
PathDetect Reporter Cell Lines
INSTRUCTION MANUAL
Catalog #800050 (PathDetect HLR Cell Line)
#800055 (HLR-Elk1 Cell Line)
#800060 (HLR-CHOP Cell Line)
#800065 (HLR-CREB Cell Line)
Revision A
BN #800050-12
For In Vitro Use Only
800050-12
LIMITED PRODUCT WARRANTY
This warranty limits our liability to replacement of this product. No other warranties of any kind,
express or implied, including without limitation, implied warranties of merchantability or fitness for
a particular purpose, are provided by Agilent. Agilent shall have no liability for any direct, indirect,
consequential, or incidental damages arising out of the use, the results of use, or the inability to use
this product.
ORDERING INFORMATION AND TECHNICAL SERVICES
United States and Canada
Agilent Technologies
Stratagene Products Division
11011 North Torrey Pines Road
La Jolla, CA 92037
Telephone (858) 373-6300
Order Toll Free (800) 424-5444
Technical Services
Internet
World Wide Web
techservices@agilent.com
Europe
Location Telephone Fax Technical Services
Austria 0800 292 499 0800 292 496 0800 292 498
(800) 894-1304
www.stratagene.com
00800 7000 7000 00800 7001 7001 00800 7400 7400 Belgium
0800 15775 0800 15740 0800 15720
00800 7000 7000 00800 7001 7001 00800 7400 7400 France
0800 919 288 0800 919 287 0800 919 289
00800 7000 7000 00800 7001 7001 00800 7400 7400 Germany
0800 182 8232 0800 182 8231 0800 182 8234
00800 7000 7000 00800 7001 7001 00800 7400 7400 Netherlands
0800 023 0446 +31 (0)20 312 5700 0800 023 0448
00800 7000 7000 00800 7001 7001 00800 7400 7400 Switzerland
0800 563 080 0800 563 082 0800 563 081
00800 7000 7000 00800 7001 7001 00800 7400 7400 United Kingdom
0800 917 3282 0800 917 3283 0800 917 3281
All Other Countries
Please contact your local distributor. A complete list of distributors is available at www.stratagene.com.

PathDetect Reporter Cell Lines
CONTENTS
Materials Provided.............................................................................................................................. 1
Storage Conditions.............................................................................................................................. 1
Additional Materials Required .......................................................................................................... 2
Safety Consideration........................................................................................................................... 2
Notices to Purchaser ........................................................................................................................... 2
Introduction......................................................................................................................................... 3
Preprotocol Considerations................................................................................................................ 5
Choosing a Transfection Method .......................................................................................... 5
Tissue Cultureware................................................................................................................ 5
Designing the Experiment ..................................................................................................... 5
Cell Culture Protocols......................................................................................................................... 7
Starting the Cells in Culture .................................................................................................. 7
Passaging the Cells................................................................................................................ 7
Freezing the Cells for Long-term Storage ............................................................................. 8
Preparing the Cells for Transfection................................................................................................. 9
Preparing Plasmids for Transfection ................................................................................................9
Studying the Effects of a Gene Product................................................................................. 9
Transfecting the Cells ......................................................................................................................... 9
Preparing Cell Lysates for Luciferase Activity Assay ................................................................... 10
Performing the Luciferase Activity Assay ...................................................................................... 10
Troubleshooting ................................................................................................................................ 11
Appendix I: Plasmid Information.................................................................................................... 12
The pFA trans-Activator Plasmids...................................................................................... 12
Preparation of Media and Reagents................................................................................................ 13
References .......................................................................................................................................... 14
Endnotes............................................................................................................................................. 14
MSDS Information............................................................................................................................ 14
Quick-Reference Protocol ................................................................................................................ 15


PathDetect Reporter Cell Lines
ATERIALS PROVIDED
M
Catalog
#800050
HLR Cell Line — 1 × 106 viable
pFA2-CREB fusion trans-activator plasmid 25 ng/μl 5 μg
pFC-PKA plasmid (positive control) 25 ng/μl 5 μg
pFC-CMV negative control plasmid 25 ng/μl 5 μg
Catalog
#800055
HLR-Elk1 trans-Reporting Cell Line — 1 × 106 viable
pFC-CMV negative control plasmid 25 ng/μl 5 μg
pFC-MEK1 plasmid (positive control) 25 ng/μl 5 μg
Catalog
#800060
HLR-CHOP trans-Reporting Cell Line — 1 × 106 viable
pFC-CMV negative control plasmid 25 ng/μl 5 μg
pFC-MEK3 plasmid (positive control) 25 ng/μl 5 μg
Catalog
#800065
HLR-CREB trans-Reporting Cell Line — 1 × 106 viable
pFC-CMV negative control plasmid 25 ng/μl 5 μg
pFC-PKA plasmid (positive control) 25 ng/μl 5 μg
Component Concentration Quantity
cells
Component Concentration Quantity
cells
Component Concentration Quantity
cells
Component Concentration Quantity
cells
STORAGE CONDITIONS
Control Plasmids: –20°C
Cell lines: Liquid Nitrogen
Revision A © Agilent Technologies, Inc. 2008.
