Page 1

Lambda ZAP-CMV RI Predigested
Vector Kit
INSTRUCTION MANUAL
Catalog #239221 (Lambda ZAP-CMV RI Predigested Vector Kit)
Revision A
For In Vitro Use Only
239221-12
Page 2
LIMITED PRODUCT WARRANTY
This warranty limits our liability to replacement of this product. No other warranties of any kind,
express or implied, including without limitation, implied warranties of merchantability or fitness for
a particular purpose, are provided by Agilent. Agilent shall have no liability for any direct, indirect,
consequential, or incidental damages arising out of the use, the results of use, or the inability to use
this product.
ORDERING INFORMATION AND TECHNICAL SERVICES
United States and Canada
Agilent Technologies
Stratagene Products Division
11011 North Torrey Pines Road
La Jolla, CA 92037
Telephone (858) 373-6300
Order Toll Free (800) 424-5444
Technical Services
Internet
World Wide Web
techservices@agilent.com
Europe
Location Telephone Fax Technical Services
Austria 0800 292 499 0800 292 496 0800 292 498
(800) 894-1304
www.stratagene.com
00800 7000 7000 00800 7001 7001 00800 7400 7400 Belgium
0800 15775 0800 15740 0800 15720
00800 7000 7000 00800 7001 7001 00800 7400 7400 France
0800 919 288 0800 919 287 0800 919 289
00800 7000 7000 00800 7001 7001 00800 7400 7400 Germany
0800 182 8232 0800 182 8231 0800 182 8234
00800 7000 7000 00800 7001 7001 00800 7400 7400 Netherlands
0800 023 0446 +31 (0)20 312 5700 0800 023 0448
00800 7000 7000 00800 7001 7001 00800 7400 7400 Switzerland
0800 563 080 0800 563 082 0800 563 081
00800 7000 7000 00800 7001 7001 00800 7400 7400 United Kingdom
0800 917 3282 0800 917 3283 0800 917 3281
All Other Countries
Please contact your local distributor. A complete list of distributors is available at www.stratagene.com.
Page 3

Lambda ZAP-CMV RI Predigested Vector Kit
CONTENTS
Materials Provided.............................................................................................................................. 1
Storage Conditions.............................................................................................................................. 1
Additional Materials Required .......................................................................................................... 1
Notice to Purchaser ............................................................................................................................. 1
Introduction......................................................................................................................................... 2
Overview of the Lambda ZAP-CMV RI Vector System....................................................... 2
Lambda ZAP-CMV Vector Map........................................................................................... 2
pCMV-Script EX Vector Map............................................................................................... 3
Bacterial Host Strains ......................................................................................................................... 4
Host Strain Genotypes...........................................................................................................4
XL1-Blue MRF´ Bacterial Strain Description....................................................................... 4
Recommended Media............................................................................................................ 5
Establishing an Agar Plate Bacterial Stock ........................................................................... 5
Preparing a –80°C Bacterial Glycerol Stock ......................................................................... 5
Growth of Cells for Plating Phage......................................................................................... 6
Helper Phage ....................................................................................................................................... 6
Storing the Helper Phage....................................................................................................... 6
Titering the Helper Phage...................................................................................................... 6
Amplifying the Helper Phage................................................................................................ 7
Ligating the Insert............................................................................................................................... 8
Packaging Reaction............................................................................................................................. 9
General Information ..............................................................................................................9
Packaging Instructions......................................................................................................... 10
Titering the Packaging Reaction ......................................................................................... 10
Amplifying the Library..................................................................................................................... 12
Performing Plaque Lifts ................................................................................................................... 13
Hybridizing and Screening............................................................................................................... 14
In Vivo Excision of the pCMV-Script EX Phagemid Vector from the Lambda ZAP-CMV RI
Vector ......................................................................................................................................... 15
In Vivo Excision Protocols Using ExAssist Helper Phage with XLOLR Strain.......................... 16
Single-Clone Excision Protocol .......................................................................................... 16
Mass Excision Protocol ....................................................................................................... 18
Page 4

Eukaryotic Screening with the Lambda ZAP-CMV RI Library.................................................. 20
Panning Assay ..................................................................................................................... 20
Functional Assay ................................................................................................................. 20
Eukaryotic Expression...................................................................................................................... 21
Appendix: Recovery of Single-Stranded DNA from Cells Containing the pCMV-Script EX
Phagemid Vector ....................................................................................................................... 22
Single-Stranded Rescue Protocol ........................................................................................ 23
Troubleshooting ................................................................................................................................ 24
Packaging ............................................................................................................................24
Excision ............................................................................................................................... 24
Preparation of Media and Reagents................................................................................................ 25
References .......................................................................................................................................... 27
Endnotes............................................................................................................................................. 27
MSDS Information............................................................................................................................ 27
Page 5
Lambda ZAP-CMV RI Predigested Vector Kit
ATERIALS PROVIDED
M
Materials provided Quantity
Lambda ZAP-CMV RI vector digested with EcoR I and CIAP treateda 10 μg
RHEO test insert digested with EcoR I 1.25 μg
XL1-Blue MRF´ strainb 0.5-ml bacterial glycerol stock
XLOLR strainb 0.5-ml bacterial glycerol stock
ExAssist interference-resistant helper phage
R408 Interference-Resistant Helper Phage
a
On arrival, store the Lambda ZAP-CMV RI vector at –20°C. After thawing, aliquot and store at –20°C. Do not pass
through more than two freeze–thaw cycles. For short-term storage, store at 4°C for 1 month.
b
Use the XLOLR strain for plating excised phagemids and the XL1-Blue MRF´ strain for all other manipulations. For host
strain shipping and storage conditions, see Bacterial Host Strains.
c
The titer of the ExAssist interference-resistant helper phage is ~1.0 × 1010 pfu/ml. This supercoiled single-stranded DNA
migrates at ~5 kb on an agarose gel. ExAssist helper phage is recommended for excision of the pCMV-Script EX
phagemid vector from the Lambda ZAP-CMV RI vector. It should not be used for single-stranded rescue.
d
Retiter after 1 month. (Take care not to contaminate the Lambda ZAP-CMV RI vector with this high-titer filamentous helper
phage.) Store at –80°C.
e
The titer of the R408 interference-resistant helper phage is ~7.5 × 1010 pfu/ml. This supercoiled single-stranded DNA
migrates at ~4 kb on an agarose gel. The R408 interference-resistant helper phage is recommended for single-stranded
rescue (see Appendix: Recovery of Single-Stranded DNA from Cells Containing the pCMV-Script EX Phagemid Vector).
c,d
1 ml
d,e
1 ml
STORAGE CONDITIONS
Lambda ZAP-CMV RI Vector: –20°C
Test Insert: –20°C
Helper Phage: –80°C
Bacterial Glycerol Stocks: –80°C
ADDITIONAL MATERIALS REQUIRED
Gigapack III Plus or Gigapack III Gold packaging extract (Stratagene Catalog #200204 and
#200201, respectively)
NOTICE TO PURCHASER
The use of the CMV Promoter is covered under U.S. Patent Nos. 5,168,062 and 5,385,839 owned by
the University of Iowa Research Foundation and licensed FOR RESEARCH USE ONLY. For
further information, please contact UIRF at 319-335-4546.
Revision A © Agilent Technologies, Inc. 2008.
Lambda ZAP-CMV RI Predigested Vector Kit 1
Page 6

