PCR
instrument) using nuclease-free PCR-grade H
instruments, use the guidelines in the Reference Dye section under
Preprotocol Considerations. When used according to the protocol
below, this will result in a final reference dye concentration of 30 nM
for the Mx3000P, Mx3005P, and Mx4000 instruments and 300 nM for
the ABI 7900HT instrument. Keep all solutions containing the
reference dye protected from light.
O. For other
2
Note If using a system other than the Mx4000, Mx3000P or
Mx3005P instruments, the use of the reference dye may be
required for optimal results.
2. Thaw the 2× Brilliant II QRT-PCR master mix and store on ice. Mix
the solution well by gentle inversion prior to pipetting.
Brilliant II QRT-PCR Master Mix Kit, 1-Step 13
Page 18

3. Prepare the experimental reactions by combining the following
components in order. Prepare a single reagent mixture for duplicate
experimental reactions and duplicate no-template controls (plus at least
one reaction volume excess), using multiples of each component listed
below.
Reagent Mixture
Nuclease-free PCR-grade H2O to adjust the final volume to 25 μl
(including experimental RNA)
12.5 μl of 2× QRT-PCR master mix
x μl of experimental probe (optimized concentration)
x μl of upstream primer (optimized concentration)
x μl of downstream primer (optimized concentration)
0.375 μl of the diluted reference dye (optional)
1.0 μl of RT/RNase block enzyme mixture
Note A total reaction volume of 50
4. Gently mix the reagents without creating bubbles (do not vortex), then
distribute the mixture to individual PCR reaction tubes.
5. Add x μl of experimental RNA to each reaction. The quantity of RNA
depends on the RNA purity and the specific mRNA abundance. As a
guideline, use 1 pg–400 ng of total RNA or 0.1 pg–1 ng of mRNA.
6. Gently mix the reactions without creating bubbles (do not vortex).
Note Bubbles interfere with fluorescence detection.
7. Centrifuge the reactions briefly.
μ
l may also be used.
14 Brilliant II QRT-PCR Master Mix Kit, 1-Step
Page 19

RT-PCR Cycling Programs
8. Place the reactions in the QPCR instrument and run the appropriate
RT-PCR program using the guidelines in the tables below. The 2-step
cycling protocol is preferred for most primer/template systems.
Two-Step Cycling Protocol
Cycles Duration of cycle Temperature
1 30 minutes 50°C
1 10 minutesa 95°C
a
Initial 10 minute incubation is required to fully activate the DNA polymerase.
b
Set the temperature cycler to detect and report fluorescence during the
annealing/extension step of each cycle.
Alternative Protocol with Three-Step Cycling
Cycles Duration of cycle Temperature
1 30 minutes 50°C
1 10 minutesa 95°C
40
a
Initial 10 minute incubation is required to fully activate the DNA polymerase.
b
Set the temperature cycler to detect and report fluorescence during the annealing
and extension step of each cycle.
c
Choose an appropriate annealing temperature for the primer set used.
15 seconds 95°C 40
1 minute
b
60°C
30 seconds 95°C
1 minuteb 50–60°Cc
30 seconds 72°C
Brilliant II QRT-PCR Master Mix Kit, 1-Step 15
Page 20