PathDetect Reporter Cell Lines 1

ADDITIONAL MATERIALS REQUIRED
Growth medium (See Preparation of Media and Reagents)
Luciferase assay kit
5-ml BD Falcon polystyrene round-bottom tubes (BD Biosciences Catalog #352054)
Calcium- and magnesium-free PBS
Tissue culture plasticware
Transfection reagents
Luminometer
Hygromycin
G418 Sulfate
SAFETY CONSIDERATION
The American Type Culture Collection (ATCC) has designated CCL-2 HeLa cell lines as Biosafety
Level 2. Appropriate caution should be used while handling any human-derived cell line. For more
information, see Biosafety in Microbiological and Biomedical Laboratories (4th edition). It is
available on the Web sites of both the National Institutes of Health at http://bmbl.od.nih.gov and the
Office of Health and Safety of the Centers for Disease Control and Prevention at
www.cdc.gov/od/ohs (Click the link to Biosafety Information).
NOTICES TO PURCHASER
The use of the CMV Promoter is covered under U.S. Patent Nos. 5,168,062 and 5,385,839 owned by
the University of Iowa Research Foundation and licensed FOR RESEARCH USE ONLY. For
further information, please contact UIRF at 319-335-4546.
A license (from Promega for research reagent products and from The Regents of the University of
California for all other fields) is needed for any commercial sale of nucleic acid contained within or
derived from this product.
The HLR, HLR-cJun, HLR-Elk1, HLR-CHOP, and HLR-CREB cell lines are proprietary to Agilent.
Purchase of these cell lines includes a limited non-exclusive license under Agilent’s proprietary
rights to use these cell lines for research purposes only. This license does not grant any rights to: (i)
modify any of these cell lines; (ii) offer any of these cell lines, or any derivatives or modifications
thereof, for resale; or (iii) otherwise distribute or transfer any of these cell lines, or any derivative or
modification thereof, to third parties for any purpose or use. No other license, express, implied or by
estoppel, is granted.
2 PathDetect Reporter Cell Lines
INTRODUCTION
The Stratagene PathDetect reporter cell lines are designed for specific,
rapid, and convenient analysis of the in vivo activation of transcriptional
activators and upstream signal transduction pathways.
1,2
Two types of stable
reporter cell lines have been developed for use with the PathDetect system
using ATCC, CCL-2 HeLa cells. The first type is the HeLa Luciferase
Reporter (HLR) cell line, which contains a stably integrated luciferase
reporter gene. The second type includes three HLR trans-reporting cell
lines, which contain both the luciferase reporter gene and the gene for a
trans-acting GAL4 fusion protein (Elk1, CHOP, or CREB). The cell lines
are useful for examining whether and where a newly cloned gene is
involved in a signal transduction pathway or to study the effects of growth
factors, drug candidates, and extracellular stimuli on signal transduction
pathways converging at these transcription factors.
The PathDetect HeLa Luciferase Reporter (HLR) cell line contains an
integrated synthetic promoter consisting of five direct repeats of the yeast
GAL4 binding site that controls expression of the luciferase gene from
Photinus pyralis [American firefly (see Appendix I: Plasmid Information)].