INTRODUCTION
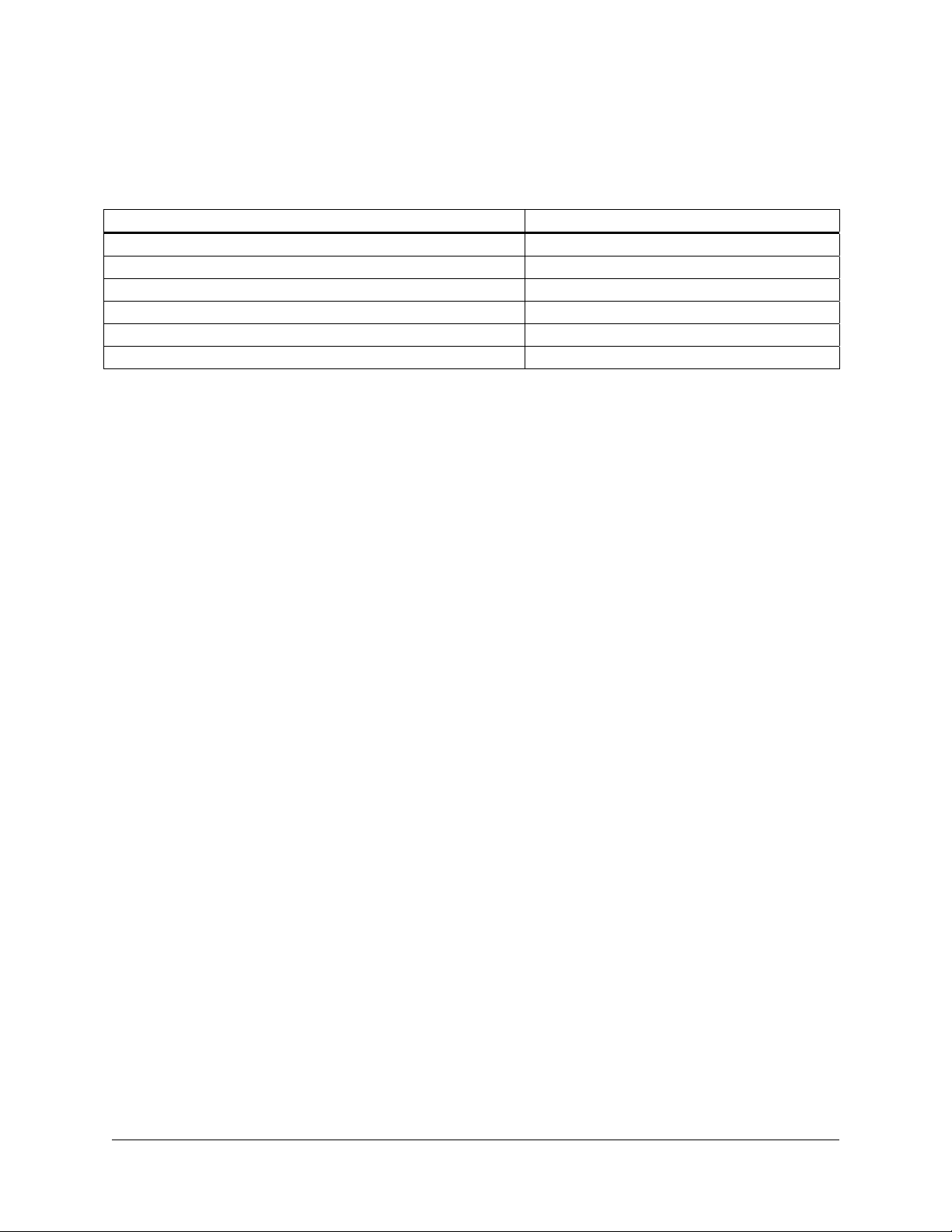
Overview of the Lambda ZAP-CMV RI Vector System
The Lambda ZAP-CMV RI vector (predigested with EcoR I) allows
eukaryotic expression.
CMV RI vector, Sac I, Not I, Srf I, EcoR I, and Xho I, accommodate DNA
inserts up to 6.5 kb in length (see Figure 1). Inserts cloned into the Lambda
ZAP-CMV RI vector can be excised out of the phage in the form of the
kanamycin-resistant pCMV-Script
the same excision mechanism used with the Lambda ZAP vectors.
Clones in the Lambda ZAP-CMV RI vector can be screened with DNA
probes and in vivo rapid excision of the pCMV-Script EX phagemid vector
allows insert characterization in a plasmid system. Alternatively, the entire
library can be mass excised for functional screening in mammalian cells.
The polylinker of pCMV-Script EX phagemid has 15 unique cloning sites
flanked by T3 and T7 promoters and has three primer sites for DNA
sequencing. The plasmid has the bacteriophage f1 origin of replication
allowing rescue of single-stranded DNA, which can be used for DNA
sequencing or site-directed mutagenesis. Unidirectional deletions can be
made using exonuclease III and mung bean nuclease by taking advantage of
the unique positioning of 5´ and 3´ restriction sites. Transcripts made from
the T3 and T7 promoters generate riboprobes useful in Southern and
Northern blotting.
1, 2
The five unique cloning sites of the Lambda ZAP-
EX phagemid vector (see Figure 2) by
1, 3, 4
Note The pCMV-Script EX vector differs from the pCMV-Script vector
by 29 bases located downstream of the f1 origin.
Eukaryotic expression of inserts in pCMV-Script EX is driven by the
cytomegalovirus (CMV) immediate early (IE) promoter with the
SV40 transcription terminator and polyadenylation signal. Stable selection
of clones in eukaryotic cells is made possible by the presence of the
neomycin-and kanamycin-resistance gene, which is driven by the
SV40 early promoter with TK transcription polyadenylation signals to
render transfectants resistant to G418 (geneticin).
The pCMV-Script EX vector does not contain an ATG initiation codon. A
translation initiation sequence must be incorporated if the DNA fragment to
be cloned does not have an initiating ATG codon or an optimal sequence for
initiating translation, such as the Kozak sequence [GCC(A/G)CCATGG].
Lambda ZAP-CMV Vector Map
FIGURE 1 Map of the Lambda ZAP-CMV vector.
2 Lambda ZAP-CMV RI Predigested Vector Kit
Page 7
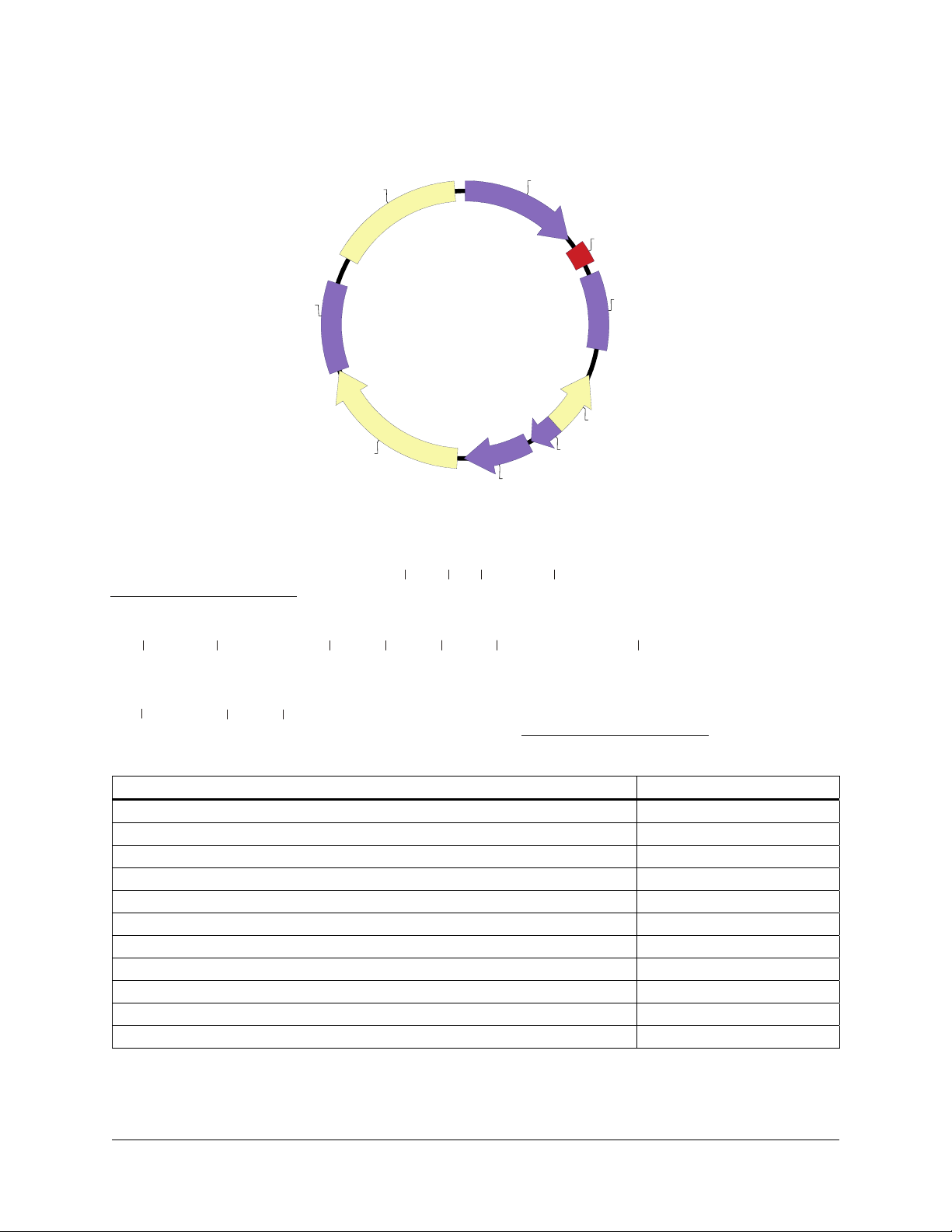
pCMV-Script EX Vector Map
A
pUC ori
TK pA
pCMV-Script EX
4.3 kb
neo/kan
pCMV-Script EX Multiple Cloning Site Region
(sequence shown 620–799)
T3 promoter
AATTAACCCTCACTAAAGGGAACAAAAGCTGGAGCTC CACCGCGGTGGCGGCCGCTCTA...
P CMV
MCS
SV40 pA
f1 ori
P bla
P SV40
Not ISac IIBstX ISac I
Srf I Pst I
...GCCCGGGCGGATCCCCCGGGCTGCAG GAATTCGATATCAAGCTTATCGATACCGTCGAC...
Xho I
...CTCGAGGGGGGGCCCGGTACCAGGTA AGTGTACCCAATTCG CCCCTATAGTGAGTCGTATTA
BamH I
pa I
Kpn I
EcoR I
EcoR V
Acc I/Sal IHind III
T7 promoter
Feature Nucleotide Position
CMV promoter 1–602
T3 promoter and T3 primer binding site 620–639
multiple cloning site 651–758
T7 promoter and T7 primer binding site 778–799
SV40 polyA signal 811–1194
f1 origin of ss-DNA replication 1332–1638
bla promoter 1692–1816
SV40 promoter 1836–2174
neomycin/kanamycin resistance ORF 2209–3000
HSV-thymidine kinase (TK) polyA signal 3001–3450
pUC origin 3588–4255
Figure 2 Circular map and polylinker sequence of the pCMV-Script EX vector.
Lambda ZAP-CMV RI Predigested Vector Kit 3
Page 8