TROUBLESHOOTING: TAQMAN
Observation Suggestion
Little or no increase in fluorescence with
cycling
Increasing fluorescence in no-template control
reactions with cycling
Ct reported for the no-template control (NTC)
sample is less than the total number of cycles
but the curve on the amplification plot is
horizontal
®
PROBES
The probe is not binding to its target efficiently because the annealing
temperature is too high. Verify the calculated melting temperature using
appropriate software.
The probe is not binding to its target efficiently because the PCR product
is too long. Design the primers so that the PCR product is <150 bp in
length.
Design a probe that is compatible with 5.5 mM MgCl2.
For multiplex PCR, the MgCl2 concentration may be increased, if desired,
by adding a small amount of concentrated MgCl2 (not provided in this kit)
to the 1× experimental reaction at the time of set up.
The probe has a nonfunctioning fluorophore. Verify that the fluorophore
functions by digesting the probe (100 nM probe in 25 μl 1× buffer with
10 U DNase or S1 nuclease) at room temperature for 30 minutes to
confirm an increase in fluorescence following digestion.
Redesign the probe.
The reaction is not optimized and no or insufficient product is formed.
Verify formation of the specific product by gel electrophoresis.
The RNA template may be degraded. Ensure that the template RNA is
stored properly (at –20°C or –80°C) and is not subjected to multiple
freeze-thaw cycles. Check the quality of the RNA in the sample by gel
electrophoresis or using an automated RNA population analysis system
such as the Agilent 2100 Bioanalyzer.
If the target RNA contains extensive secondary structure, increase the
incubation temperature used during the first step of the RT-PCR program
to up to 55°C.
For low-abundance targets or long amplicons, increase the duration of
the cDNA synthesis step to 60-minutes while lowering the incubation
temperature down to 42°C.
Verify that all reagents and supplies are RNase-free.
Where possible, increase the amount of template RNA. (Do not exceed
the recommended amount of template.)
For multiplex PCR of more than two targets, reactions may need to be
supplemented with additional polymerase and dNTPs (not provided).
The reaction has been contaminated. Follow the procedures outlined in
reference 12 to minimize contamination.
Variation in fluorescence intensity. Review the amplification plot and, if
appropriate, adjust the threshold accordingly.
16 Brilliant II QRT-PCR Master Mix Kit, 1-Step
Page 21

TROUBLESHOOTING: MOLECULAR BEACONS
Observation Suggestion
Little or no increase in fluorescence with
cycling
Increasing fluorescence in no-template
control reactions with cycling
Ct reported for the no-template control
(NTC) sample is less than the total number
of cycles but the curve on the amplification
plot is horizontal
The molecular beacon is not binding to its target efficiently because the
loop portion is not completely complementary. Perform a melting curve
analysis to determine if the probe binds to a perfectly complementary
target.
The molecular beacon is not binding to its target efficiently because the
annealing temperature is too high. Perform a melting curve analysis to
determine the optimal annealing temperature.
The molecular beacon is not binding to its target efficiently because the
PCR product is too long. Design the primers so that the PCR product is
<150 bp in length.
Design the molecular beacon with a stem that is compatible with 5.5 mM
MgCl
.
2
For multiplex PCR, the MgCl2 concentration may be increased, if desired,
by adding a small amount of concentrated MgCl
to the 1× experimental reaction at the time of set up.
The molecular beacon has a nonfunctioning fluorophore. Verify that the
fluorophore functions by detecting an increase in fluorescence in the
denaturation step of thermal cycling or at high temperatures in a melting
curve analysis. If there is no increase in fluorescence, resynthesize the
molecular beacon.
Resynthesize the molecular beacon using a different fluorophore.
Redesign the molecular beacon.
The reaction is not optimized and no or insufficient product is formed.
Verify formation of the specific product by gel electrophoresis.
The RNA template may be degraded. Ensure that the template RNA is
stored properly (at –20°C or –80°C) and is not subjected to multiple
freeze-thaw cycles. Check the quality of the RNA in the sample by gel
electrophoresis or using an automated RNA population analysis system
such as the Agilent 2100 Bioanalyzer.
If the target RNA contains extensive secondary structure, increase the
incubation temperature used during the first step of the RT-PCR program
up to 55°C.
For low-abundance targets or long amplicons, increase the duration of the
cDNA synthesis step to 60-minutes while lowering the incubation
temperature down to 42°C.
Verify that all reagents and supplies are RNase-free.
Where possible, increase the amount of template RNA. (Do not exceed the
recommended amount of template.)
For multiplex PCR of more than two targets, reactions may need to be
supplemented with additional polymerase and dNTPs (not provided).
The reaction has been contaminated. Follow the procedures outlined in
reference 12 to minimize contamination.
Variation in fluorescence intensity. Review the amplification plot and, if
appropriate, adjust the threshold accordingly.
(not provided in this kit)
2
Brilliant II QRT-PCR Master Mix Kit, 1-Step 17
Page 22