The HLR cell line also contains an expression cassette for hygromycinresistance. To assay for involvement of a gene in a particular pathway, a
plasmid encoding a fusion trans-activator protein consisting of the GAL4
DNA binding domain (DBD) fused to a target activation domain for the
gene of interest is cotransfected into cells with an expression vector
containing the gene of interest. If the gene of interest directly or indirectly
phosphorylates the activation domain of the fusion trans-activator protein,
this will activate transcription of luciferase. Expression (or activity) levels of
luciferase reflect the activation status of the signaling events. The HLR cell
line can also serve as the reporter cell line for many other applications using
the GAL4 DNA binding domain, such as testing protein-protein interactions
with mammalian two-hybrid vectors.
Each PathDetect HLR trans-reporting cell line contains the luciferase
reporter cassette found in the HLR cell line and also expresses a unique,
stably integrated, trans-acting fusion protein. Each fusion protein consists of
the activation domain of one of three transcriptional activators: Elk1,
CREB,
1–147).
6,7
or CHOP,
9,10
The transcriptional activator domains of Elk1, CHOP, and CREB
2,8
that are fused to the yeast GAL4 DBD (residues
3–6
are activated upon phosphorylation by mitogen-activated protein kinase
(MAPK), p38 MAP kinase, or cyclic AMP-dependent kinase (PKA),
respectively. Transcriptional activation of the reporter in these cell lines thus
reflects in vivo activation of these kinases and their corresponding signal
transduction pathways. Constitutive expression of the trans-acting fusion
genes in the PathDetect cell lines is driven by the human cytomegalovirus
(CMV) immediate early promoter. The trans-reporting cell lines are
resistant to both G418 and hygromycin.
PathDetect Reporter Cell Lines 3
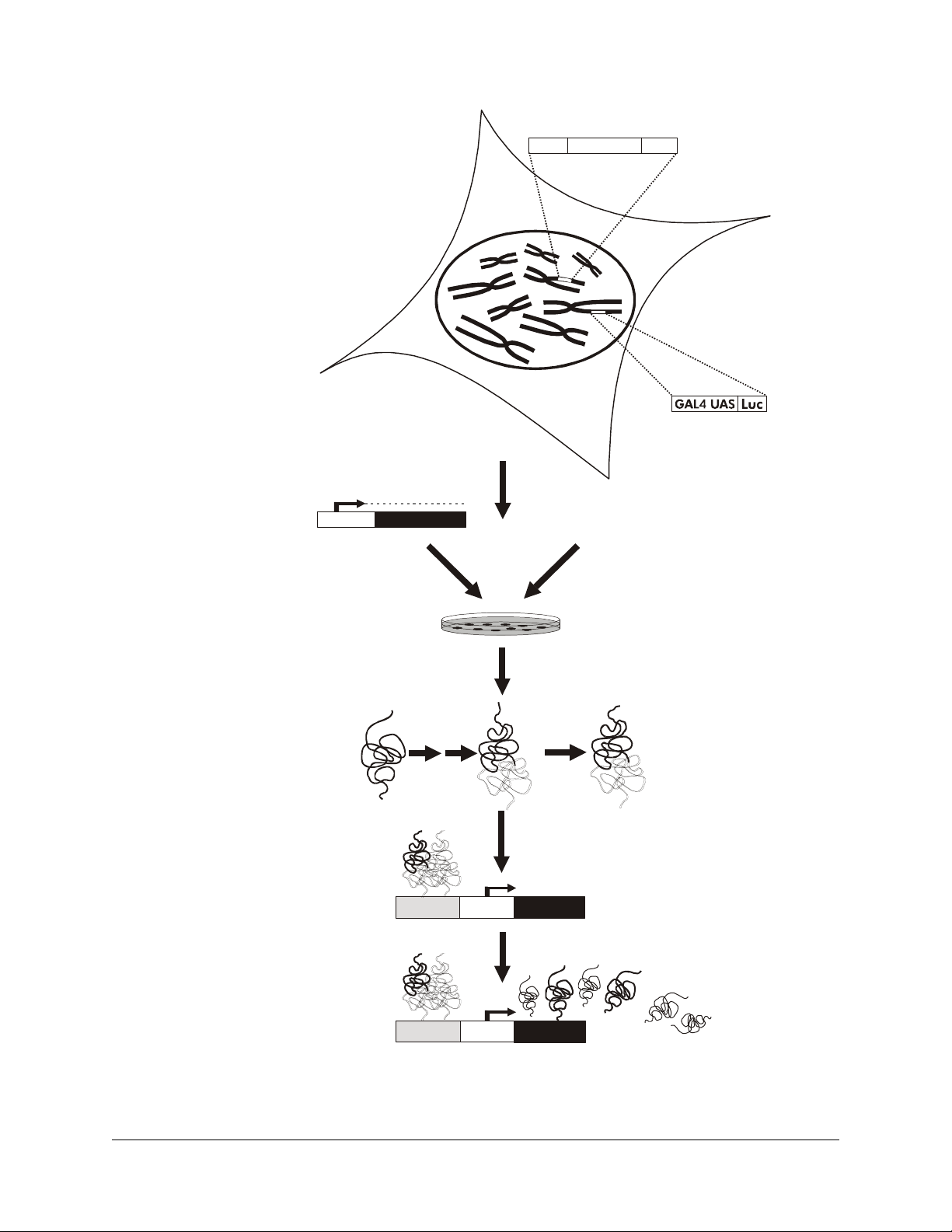
HLR double-stable cell
Double-stable HLR cell lines
have both the pFR-Luc plasmid
and either the pFA2-Elk1, pFA2-CREB, or
pFA-CHOP, fusion -activator
plasmid stably integrated into the genome
trans
Transfection/Extracellular stimulus
Fusion -activator expression cassette
trans
CMV GAL4 DBD
AD
Luciferase expression cassette
Pro moter
Gene of interest
Cells are transfected with a gene of interest
or treated with an extracellular stimulus
Protein