BACTERIAL HOST STRAINS
Host Strain Genotypes
Host strain Genotype
XL1-Blue MRF´ strain Δ(mcrA)183 Δ(mcrCB-hsdSMR-mrr)173 endA1 supE44 thi-1
recA1 gyrA96 relA1 lac [F´ proAB lacI
XLOLR straina Δ(mcrA)183 Δ(mcrCB-hsdSMR-mrr)173 endA1 thi-1 recA1
gyrA96 relA1 lac [F´ proAB lacIqZΔM15 Tn10 (Tetr)] Su–
(nonsuppressing) λ
a
Use the XLOLR strain for excision only.
XL1-Blue MRF´ Bacterial Strain Description
The RecA– E. coli host strain XL1-Blue MRF´ is supplied with the Lambda
ZAP-CMV RI predigested vector kit.
phagemid vector does not require a supF genotype, the amplified library
grows very efficiently on the XL1-Blue MRF´ strain. In addition, use of the
correct host strain is important when working with the pCMV-Script EX
phagemid vector due to the F´ episome present in the XL1-Blue MRF´
strain.
The F´ episome expresses the genes forming the F´ pili found on the surface
of the bacteria. Without pili formation, filamentous phage (i.e., M13 or f1)
infection could not occur. Because the conversion of a recombinant
ZAP-CMV RI clone to a pCMV-Script EX phagemid vector requires
superinfection with a filamentous helper phage, the F´ episome is required
for in vivo excision (see In Vivo Excision of the pCMV-Script EX Phagemid
Vector from the Lambda ZAP-CMV RI Vector).
q
ZΔM15 Tn10 (Tetr)]
r
(lambda resistant)
2
Because the pCMV-Script EX
Note The strains Y1088, Y1089, and Y1090 are not suitable for use with
the Lambda ZAP-CMV RI vector as these strains contain the
plasmid pMC9, a pBR322 derivative, which contains many of the
same sequences as those found in the phagemid portion of the
Lambda ZAP-CMV RI vector. Using these strains with the Lambda
ZAP-CMV RI vector could result in recombination between the
homologous sequences. The SURE and SOLR strains are not
compatible with the Lambda ZAP-CMV RI system since these
strains contain the kanamycin-resistance gene found in the
pCMV-Script EX phagemid vector.
4 Lambda ZAP-CMV RI Predigested Vector Kit
Page 9
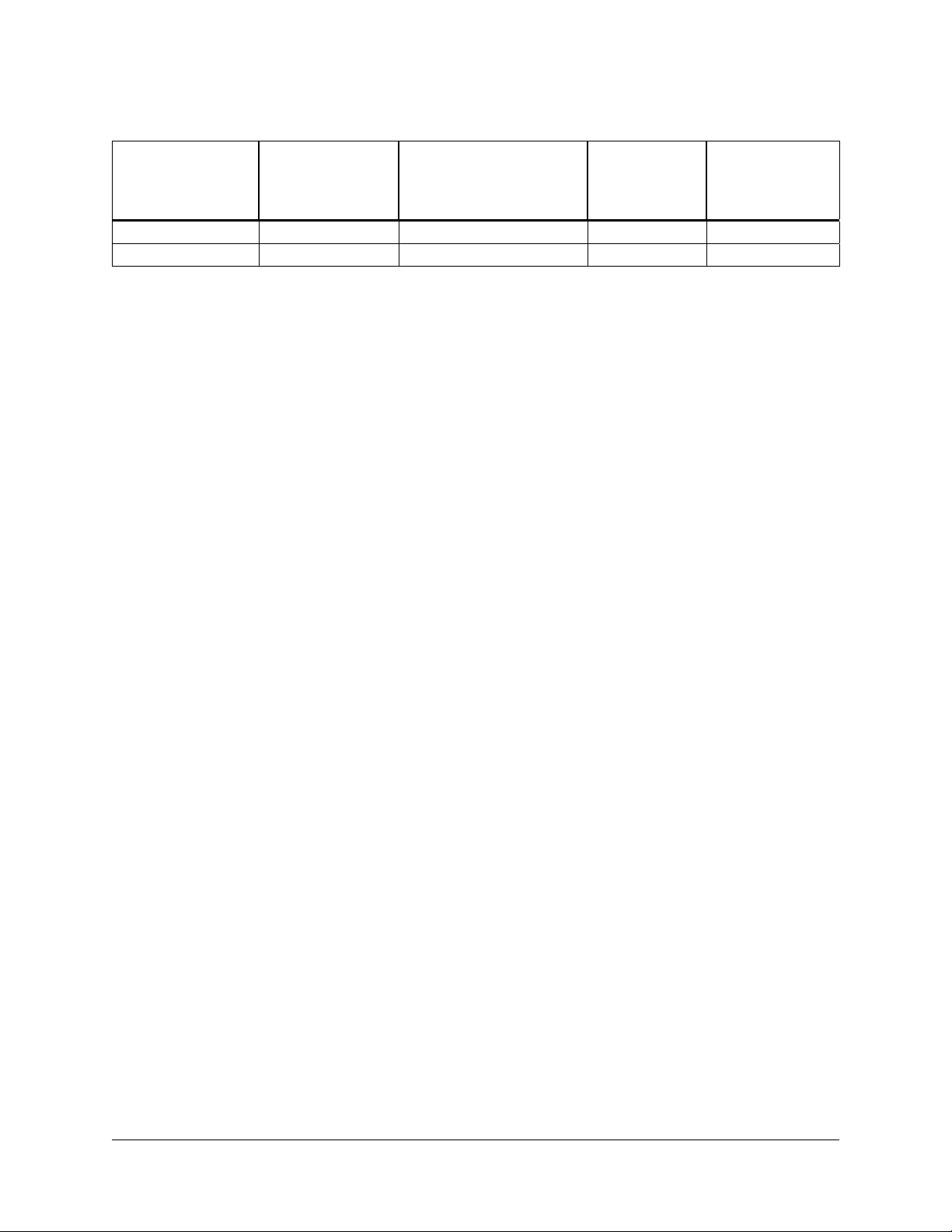
Recommended Media
Host strain
XLOLR strain LB-tetracyclinea LB broth with supplements
XL1-Blue MRF´ strain LB-tetracyclinea LB broth with supplements
a
See Preparation of Media and Reagents.
b
LB broth with 0.2% (w/v) maltose and 10 mM MgSO4.
c
Maltose and magnesium supplements are required for optimal lambda phage receptor expression on the surface of the
XL1-Blue MRF´ host cell. The media supplements are not required for helper phage infection, but are included in both
protocols for simplified media preparation.
Agar plates and
liquid medium for
bacterial streak
and glycerol stock
Liquid medium for
bacterial cultures prior to
phage attachment
a-c
a-c
Agar plates
and top agar
for plaque
formation
— LB-kanamycina
NZYa —
Agar plates for
excision protocol
Establishing an Agar Plate Bacterial Stock
The bacterial host strains are shipped as bacterial glycerol stocks. On arrival,
prepare the following plates from the bacterial glycerol stocks.
Note The host strains may thaw during shipment. The vials should be
stored immediately at –20° or –80°C, but most strains remain
viable longer if stored at –80°C. It is best to avoid repeated
thawing of the host strains in order to maintain extended viability.
1. Revive the stored cells by scraping off splinters of solid ice with a
sterile wire loop.
2. Streak the splinters onto an LB agar plate containing the appropriate
antibiotic (see Recommended Media), if one is necessary.
3. Incubate the plate overnight at 37°C.
4. Seal the plate with Parafilm
®
for up to 1 week.
5. Restreak the cells onto a fresh plate every week.
Preparing a –80°C Bacterial Glycerol Stock
1. In a sterile 50-ml conical tube, inoculate 10 ml of LB broth with the
appropriate antibiotic (see Recommended Media) with one colony from
the plate. Grow the cells to late log phase.
2. Add 4.5 ml of a sterile glycerol-liquid medium solution (prepared by
mixing 5 ml of glycerol + 5 ml of the appropriate medium) to the
bacterial culture from step 1. Mix well.
3. Aliquot into sterile centrifuge tubes (1 ml/tube).
This preparation may be stored at –20°C for 1–2 years or at –80°C for more
than 2 years.
laboratory film and store the plate at 4°C
Lambda ZAP-CMV RI Predigested Vector Kit 5
Page 10