REFERENCES
1. Kostrikis, L. G., Tyagi, S., Mhlanga, M. M., Ho, D. D. and Kramer, F. R. (1998)
Science 279(5354):1228-9.
2. Padmabandu, G., Grismer, L. and Mueller, R. (2000) Strategies 13(3):88-92.
3. Piatek, A. S., Tyagi, S., Pol, A. C., Telenti, A., Miller, L. P. et al. (1998) Nat
Biotechnol 16(4):359-63.
4. Tyagi, S., Bratu, D. P. and Kramer, F. R. (1998) Nat Biotechnol 16(1):49-53.
5. Tyagi, S. and Kramer, F. R. (1996) Nat Biotechnol 14(3):303-8.
6. Higuchi, R., Fockler, C., Dollinger, G. and Watson, R. (1993) Biotechnology (N Y)
11(9):1026-30.
7. Holland, P. M., Abramson, R. D., Watson, R. and Gelfand, D. H. (1991) Proc Natl
Acad Sci U S A 88(16):7276-80.
8. Bustin, S. A. (2000) Journal of Molecular Endocrinology 25:169-193.
9. Edwards, M. and Gibbs, R. (1995). Multiplex PCR. In PCR Primer: A Laboratory
Manual,C. W. Dieffenbach and G. S. Dveksler (Eds.), pp. 157-171. Cold Spring Harbor
Laboratory Press, Plainview, NY.
10. McBride, L., Livak, K., Lucero, M., Goodsaid, F., Carlson, D. et al. (1998).
Quantitative PCR Technology. In Gene Quantification,F. Ferré (Ed.), pp. 97-110.
Birkhauser Boston Press, Boston.
11. Marras, S. A., Kramer, F. R. and Tyagi, S. (1999) Genet Anal 14(5-6):151-6.
12. Kwok, S. and Higuchi, R. (1989) Nature 339(6221):237-8.
ENDNOTES
ABI PRISM® is a registered trademark of Applied Biosystems.
Primer Express
TaqMan
®
is a registered trademark of The Perkin-Elmer Corporation.
®
is a registered trademark of Roche Molecular Systems, Inc.
MSDS INFORMATION
The Material Safety Data Sheet (MSDS) information for Stratagene products is provided on the web at
http://www.genomics.agilent.com. MSDS documents are not included with product shipments.
18 Brilliant II QRT-PCR Master Mix Kit, 1-Step
Page 23

BRILLIANT II QRT-PCR MASTER MIX, 1-STEP
Catalog #600809, #600818
QUICK-REFERENCE PROTOCOL
Note This protocol has been optimized for the Stratagene Mx3000P, Mx3005P, and Mx4000
instruments and the ABI 7900HT instrument. The protocol may be adapted for use with
most other instruments by changing the reference dye dilution according to the guidelines
in the manual and following the instrument manufacturer’s recommendations for RT-PCR
cycling programs.
1. If the passive reference dye will be included in the reaction (optional), dilute 1:500 (Mx3000P,
Mx3005P, or Mx4000 instrument) or 1:50 (ABI 7900HT instrument). Keep all solutions
containing the reference dye protected from light.
Note If using a system other than the Mx4000, Mx3000P or Mx3005P instruments, the use of
the reference dye may be required for optimal results.
2. Thaw the 2× QRT-PCR master mix and store on ice. Following initial thawing of the master mix,
store the unused portion at 4°C.
Note Multiple freeze-thaw cycles should be avoided.
3. Prepare the experimental reactions by adding the following components in order. Prepare a
single reagent mixture for multiple reactions using multiples of each component listed below.
Reagent Mixture
Nuclease-free PCR-grade H2O to bring the final volume to 25 μl (including experimental RNA)
12.5 μl of 2× QRT-PCR master mix
x μl of experimental probe (optimized concentration)
x μl of upstream primer (optimized concentration)
x μl of downstream primer (optimized concentration)
0.375 μl of diluted reference dye from step 1 (optional)
1.0 μl of RT/RNase block mixture
Note A total reaction volume of 50
4. Gently mix the reagents without creating bubbles (do not vortex), then distribute the mixture to
individual PCR reaction tubes.
5. Add x μl of experimental RNA to each reaction.
6. Gently mix the reactions without creating bubbles (do not vortex).
μ
l may also be used.
19
Page 24

7. Centrifuge the reactions briefly.
8. Place the reactions in the instrument and run the appropriate PCR program below.
Two-Step Cycling Protocol
Cycles Duration of cycle Temperature
1 30 minutes 50°C
1 10 minutesb 95°C
15 seconds 95°C 40
1 minute
a
A protocol for three-step cycling is provided in the Protocol section.
b
Initial 10 minute incubation is required to fully activate the DNA polymerase.
c
Set the temperature cycler to detect and report fluorescence during the annealing/extension step of each cycle.
a
c
60°C
20
 Loading...
Loading...