of interest
Pathway-specific fusion
-activator protein
trans
Assay for luciferase
or
Activation
domain
GAL4
dbd
P
O
4
P
O
4
GAL4 UAS TATATA
P
O
4
P
O
4
Extracellular stimulus
(Growth factors,
drug candidates,
signaling molecules)
H
O
Phosphorylated pathway-specific
fusion -activator protein binds as a
trans
dimer with GAL4 UAS and activates
transcription of luciferase
P
O
4
Luciferase
TATATAGAL4 UAS
Luciferase
Figure 1 Schematic diagram for use of the PathDetect double-stable trans-reporting cell lines.
4 PathDetect Reporter Cell Lines

PREPROTOCOL CONSIDERATIONS
Choosing a Transfection Method
As with all transfection assays, the sensitivity of an assay using a PathDetect
signal transduction pathway stable reporter cell line is greatly influenced by
the transfection efficiency. A high transfection efficiency generally provides
a more sensitive assay that requires a smaller volume of sample.
Transfection conditions should be optimized with the provided positive
control plasmid before performing the assays. Sufficient amounts of control
plasmids are included for several optimization experiments. Because the
luciferase assay is very sensitive, various transfection methods, such as
calcium phosphate precipitation and lipid-mediated transfection, may be
used.
Tissue Cultureware
The protocols given are based on 24-well tissue culture dishes with a well
diameter of ~15 mm and a surface area of ~1.88 cm
Designing the Experiment
Studying the Effects of a Gene Product using HLR trans-
reporting Cell Lines
The Elk1, CHOP, and CREB reporter cell lines include known upstream
activators that can be used as positive controls for transactivation. The
experimental plasmid minus the gene of interest can be used as a negative
control to ensure that the introduction of viral promoters (e.g., CMV, RSV,
or SV40) or other proteins expressed from the plasmid does not activate the
fusion trans-activator protein. Depending on the purpose of the experiment,
other controls such as a nonactivatable mutant of the fusion trans-activator
protein might be required.
Typical initial experimental parameters for studying the effects of a gene
product using the PathDetect trans-reporting cell lines are outlined in
Table I. As all assays are to be run in triplicate, eight samples will utilize
one 24-well tissue culture dish. Sample numbers are indicated in Column A.
2
.