Growth of Cells for Plating Phage
Bacterial cultures for plating phage should be started from a fresh plate
using a single colony and should be grown overnight with vigorous shaking
at 30°C in 50 ml of LB broth supplemented with 0.2% (w/v) maltose and
10 mM MgSO
The lower temperature ensures that the cells will not overgrow. The cells
should be spun at 1000 × g for 10 minutes then gently resuspended in
10 ml of 10 mM MgSO
10 mM MgSO
phage manipulations described within the manual. Highest efficiencies are
obtained from freshly prepared cells.
HELPER PHAGE
Two different helper phages are provided with the Lambda ZAP-CMV RI
predigested vector kit: (1) the ExAssist interference-resistant helper phage
with XLOLR strain and (2) the R408 helper phage. The ExAssist
interference-resistant helper phage with XLOLR strain is designed to allow
efficient in vivo excision of the pCMV-Script EX phagemid vector from the
Lambda ZAP-CMV RI vector while preventing problems associated with
helper phage co-infection. The ExAssist helper phage contains an amber
mutation that prevents replication of the phage genome in a nonsuppressing
E. coli strain (e.g., XLOLR cells). Only the excised phagemid can replicate
in the host, removing the possibility of co-infection from the ExAssist
helper phage. Because ExAssist helper phage cannot replicate in the
XLOLR strain, single-stranded rescue cannot be performed in this strain
using ExAssist helper phage. XLOLR cells are also resistant to lambda
infection, preventing lambda DNA contamination after excision.
. (Do not use tetracycline in the presence of magnesium.)
4
. Before use, dilute cells to an OD
4
. Bacterial cells prepared in this manner can be used for all
4
of 0.5 with
600
Storing the Helper Phage
The ExAssist helper phage and the R408 helper phage are supplied in
7% dimethylsulfoxide (DMSO) and should be stored at –80°C. The helper
phage may be stored for short periods of time at –20°C or 4°C. It is
important to titer the helper phage prior to each use. Expect titers of
approximately 10
the R408 helper phage. If the titer drops over time, prepare a fresh high-titer
stock of the helper phage as outlined in Amplifying the Helper Phage.
Titering the Helper Phage
1. Transfer a colony of XL1-Blue MRF´ cells into 10 ml of LB broth with
supplements in a 50-ml conical tube. Incubate the conical tube with
shaking at 37°C until growth reaches an OD
2. Dilute the phage (10
and Reagents) and combine 1 μl of each dilution with 200 μl of
XL1-Blue MRF´ cells (OD
3. Incubate the helper phage and the XL1-Blue MRF´ cells for 15 minutes
at 37°C to allow the phage to attach to the cells.
10
pfu/ml for the ExAssist helper phage or 1010 pfu/ml for
of 1.0.
600
–4
–10–7) in SM buffer (See Preparation of Media
= 1.0).
600
6 Lambda ZAP-CMV RI Predigested Vector Kit
Page 11

4. Add 3 ml of NZY top agar, melted and cooled to ~48°C, and plate
(
immediately onto dry, prewarmed NZY agar plates. Allow the plates to
set for 10 minutes.
5. Invert the plates and incubate overnight at 37°C.
Note ExAssist and R408 plaques will have a cloudier appearance
than lambda phage plaques.
6. To determine the titer [in plaque-forming units per milliliter (pfu/ml)],
use the following formula:
⎡
Number of plaques pfu dilution factor
⎢
⎣
where the volume plated (in microliters) refers to the volume of the
helper phage solution added to the cells.
Amplifying the Helper Phage
1. Transfer a colony of XL1-Blue MRF´ cells into 10 ml of LB broth with
supplements in a 50-ml conical tube. Incubate the conical tube with
shaking at 37°C until growth reaches an OD
×
)
Volume plated l
μ
(
⎤
1000 l / ml
×
)
⎥
⎦
of 0.3.
600
μ
Note An OD
of 0.3 corresponds to 2.5 × 108 cells/ml.
600
2. Add the helper phage at a multiplicity of infection (MOI) of 20:1
(phage-to-cells ratio).
3. Incubate the conical tube at 37°C for 15 minutes to allow the phage to
attach to the cells.
4. Incubate the conical tube with shaking at 37°C for 8 hours.
5. Heat the conical tube at 65°C for 15 minutes.
6. Spin down the cell debris and transfer the supernatant to a fresh conical
tube.
10
7. The titer of the supernatant should be between 7.5 × 10
1.0 × 10
1.0 × 10
12
pfu/ml for ExAssist helper phage or between 1.0 × 1011 and
12
pfu/ml for R408 helper phage.
and
Note ExAssist and R408 plaques will have a cloudier appearance
than lambda phage plaques.
8. Add dimethylsulfoxide (DMSO) to a final concentration of 7% (v/v)
and store at –80°C.
9. For further details about helper phage titering or amplification, please
see
Titering the Helper Phage or Reference 5.
Lambda ZAP-CMV RI Predigested Vector Kit 7
Page 12

LIGATING THE INSERT
Notes In all ligations, the final glycerol content should be less than
The Lambda ZAP-CMV RI vector is shipped in 10 mM Tris-HCl (pH 7.0)
and 0.1 mM EDTA and can be stored up to 1 month at 4°C or frozen in
aliquots at –20°C for longer storage. The RHEO test insert should be stored
at –20°C. However, do not put samples through multiple freeze–thaw
cycles.
When ligating the sample insert, use a volume up to 2.5 μl. Use an equal
molar ratio (or less to prevent multiple inserts) of the insert. The
Lambda ZAP-CMV vector can accommodate inserts up to 6.5 kb. The
Lambda ZAP-CMV vector is ~42 kb in length. If ligating a 4000-bp insert to
the vector, use 0.1 μg of insert for every 1 μg of vector.
5% (v/v). Do not exceed 5% (v/v) glycerol. Due to the high
molecular weight of the lambda vector, the contents may be very
viscous. It is important to microcentrifuge the contents of the
lambda vector tube briefly at 11,000 × g, then gently mix the
solution by stirring with a yellow pipet tip prior to pipetting.
Polyethylene glycol (PEG), which is present in some ligase
buffers, can inhibit packaging.
Note A general rule when constructing cDNA libraries is to add
between 50–150 ng of cDNA/
1. Set up a control ligation to ligate the test insert into the
Lambda ZAP-CMV RI vector. Add the following components in order:
1.0 μl of the predigested Lambda ZAP-CMV RI vector (1 μg)
0.8 μl of RHEO test insert (0.2 μg)
0.5 μl of 10× ligase buffer
0.5 μl of 10 mM rATP (pH 7.5)
X μl of water for a final volume of 5 μl
X μl of T4 DNA ligase (2 Weiss U)
Prepare the sample ligation in a separate tube, using equal molar ratios
2.
of insert and vector. Add the following components in order:
1.0 μl of the predigested Lambda ZAP-CMV RI vector (1 μg)
X μl of the sample insert
0.5 μl of 10
0.5 μl of 10 mM rATP (pH 7.5)
X μl of water for a final volume of 5 μl
X μl of T4 DNA ligase (2 Weiss U)
× ligase buffer
μ
g of lambda arms.
3.
Incubate the reaction tubes overnight at 4°C.
8 Lambda ZAP-CMV RI Predigested Vector Kit
Page 13

After ligation is complete, package each ligation, including the control
ligation. If the insert used is free from contaminants and contains a high
percentage of ligatable ends, expect ~1 × 10
6
–1.5 × 107 recombinant
plaques/μg vector when using high-efficiency packaging extracts, such as
Gigapack III Plus or Gigapack III Gold packaging extracts (Stratagene
Catalog #200204 and #200201, respectively). Protocols are provided below
for packaging ligation reactions using these recommended extracts. See the
appropriate Gigapack III packaging extract instruction manual for additional
details and recommended packaging control reactions.
Note We recommend using the high-efficiency Gigapack III Gold
PACKAGING REACTION
General Information
Packaging extracts are used to package recombinant lambda phage with
high efficiency, which increases the size of gene libraries.
Gigapack III Gold packaging extract increases the efficiency and
representation of libraries constructed from highly methylated DNA. The
packaging extracts are restriction minus (HsdR
McrF
When used in conjunction with restriction-deficient plating cultures,
Gigapack III Gold packaging extract should improve the quality of DNA
libraries constructed from methylated DNA.
Optimal packaging efficiencies are obtained with lambda DNAs that are
concatemeric. Ligations should be carried out at DNA concentrations of
0.2 μg/μl or greater, which favors concatemers and not circular DNA
molecules that only contain one
relatively free from contaminants. DNA may be used directly from ligation
reactions in most cases; however, polyethylene glycol (PEG), which is
contained in some ligase buffers, has been shown to inhibit packaging. The
volume of DNA added to each extract should be <5 μl.
Undigested wild-type lambda DNA will be packaged with efficiencies
exceeding 2 × 10
packaging extract. Predigested arms, when ligated to a test insert, will yield
~1.0 × 10
6
packaging extract
and Mrr
–
. Other commercially available packaging extracts can
since this packaging extract is McrA–, McrB–,
restrict hemimethylated DNA, therefore producing low-titer
libraries.
–
–
Mrr–) to optimize packaging efficiency and library representation.
6, 7, 8, 9
McrA– McrBC–
cos site. DNA to be packaged should be
9
plaques/μg of vector when using Gigapack III Gold
7
recombinant plaques/μg of vector.
Lambda ZAP-CMV RI Predigested Vector Kit 9
Page 14

Packaging Instructions
Packaging Protocol
Note Polyethylene glycol, which is present in some ligase buffers, can
1. Remove the appropriate number of packaging extracts from the
−80°C freezer and place the extracts on dry ice.
2. Quickly thaw the packaging extract by holding the tube between your
fingers until the contents of the tube just begins to thaw.
inhibit packaging.
3. Add the experimental DNA
0.1–1.0 μg of ligated DNA) to the packaging extract. (Add 1 μl of the
control ligation to a separate tube of packaging extract.)
4. Stir the mixture with a pipet tip to mix well.
allowable provided that air bubbles are not introduced.
5. Spin the tube quickly (for 3–5 seconds), if desired, to collect the liquid
at the bottom of the tube.
6. Incubate the tube at room temperature (22°C) for 2 hours.
7. Add 500 μl of SM buffer to the tube.
8. Add 20 μl of chloroform and mix the contents of the tube gently.
9. Spin the tube briefly to sediment the debris.
10.
The supernatant containing the phage is ready for titering. The
supernatant may be stored at 4°C for up to 1 month.
Titering the Packaging Reaction
Preparing the Host Bacteria
immediately (1–4 μl containing
Gentle pipetting is
1. Streak the XL1-Blue MRF´ cells onto LB agar plates containing the
appropriate antibiotic (See
overnight at 37°C.
Note Do not add antibiotic to the medium in the following step.
The antibiotic will bind to the bacterial cell wall and will
inhibit the ability of the phage to infect the cell.
2. Prepare a separate 50-ml culture of XL1-Blue MRF´ cells in LB broth
with supplements.
10 Lambda ZAP-CMV RI Predigested Vector Kit
Recommended Media). Incubate the plates
Page 15

3. Incubate with shaking at 37°C for 4–6 hours (do not grow past an
of 1.0). Alternatively, grow overnight at 30°C, shaking at
OD
600
200 rpm.
Note The lower temperature keeps the bacteria from overgrowing,
thus reducing the number of nonviable cells. Phage can
adhere to nonviable cells resulting in a decreased titer.
4. Pellet the bacteria at 1000 × g for 10 minutes.
5. Gently resuspend the cell pellet in 25 ml sterile 10 mM MgSO
.
4
Note For later use, store the cells at 4°C overnight in
10 mM MgSO
.
4
Titering Protocol
1. Dilute the XL1-Blue MRF´ cells (from step 5 of Preparing the Host
Bacteria)
Note The bacteria should be used immediately following dilution.
2. To determine the titer of the packaged ligation product, mix the
following components:
and
3. Incubate the phage and the bacteria at 37°C for 15 minutes to allow the
phage to attach to the cells.
to an OD
of 0.5 with sterile 10 mM MgSO4.
600
1 μl of the final packaged reaction
200 μl of XL1-Blue MRF´ cells at an OD
of 0.5
600
1 μl of a 1:10 dilution of the final packaged reaction
200 μl of XL1-Blue MRF´ cells at an OD
of 0.5
600
4. Add 3 ml of NZY top agar, melted and cooled to ~48°C, and plate
immediately onto dry, prewarmed NZY agar plates. Allow the plates to
set for 10 minutes. Invert the plates and incubate at 37°C.
5. Plaques should be visible after 6–8 hours. Count the plaques and
determine the titer in plaque-forming units per milliliter (pfu/ml).
Note If insert size is crucial, one may excise a few clones at this
step or a few clones prior to single-clone amplification (see
Single-Clone Excision Protocol in In Vivo Excision Protocols
Using ExAssist Interference-Resistant Helper Phage with
XLOLR Strain
).
Lambda ZAP-CMV RI Predigested Vector Kit 11
Page 16

AMPLIFYING THE LIBRARY
Note Primary libraries can be unstable; therefore, amplification of the
It is usually desirable to amplify libraries prepared in lambda vectors to
make a large, stable quantity of a high-titer stock of the library. However,
more than one round of amplification is not recommended, since slower
growing clones may be significantly underrepresented. The following
protocol is recommended for amplifying the Lambda ZAP-CMV RI library.
Day 1
1. Grow a 50-ml overnight culture of XL1-Blue MRF´ cells in LB broth
with supplements at 30°C with shaking.
Day 2
2. Gently spin down the XL1-Blue MRF´ cells (1000 × g). Resuspend the
cell pellet in 25 ml of 10 mM MgSO
suspension, then dilute the cells to an OD
3. Combine aliquots of the packaged mixture or library suspension
containing ~5 × 10
XL1-Blue MRF´ cells at an OD
polypropylene tubes. To amplify 1 × 10
20 aliquots (each aliquot contains 5 × 10
libraries is recommended immediately.
. Measure the OD
4
600
4
pfu of bacteriophage with 600 μl of
of 0.5 in Falcon® 2059
600
4
plaques/150-mm plate).
of the cell
600
of 0.5 in 10 mM MgSO4.
6
plaques, use a total of
Note Do not add more than 300 μl of phage/600 μl of cells.
4. Incubate the tubes containing the phage and host cells for 15 minutes at
37°C to allow the phage to attach to the cells.
5. Mix 6.5 ml of NZY top agar, melted and cooled to ~48°C, with each
aliquot of infected bacteria and spread evenly onto a freshly poured
150-mm NZY agar plate. Allow the plates to set for 10 minutes.
6. Invert the plates and incubate at 37°C for 6–8 hours. Do not allow the
plaques to get larger than 1–2 mm. On completion, the plaques should
be touching.
7. Overlay the plates with ~8–10 ml of SM buffer. Store the plates at 4°C
overnight (with
gentle rocking if possible). This allows the phage to
diffuse into the SM buffer.
12 Lambda ZAP-CMV RI Predigested Vector Kit
Page 17

Day 3
8. Recover the bacteriophage suspension from each plate and pool it into a
sterile polypropylene container. Rinse the plates with an additional
2 ml of SM buffer and pool. Add chloroform to a 5% (v/v) final
concentration. Mix well and incubate for 15 minutes at room
temperature.
9. Remove the cell debris by centrifugation for 10 minutes at 500 ×
10. Recover the supernatant and transfer it to a sterile polypropylene
container. If the supernatant appears cloudy or has a high amount of
cell debris, repeat steps 8 and 9. If the supernatant is clear, add
chloroform to a 0.3% (v/v) final concentration and store at 4°C. Store
aliquots of the amplified library in 7% (v/v) DMSO at –80°C.
11.
Check the titer of the amplified library using host cells and serial
dilutions of the library. (Assume ~10
Note Briefly spin the lambda phage stock to ensure that the
PERFORMING PLAQUE LIFTS
1. Titer the amplified mixture or library suspension to determine the
concentration using XL1-Blue MRF´ cells.
2. Combine the equivalent of 5 × 10
prepared XL1-Blue MRF´ cells at an OD
g.
9
–1011 pfu/ml.)
chloroform is separated completely before removing the
aliquot for titering.
4
pfu/plate and 600 μl of freshly
of 0.5.
600
3. Incubate the bacteria and phage mixture at 37°C for 15 minutes to
allow the phage to attach to the cells.
4. Add 6.5 ml of NZY top agar (~48°C) to the bacteria and phage mixture.
5. Quickly pour the plating culture onto a dry, prewarmed 150-mm NZY
agar plate, which is at least 2 days old. Carefully swirl the plate to
distribute the cells evenly. Allow the plates to set for 10 minutes. (Use
20 plates to screen 1 × 10
6
pfu.)
6. Invert the plates and incubate at 37°C for ~8 hours.
7. Chill the plates for 2 hours at 4°C to prevent the top agar from sticking
to the nitrocellulose membrane.
Note Use forceps and wear gloves for the following steps.
Lambda ZAP-CMV RI Predigested Vector Kit 13
Page 18

8. Place a nitrocellulose membrane onto each NZY agar plate for
2 minutes to allow the transfer of the phage particles to the membrane.
Use a needle to prick through the membrane and agar for orientation.
(If desired, waterproof ink in a syringe needle may be used.)
Notes If making duplicate nitrocellulose membranes, allow the
second membrane to transfer for ~4 minutes.
®
Pyrex
dishes are convenient for the following steps. All
solutions should be at room temperature.
a. Denature the nitrocellulose-bound DNA after lifting by
submerging the membrane in a 1.5 M NaCl and 0.5 M NaOH
denaturation solution for 2 minutes.
Note If using charged nylon, wash with gloved fingertips to remove
the excess top agar.
b. Neutralize the nitrocellulose membrane for 5 minutes by
submerging the membrane in a 1.5 M NaCl and 0.5 M Tris-HCl
(pH 8.0) neutralization solution.
c. Rinse the nitrocellulose membrane for no more than 30 seconds by
submerging the membrane in a 0.2 M Tris-HCl (pH 7.5) and
2× SSC buffer solution (see
Preparation of Media and Reagents).
9. Blot briefly on a Whatman
10. Crosslink the DNA to the membranes using the autocrosslink setting on
the Stratalinker UV crosslinker* (120,000 μJ of UV energy) for
~30 seconds. Alternatively, oven bake at 80°C for ~1.5–2 hours.
11. Store the stock agar plates of the transfers at 4°C to use after screening.
HYBRIDIZING AND SCREENING
Following the preparation of the membranes for hybridization, perform
prehybridization, probe preparation, hybridization, and washes for either
oligonucleotide probes or double-stranded probes and then expose the
membranes to film as outlined in standard methodology texts.
these procedures, perform secondary and tertiary screenings also as outlined
in the standard methodology texts.
RI vector, perform in vivo excision on the isolates to obtain the insertcontaining pCMV-Script EX phagemid vector (see
pCMV-Script EX from the Lambda ZAP-CMV RI Vector
Excision Protocols Using ExAssist Helper Phage with XLOLR Strain).
*
Stratagene Catalog #400071 (1800 model) or #400075 (2400 model).
®
3MM paper.
5, 10
When using the Lambda ZAP-CMV
5, 10
Following
In Vivo Excision of the
and In Vivo
14 Lambda ZAP-CMV RI Predigested Vector Kit
Page 19