PathDetect Reporter Cell Lines 5

TABLE I
Sample Experiment to Study the Effects of a Gene Product
A B C D E F
Negative
#
control plasmid Positive control
1a — — — 10 ng 240 ng
2b — — — 20 ng 230 ng
3c — — — 100 ng 150 ng
4d — — 10 ng — 240 ng
5e — — 20 ng — 230 ng
6f — — 100 ng — 150 ng
7g —
25 ng (1 μ
8h
a
Sample 1 lacks the gene of interest and, therefore, controls for sample 4.
b
Sample 2 lacks the gene of interest and, therefore, controls for sample 5.
c
Sample 3 lacks the gene of interest and, therefore, controls for sample 6.
d
Sample 4 measures the effect of the gene product on the signal transduction pathway involved.
e
Sample 5 measures the effect of the gene product on the signal transduction pathway involved.
f
Sample 6 measures the effect of the gene product on the signal transduction pathway involved.
g
Sample 7 measures the efficacy of the assay for the cell line chosen.
h
Sample 8 does not contain an activation domain and should show results similar to samples 1–3.
l)
25 ng (1 μ
— — — 225 ng
l)
Experimental plasmid
with gene of interest
— — 225 ng
Experimental plasmid
without insert
Carrier DNA
Studying the Effects of Extracellular Stimuli using HLR
trans-reporting Cell Lines
The PathDetect trans-reporting cell lines may be used to study the effects of
extracellular stimuli, such as growth factors or drug candidates, on
corresponding signal transduction pathways (see Table II). To perform such
assays, cells are first treated with the compound of interest. Luciferase
expression from the reporter indicates activation of the fusion transactivator protein and, therefore, the presence of the endogenous protein
kinase (e.g., MAPK, p38 MAPK, PKA, or an uncharacterized upstream
activator).
ABLE II
T
Sample Experiment to Study the Effects of Extracellular Stimuli
# Extracellular stimulus
1 Serum (10%)
2 PMA (10 ng/ml)
3 PMA (60 ng/ml)
4 EGF (100 ng/ml)
5 AC Toxin (10 μg/ml)
6 Medium (negative control)
6 PathDetect Reporter Cell Lines

CELL CULTURE PROTOCOLS
Starting the Cells in Culture
Note All procedures from this point should be performed using
1. Prepare the growth medium
hygromycin (100 μg/ml), depending on cell type, and transfer 10 ml to
a 15-ml conical tube.
2. Thaw the frozen cryovial of cells within 40–60 seconds by rapid
agitation in a 37°C water bath. Remove the cryovial from the water and
immediately immerse the cryovial in 70% (v/v) ethanol at room
temperature.
3. Transfer the cell suspension to the conical tube containing the growth
medium.
4. Centrifuge the cells at 200 × g for 5 minutes at room temperature and
remove the growth medium from the conical tube by aspiration.
5. Resuspend the cells in the conical tube in 5 ml of growth medium
containing the appropriate selective agent.
sterile technique.
§
with either G418 (250 μg/ml) and/or
6. Add 10 ml of growth medium containing the appropriate selective
Passaging the Cells
Note Split the cells once the cell monolayer is ~80–90% confluent or
1. Remove the medium by aspiration, rinse the cells with 1× PBS,
2. Add 1.5 ml of trypsin–EDTA
§
See Preparation of Media and Reagents.
2
agent to a 75-cm
the tissue culture flask. Place the cells in a 37°C incubator at 5% CO
tissue culture flask. Transfer the cell suspension to
.
2
approximately every 3–5 days using the following protocol.
§
and
remove the PBS by aspiration.
§
to each 75-cm2 tissue culture flask.
Incubate the cells at room temperature for ~2–5 minutes. Tap the flask
to release any adherent cells.
Note Incubate the cells in the trypsin–EDTA for the minimum time
necessary for the adherent cells to release from the flask.
Overtrypsinization can kill the cells.
PathDetect Reporter Cell Lines 7

3. If using HLR cells, perform step 3a. If using double stable cells,
perform step 3b.
a. Dilute the cells with growth medium containing hygromycin
(100 μg/ml) to a final volume of 5–10 ml.
b. Add growth medium containing G418 (250 μg/ml) and
hygromycin (100 μg/ml) to a final volume of 5–10 ml.