IN VIVO EXCISION OF THE pCMV-SCRIPT EX PHAGEMID VECTOR
FROM THE
LAMBDA ZAP-CMV RI VECTOR
The Lambda ZAP-CMV RI vector is designed to allow simple, efficient in
vivo excision and recircularization of any cloned insert contained within the
lambda vector to form a phagemid containing the cloned insert.
vivo excision depends on the placement of the DNA sequences within the
lambda phage genome and on the presence of a variety of proteins,
including filamentous (e.g., M13) bacteriophage-derived proteins. The M13
phage proteins recognize a region of DNA normally serving as the f1
bacteriophage "origin of replication." This origin of replication can be
divided into two overlying parts: (1) the site of initiation and (2) the site of
termination for DNA synthesis.
separately into the Lambda ZAP-CMV RI vector. The lambda phage (target)
is made accessible to the M13-derived proteins by simultaneously infecting
a strain of
Inside
recognize the initiator DNA that is within the lambda vector. One of these
proteins then nicks one of the two DNA strands. At the site of this nick, new
DNA synthesis begins and duplicates whatever DNA exists in the lambda
vector "downstream" (3´) of the nicking site. DNA synthesis of a new single
strand of DNA continues through the cloned insert until a termination
signal, positioned 3´ of the initiator signal, is encountered within the
constructed lambda vector. The ssDNA molecule is circularized by the gene
II product from the M13 phage, forming a circular DNA molecule
containing the DNA between the initiator and terminator. In the case of the
Lambda ZAP-CMV RI vector, this includes all sequences of the pCMVScript EX phagemid vector and the insert, if one is present. This conversion
is the "subcloning" step, since all sequences associated with normal lambda
vectors are positioned outside of the initiator and terminator signals and are
not contained within the circularized DNA. In addition, the circularizing of
the DNA automatically recreates a functional f1 origin as found in
f1 bacteriophage or phagemids.
Signals for "packaging" the newly created phagemid are linked to the
f1 origin sequence. The signals permit the circularized ssDNA to be
"packaged" into phagemid particles and secreted from the
secretion of the phagemid particle, the
of the cloned DNA are killed, and the lambda phage is lysed by heat
treatment at 70°C. The phagemid is not affected by the heat treatment.
Escherichia coli is infected with the phagemid and can be plated on
selective media to form colonies. DNA from excised colonies can be used
for analysis of insert DNA, including DNA sequencing, subcloning, and
mapping. Colonies from the excised pCMV-Script EX phagemid vector can
also be used for subsequent production of ssDNA suitable for dideoxysequencing and site-specific mutagenesis.
E. coli with both the lambda vector and the M13 helper phage.
E. coli, the "helper" proteins (i.e., proteins from M13 phage)
3, 4
This in
11
These two regions are subcloned
E. coli. Following
E. coli cells used for in vivo excision
Lambda ZAP-CMV RI Predigested Vector Kit 15
Page 20

In Vivo EXCISION PROTOCOLS USING EXASSIST HELPER PHAGE WITH
XLOLR STRAIN
The ExAssist helper phage with XLOLR strain is designed to efficiently
excise the pCMV-Script EX phagemid vector from the Lambda ZAP-CMV
RI vector, while eliminating problems associated with helper phage coinfection. The ExAssist helper phage contains an amber mutation that
prevents replication of the helper phage genome in a nonsuppressing
E. coli strain such as XLOLR cells. This allows only the excised phagemid
to replicate in the host, removing the possibility of co-infection from the
ExAssist helper phage. Since the ExAssist helper phage cannot replicate in
the XLOLR strain, single-stranded rescue cannot be performed in this strain
using this helper phage.
Note It is important not to contaminate the Lambda ZAP-CMV RI
library with the filamentous helper phage, since small amounts of
contaminating helper phage are sufficient to convert Lambda
ZAP-CMV RI phage into pCMV-Script EX phagemids. If
contamination should occur, contaminating filamentous phage can
be removed without harming the library by adding a few
microliters of chloroform to the Lambda ZAP-CMV RI phage
stock.
Mass excision can be used to generate subtraction libraries and subtracted
DNA probes. Converting the library to the phagemid form also allows
screening of the phagemid library in eukaryotic cells by transformation of
eukaryotic cells with supercoiled plasmid DNA.
Single-Clone Excision Protocol
Day 1
1. Core the plaque of interest from the agar plate and transfer the plaque
to a sterile microcentrifuge tube containing 500 μl of SM buffer and
20 μl of chloroform. Vortex the microcentrifuge tube to release the
phage particles into the SM buffer. Incubate the microcentrifuge tube
for 1–2 hours at room temperature or overnight at 4°C. (This phage
stock is stable for up to 6 months at 4°C.)
2. Grow separate 50-ml overnight cultures of XL1-Blue MRF´ and
XLOLR cells in LB broth with supplements at 30°C.
Day 2
3. Gently spin down the XL1-Blue MRF´ and XLOLR cells (1000 × g).
Resuspend each of the cell pellets in 25 ml of 10 mM MgSO
the OD
cells to an OD
of the cell suspensions, then adjust the concentration of the
600
of 1.0 (8 × 108 cells/ml) in 10 mM MgSO4.
600
. Measure
4
16 Lambda ZAP-CMV RI Predigested Vector Kit
Page 21

4. Combine the following components in a Falcon 2059 polypropylene
tube:
200 μl of XL1-Blue MRF´ cells at an OD
250 μl of phage stock (containing >1 × 10
1 μl of the ExAssist helper phage (>1 × 10
of 1.0
600
5
phage particles)
6
pfu/μl)
Note Briefly spin the lambda phage stock to ensure that the
chloroform is separated completely before removing the
aliquot used in the excision reaction.
5. Incubate the Falcon 2059 polypropylene tube at 37°C for 15 minutes to
allow the phage to attach to the cells.
6. Add 3 ml of LB broth with supplements and incubate the Falcon 2059
polypropylene tube for 2.5–3 hours at 37°C with shaking. Because
clonal representation is not relevant, single-clone excision reactions can
be safely performed overnight.
Note The turbidity of the media is not indicative of the success of
the excision.
7. Heat the Falcon 2059 polypropylene tube at 65–70°C for 20 minutes to
lyse the lambda phage particles and the cells. Spin the tube at 1000 ×
g
for 15 minutes to pellet the cell debris.
8. Decant the supernatant into a sterile Falcon 2059 polypropylene tube.
This stock contains the excised pCMV-Script EX phagemid packaged
as filamentous phage particles. (This stock may be stored at 4°C for
1–2 months.)
9. To plate the excised phagemids, add 200 μl of freshly grown
XLOLR cells from step 3 (OD
= 1.0) to two 1.5-ml microcentrifuge
600
tubes. Add 100 μl of the phage supernatant (from step 8 above) to one
microcentrifuge tube and 10 μl of the phage supernatant to the other
microcentrifuge tube.
10.
Incubate the microcentrifuge tubes at 37°C for 15 minutes.
11. Add 300 μl of NZY broth and incubate the tubes at 37°C for
45 minutes to allow sufficient expression of the kanamycin-resistance
gene product prior to plating on selective medium.
12. Plate 200 μl of the cell mixture from each microcentrifuge tube on
LB-kanamycin agar plates (50 μg/ml) and incubate the plates overnight
at 37°C.
Lambda ZAP-CMV RI Predigested Vector Kit 17
Page 22

Due to the high-efficiency of the excision process, it may be necessary to
titrate the supernatant to achieve single-colony isolation.
Colonies appearing on the plate contain the pCMV-Script EX doublestranded phagemid vector with the cloned DNA insert. Helper phage will
not grow, since helper phage is unable to replicate in Su
XLOLR strain and does not contain kanamycin-resistance genes.
XLOLR cells are also resistant to lambda phage infection, thus preventing
lambda phage contamination after excision.
To maintain the pCMV-Script EX phagemid vector, streak the colony on a
new LB–kanamycin agar plate. For long-term storage, prepare a bacterial
glycerol stock and store at –80°C.
R408 helper phage is recommended for the single-stranded rescue
procedure. The single-stranded rescue procedure can be found in
Recovery Of Single-Stranded DNA From Cells Containing the
pCMV-Script EX Phagemid Vector
Mass Excision Protocol
Day 1
1. Grow separate 50-ml overnight cultures of XL1-Blue MRF´ and
XLOLR cells in LB broth with supplements at 30°C.
–
(nonsuppressing)
Appendix:
.
Day 2
2. Gently spin down the XL1-Blue MRF´ and XLOLR cells (1000 × g).
Resuspend each of the cell pellets in 25 ml of 10 mM MgSO
the OD
cells to an OD
of the cell suspensions, then adjust the concentration of the
600
of 1.0 (8 × 108 cells/ml) in 10 mM MgSO4.
600
3. In a 50-ml conical tube, combine a portion of the amplified lambda
bacteriophage library with XL1-Blue MRF´ cells at a MOI of
1:10 lambda phage-to-cell ratio. Excise 10- to 100-fold more lambda
phage than the size of the primary library to ensure statistical
representation of the excised clones. Add ExAssist helper phage at a
10:1 helper phage-to-cells ratio to ensure that every cell is co-infected
with lambda phage and helper phage.
For example, use
7
10
pfu of the lambda phage (i.e., 10- to 100-fold above the
primary library size)
8
XL1-Blue MRF´ cells (1:10 lambda phage-to-cell ratio, noting
10
that an OD
9
10
pfu of ExAssist helper phage (10:1 helper phage-to-cells ratio)
of 1.0 corresponds to 8 × 108 cells/ml)
600
Note Briefly spin the lambda phage stock to ensure that the
chloroform is separated completely before removing the
aliquot used in the excision reaction.
. Measure
4
18 Lambda ZAP-CMV RI Predigested Vector Kit
Page 23

4. Incubate the conical tube at 37°C for 15 minutes to allow the phage to
attach to the cells.
5. Add 20 ml of LB broth with supplements and incubate the conical tube
for 2.5–3 hours at 37°C with shaking.
Notes Incubation times for mass excision in excess of 3 hours may
alter the clonal representation.
The turbidity of the media is not indicative of the success of
the excision.
6. Heat the conical tube at 65–70°C for 20 minutes to lyse the lambda
phage particles and the cells.
7. Spin the conical tube at 1000 ×
g for 10 minutes to pellet the cell debris
and then decant the supernatant into a sterile conical tube.
8. To titer the excised phagemids, combine 1 μl of this supernatant with
200 μl of XLOLR cells from step 2 in a 1.5-ml microcentrifuge tube.
9.
Incubate the microcentrifuge tube at 37°C for 15 minutes.
10. Add 40 μl of 5× NZY broth (for a final concentration of 1×) and
incubate the tube at 37°C for 45 minutes to allow sufficient expression
of the kanamycin-resistance gene product prior to plating on selective
medium.
11. Plate 100 μl of the cell mixture onto LB–kanamycin agar plates
(50 μg/ml) and incubate the plates overnight at 37°C.
Note It may be necessary to further dilute the cell mixture to
achieve single-colony isolation.
Colonies may now be selected for plasmid preps, or the cell mixture may be
plated directly onto filters for colony screening.
Lambda ZAP-CMV RI Predigested Vector Kit 19
Page 24

EUKARYOTIC SCREENING WITH THE LAMBDA ZAP-CMV RI LIBRARY
Screening libraries in eukaryotic cells has proved to be an effective way of
identifying clones otherwise nonidentifiable in prokaryotic screening
systems. The screening technique used will depend on the clone of interest
and on the type of assay available. An appropriate cell line for screening
must be obtained, and an assay or reagent capable of identifying the cell or
cells expressing the desired target protein must be developed. The panning
assay and functional analysis of clone pools are two available techniques.
Panning Assay
Clone identification by "panning" requires the transfection of a library into a
cell line deficient in the desired surface protein. When the clone of interest
is translated and expressed on the surface of eukaryotic cells, the translated
protein product is made accessible to an antibody, ligand, or receptor
coupled either directly or indirectly to a solid-phase matrix. Eukaryotic
transfectant clones expressing the appropriate insert will bind to the affinity
matrix, while cells not adhering are washed away. Either transient or stable
transfection protocols can be used.
Functional Assay
Functional assay screening can also be performed on either transiently or
stably transfected cells. Transient expression will likely require subdividing
the amplified library into smaller pools of clones to prevent the dilution of a
positive cell signal with an excess of negative clones. Each clone pool is
amplified separately and transfected into the eukaryotic cells. The
transfected cells are then tested for the expression of the desired clone. Once
a pool is identified as containing the clone of interest, it is subdivided into
smaller pools for a second round of prokaryotic amplification, eukaryotic
transfection, and screening. After several rounds of enriching for the desired
clone, a single clone can be isolated. The initial pool size is determined
according to the sensitivity of the available assay so that a single clone
within the pool is still theoretically detectable in the transfected cells. For
example, if a positive assay signal is 1000-fold above background, pools
containing 500–1000 members should still give a signal above background.
The sensitivity of the assay dictates the initial size of the pools, as well as
the number of pools required to screen. If stable transformants are created
using G418 selection, pools of stable clones can be assayed. This simplifies
the identification of isolated positive eukaryotic clones, because the
eukaryotic colonies can be picked or diluted in microtiter tissue culture
plates.
12
20 Lambda ZAP-CMV RI Predigested Vector Kit
Page 25

After a clone has been identified within the eukaryotic cells, the clone can
be retrieved by several methods. Plasmid DNA within the tissue culture
cells can be collected using the Hirt, Birnboim, and Doly procedures,
then transferred into
preparations. Simmons
E. coli cells for amplification and plasmid DNA
15
et al.
were able to screen libraries in COS cells,
13, 14
where the presence of the SV40 T antigen increases the copy number of
phagemids containing the SV40 origin of replication. This results in a higher
episomal copy number, which may help in the retrieval of the plasmids.
Inserts can also be isolated by polymerase chain reaction (PCR)
amplification of the tissue culture cells using T3/T7 primer sets. The
resulting PCR fragment can be digested using restriction sites flanking the
insert, then recloned into pCMV-Script EX phagemid DNA for further
analysis.
Note Screening libraries in eukaryotic cells can be extremely laborious.
Many functional assays are not sensitive enough to detect a clone
from pools of nonrelated clones. Therefore, it is worth considering
the use of techniques, such as differential PCR,
hybridization
17
and degenerate oligonucleotides, to develop DNA
16
selective
probes for initial screening using prokaryotic plaques. Positive
clones can then be screened by eukaryotic transfection and
expression.
EUKARYOTIC EXPRESSION
The CMV promoter is considered to be a strong promoter and to function in
many different cell lines.
sensitive to many factors. If little or no expression is observed in eukaryotic
cells, several factors can be considered.
♦ Methylation of some insert DNA can prevent expression in some cell
lines.
♦ The promoter may not be functional in some cell lines and should be
tested before screening a library.
♦ The pCMV-Script EX vector does not contain an ATG initiation codon.
A translation initiation sequence must be incorporated if the DNA
fragment to be cloned does not have an initiating ATG codon or an
optimal sequence for initiating translation, such as the Kozak sequence
[GCC(A/G)CCATGG].
18
However, expression in eukaryotic cells is
19
20
Lambda ZAP-CMV RI Predigested Vector Kit 21
Page 26

APPENDIX: RECOVERY OF SINGLE-STRANDED DNA FROM CELLS
CONTAINING THE PCMV-SCRIPT EX PHAGEMID VECTOR
The pCMV-Script EX vector is a phagemid that can be secreted as singlestranded DNA (ssDNA) in the presence of M13 helper phage. These
phagemids contain the intergenic (IG) region of a filamentous f1 phage.
This region encodes all of the
packaging and replication. In
F´ episome), pCMV-Script EX phagemid vectors will be secreted as singlestranded f1 "packaged" phage when the bacteria have been infected by a
helper phage. Because these filamentous helper phages (M13, f1) will not
infect
E. coli without an F´ episome coding for pili, it is essential to use the
XL1-Blue MRF´ strain or a similar strain containing the
F´ episome.
We offer helper phage that
phagemid vector. Typically, 30–50 pCMV-Script EX molecules are
packaged per helper phage DNA molecule. The pCMV-Script EX phagemid
vector is offered with the IG region in the minus orientation.
Yields of ssDNA can depend on the specific insert sequence, but for most
inserts >1 μg of ssDNA can be obtained from a 1.5-ml miniprep if grown in
XL1-Blue MRF´. A faint single-stranded helper phage band may appear on
a gel at ~4 kb for R408 helper phage. This DNA mixture can be sequenced
with primers that are specific for the pCMV-Script EX phagemid vectors
and do not hybridize to the helper phage genome.
R408 helper phage can be used to produce a large amount of single-stranded
pCMV-Script EX phagemid vector. Use the ExAssist interference-resistant
helper phage with XLOLR strain for the excision of the pCMV-Script EX
phagemid vector from the Lambda ZAP-CMV RI vector and use the R408
helper phage for single-stranded rescue.
21, 22
cis-acting functions of the phage required for
E. coli with the F
preferentially package the pCMV-Script EX
+
phenotype (containing an
22 Lambda ZAP-CMV RI Predigested Vector Kit
Page 27

Single-Stranded Rescue Protocol
1. Inoculate a single colony into 5 ml of 2× YT broth§ containing
50 μg/ml kanamycin and R408 helper phage at 10
(MOI ~10).
2. Grow the culture at 37°C with vigorous aeration for 16–24 hours, or
until growth has reached saturation.
3. Centrifuge 1.5 ml of the cell culture for 5 minutes in a microcentrifuge.
4. Remove 1 ml of the supernatant to a fresh tube, then add 150 μl of a
solution containing 20% PEG8000 and 2.5 M NaCl. Allow phage
particles to precipitate on ice for 15 minutes.
Note For increased yield, perform the PEG precipitation overnight
at 4°C.
5. Centrifuge for 5 minutes in a microcentrifuge. (A pellet should
be obvious.)
6. Remove supernatant. Centrifuge the PEG pellets a few seconds more to
collect residual liquid, then remove and discard the residual liquid.
7
–108 pfu/ml
7. Resuspend the pellet in 400 μl of 0.3 M NaOAc (pH 6.0) and
1 mM EDTA by vortexing vigorously.
8. Extract with 1 volume phenol–chloroform and centrifuge for
1–2 minutes to separate phases.
9. Transfer the aqueous phase to a fresh tube and add 1 ml of ethanol.
Centrifuge for 5 minutes.
10. Remove ethanol and dry the DNA pellet.
§
11. Dissolve the pellet in 25 μl of TE buffer.
12.
Analyze 1–2 μl on an agarose gel.
§
See Preparation of Media and Reagents.
Lambda ZAP-CMV RI Predigested Vector Kit 23
Page 28

TROUBLESHOOTING
Packaging
Observations Suggestions
Packaging efficiency is too low
Neither a bacterial lawn nor
plaques is observed on the plate
when titering or amplifying the
library
Excision
Observations Suggestions
The number of colonies is too low
Gigapack III packaging extract is very sensitive to slight variations in temperature;
therefore, store the packaging extracts at the bottom of a –80°C freezer and avoid
transferring tubes from one freezer to another
Do not allow the packaging extracts to thaw
Avoid use of ligase buffers containing PEG, which can inhibit packaging
Ensure the DNA concentration is sufficient. Ligate at DNA concentrations of
0.2 μg/μl or greater and package between 1 and 4 μl of the ligation reaction
Never package >4 μl of the ligation reaction, which causes dilution of the
proteins contained within the packaging extract
The lambda phage stock aliquot used when determining titer and amplifying the
library cannot contain chloroform, as chloroform lyses the E. coli cells. Briefly spin
the lambda phage stock to ensure that the chloroform is separated completely
before removing the aliquot
Verify that the titer on the tubes is current and correct and use only calibrated
pipettors. The molar ratios of lambda phage to cells to helper phage is critical
If an excision is unsuccessful, prepare a high-titer stock of the phage and repeat
the excision procedure, as excision efficiencies are directly related to the lambda
phage titer
Ensure that the platings are performed using agar plates containing kanamycin
The lambda phage stock aliquot used for in vivo excision cannot contain
chloroform, as chloroform lyses the E. coli cells. Briefly spin the lambda phage
stock to ensure that the chloroform is separated completely before removing the
aliquot
24 Lambda ZAP-CMV RI Predigested Vector Kit
Page 29

PREPARATION OF MEDIA AND REAGENTS
LB Agar (per Liter)
10 g of NaCl
10 g of tryptone
5 g of yeast extract
20 g of agar
Add deionized H
O to a final volume
2
of 1 liter
Adjust pH to 7.0 with 5 N NaOH
Autoclave
Pour into petri dishes
(~25 ml/100-mm plate)
LB Broth with Supplements
Prepare 1 liter of LB broth
Autoclave
Add the following filter-sterilized
supplements prior to use
10 ml of 1 M MgSO
4
3 ml of a 2 M maltose solution or 10 ml
of 20% (w/v) maltose
LB–Tetracycline Agar (per Liter)
1 liter of LB agar
Autoclave
Cool to 55°C
Add 1.5 ml of 10 mg/ml tetracycline (filter-
sterilized)
Pour into petri dishes (~25 ml/100-mm
plate)
Store plates in a dark, cool place or cover
plates with foil if left out at room
temperature for extended time periods
as tetracycline is light-sensitive
LB Broth (per Liter)
10 g of NaCl
10 g of tryptone
5 g of yeast extract
Add deionized H
O to a final volume
2
of 1 liter
Adjust to pH 7.0 with 5 N NaOH
Autoclave
LB–Kanamycin Agar (per Liter)
1 liter of LB agar
Autoclave
Cool to 55°C
Add 7.5 ml of 10 mg/ml kanamycin (filter-
sterilized)
Pour into petri dishes
(~25 ml/100-mm plate)
LB–Tetracycline Broth (per Liter)
Prepare 1 liter of LB broth
Autoclave
Cool to 55°C
Add 1.25 ml of 10 mg/ml-filter-sterilized
tetracycline
Store broth in a dark, cool place as
tetracycline is light-sensitive
NZY Agar (per Liter)
5 g of NaCl
2 g of MgSO
5 g of yeast extract
10 g of NZ amine (casein hydrolysate)
15 g of agar
Add deionized H
of 1 liter
Adjust the pH to 7.5 with NaOH
. 7H2O
4
O to a final volume
2
NZY Broth (per Liter)
5 g of NaCl
. 7H2O
4
O to a final volume
2
2 g of MgSO
5 g of yeast extract
10 g of NZ amine (casein hydrolysate)
Add deionized H
of 1 liter
Adjust the pH to 7.5 with NaOH
Autoclave
Autoclave
Pour into petri dishes
(~80 ml/150-mm plate)
Lambda ZAP-CMV RI Predigested Vector Kit 25
Page 30

NZY Top Agar (per Liter)
1 liter of NZY broth
Add 0.7% (w/v) agarose
Autoclave
SM Buffer (per Liter)
5.8 g of NaCl
2.0 g of MgSO
50.0 ml of 1 M Tris-HCl (pH 7.5)
5.0 ml of 2% (w/v) gelatin
Add deionized H
of 1 liter
· 7H2O
4
O to a final volume
2
20× SSC Buffer (per Liter)
175.3 g of NaCl
88.2 g of sodium citrate
800.0 ml of deionized H
O
2
10.0 N NaOH
Adjust to pH 7.0 with a few drops of
10.0 N NaOH
Add deionized H
of 1 liter
O to a final volume
2
TE Buffer
10 mM Tris-HCl (pH 7.5)
1 mM EDTA
2× YT Broth (per Liter)
10 g of NaCl
10 g of yeast extract
16 g of tryptone
Add deionized H
of 1 liter
Adjust to pH 7.5 with NaOH
Autoclave
O to a final volume
2
26 Lambda ZAP-CMV RI Predigested Vector Kit
Page 31

REFERENCES
1. Alting-Mees, M., Hoener, P., Ardourel, D., Sorge, J. and Short, J. M. (1992) Strategies
5(3):58-61.
2. Jerpseth, B., Greener, A., Short, J. M., Viola, J. and Kretz, P. L. (1992) Strategies
5(3):81–83.
3. Short, J. M. and Sorge, J. A. (1992) Methods Enzymol 216:495-508.
4. Short, J. M., Fernandez, J. M., Sorge, J. A. and Huse, W. D. (1988) Nucleic Acids Res
16(15):7583-600.
5. Sambrook, J., Fritsch, E. F. and Maniatis, T. (1989). Molecular Cloning: A Laboratory
Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
6. Kretz, P. L., Danylchuk, T., Hareld, W., Wells, S., Provost, G. S. et al. (1994)
Strategies 7(2):44-45.
7. Kohler, S. W., Provost, G. S., Kretz, P. L., Dycaico, M. J., Sorge, J. A. et al. (1990)
Nucleic Acids Res 18(10):3007-13.
8. Kretz, P. L., Kohler, S. W. and Short, J. M. (1991) J Bacteriol 173(15):4707-16.
9. Kretz, P. L., Reid, C. H., Greener, A. and Short, J. M. (1989) Nucleic Acids Res
17(13):5409.
10. Ausubel, F. M., Brent, R., Kingston, R. E., Moore, D. D., Seidman, J. G. et al. (1987).
Current Protocols in Molecular Biology. John Wiley and Sons, New York.
11. Dotto, G. P., Horiuchi, K. and Zinder, N. D. (1984) J Mol Biol 172(4):507-21.
12. Seed, B. and Aruffo, A. (1987) Proc Natl Acad Sci U S A 84(10):3365-9.
13. Hirt, B. (1967) J Mol Biol 26(2):365-9.
14. Birnboim, H. C. and Doly, J. (1979) Nucleic Acids Res 7(6):1513-23.
15. Simmons, D. L., Satterthwaite, A. B., Tenen, D. G. and Seed, B. (1992) J Immunol
148(1):267-71.
16. Liang, P. and Pardee, A. B. (1992) Science 257(5072):967-71.
17. Lee, S. W., Tomasetto, C. and Sager, R. (1991) Proc Natl Acad Sci U S A
88(7):2825-9.
18. Foecking, M. K. and Hofstetter, H. (1986) Gene 45(1):101-5.
19. MacGregor, G. R., Mogg, A. E., Burke, J. F. and Caskey, C. T. (1987) Somat Cell Mol
Genet 13(3):253-65.
20. Kozak, M. (1986) Cell 44(2):283-92.
21. Dente, L., Cesareni, G. and Cortese, R. (1983) Nucleic Acids Res 11(6):1645-55.
22. Mead, D. A., Skorupa, E. S. and Kemper, B. (1985) Nucleic Acids Res 13(4):1103-18.
ENDNOTES
Falcon® is a registered trademark of Becton Dickinson and Company.
Parafilm
Pyrex
Whatman
®
is a registered trademark of American National Can.
®
is a registered trademark of Corning Glass Works.
®
is a registered trademark of Whatman Ltd.
MSDS INFORMATION
The Material Safety Data Sheet (MSDS) information for Stratagene products is provided on the web at
http://www.stratagene.com/MSDS/. Simply enter the catalog number to retrieve any associated MSDS’s
in a print-ready format. MSDS documents are not included with product shipments.
Lambda ZAP-CMV RI Predigested Vector Kit 27
 Loading...
Loading...