4. The serum in the medium will inactivate the trypsin. Transfer the cells
to the desired number of 75-cm
Depending on the experiment, use an inoculum of
4
1 × 10
–1 × 106 cells. Add growth medium to the flasks to a final
volume of 15 ml for 75-cm
30 ml for 175-cm
incubator at 5% CO
2
tissue culture flasks. Place the cells in a 37°C
.
2
Freezing the Cells for Long-term Storage
1. Remove the growth medium by aspiration, rinse the cells with 1× PBS,
and remove the PBS by aspiration.
2. Add 1.5 ml of trypsin–EDTA to each 75-cm
Incubate the cells at room temperature for ~2–5 minutes. Tap the flask
to release the adherent cells.
Note Incubate the cells in the trypsin–EDTA for the minimum time
necessary for the adherent cells to release from the flask.
Overtrypsinization can kill the cells.
3. Dilute the cells in 8.5 ml of growth medium. The serum in the medium
inactivates the trypsin.
2
or 175-cm2 tissue culture flasks.
2
tissue culture flasks or to a final volume of
2
tissue culture flask.
4. Transfer the cell suspension to a 15-ml conical tube. Centrifuge the
cells at 200 × g for 5 minutes at room temperature. Remove the growth
medium from the tube by aspiration.
5. Resuspend the cells in 1–2 ml of freezing medium.
§
6. Transfer the cell suspension to cryovials and place the cryovials in a
chilled controlled-freezing container. Incubate the cryovials at –80°C
overnight.
7. The following day, transfer the cryovials to liquid nitrogen for longterm storage. If properly stored, the cells should remain stable for
>1 year.
§
See Preparation of Media and Reagents.
8 PathDetect Reporter Cell Lines

PREPARING THE CELLS FOR TRANSFECTION
1. Seed 5 × 10
each well of a 24-well tissue culture dish.
4
cells in 1 ml of growth medium without selective agents in
2. Incubate the cells at 37°C in a 5% CO
PREPARING PLASMIDS FOR TRANSFECTION
Note The DNA used for transfections must be of high quality (i.e.,
double cesium chloride banded). Ensure that the plasmid DNA has
an OD
of ~1.8–2.0 and is endotoxin free.
260/280
Studying the Effects of a Gene Product
Prepare plasmid DNA mixtures in sterile BD Falcon polystyrene tubes as
indicated by samples 1–8 in Table I. As each assay is run in triplicate, the
amount of plasmid DNA in each tube should be sufficient for four
transfections. See Table I for the appropriate amounts. For example, to
prepare sample #1 in triplicate as indicated in Table I, combine the
following components in a microcentrifuge tube and then proceed to
Transfecting the Cells:
30 ng of experimental plasmid without an insert
720 μg of unrelated plasmid DNA
TRANSFECTING THE CELLS
incubator for 24 hours.
2
A number of transfection methods, including calcium phosphate
precipitation and lipid-mediated transfection may be used. Transfection
efficiencies vary between cell lines and according to experimental
conditions. Transfection procedures should be optimized for the cell line
chosen.
Notes Due to the possible induction of pathways by unknown factors in
the serum, use low serum concentrations; however, the use of 10%
serum has also yielded satisfactory results in some cases.
If studying the effects of extracellular stimuli, replace the medium
with fresh medium containing the appropriate extracellular stimuli
(e.g., EGF) 18–24 hours after the beginning of transfection. After
incubating an additional 5–7 hours, proceed to Preparing Cell
Lysates for Luciferase Activity Assay.
PathDetect Reporter Cell Lines 9

PREPARING CELL LYSATES FOR LUCIFERASE ACTIVITY ASSAY
1. Remove the medium from the cells and carefully wash the cells twice
with 2 ml of 1× PBS buffer.
2. Remove as much 1× PBS as possible from the wells with a Pasteur
pipet. Add 100 μl of 1× cell lysis buffer
dishes gently to ensure uniform coverage of the cells.
3. Incubate the dishes for 15 minutes at room temperature. Swirl the
dishes gently midway through the incubation.
4. Assay for luciferase activity directly from the wells within 2 hours.
5. To store for later analysis, transfer the solutions from each well into a
separate microcentrifuge tube. Spin the samples in a microcentrifuge at
12,000 × g for 5 minutes at 4°C. Remove the supernatant and store at
–80°C. Each freeze–thaw cycle results in a significant loss of
luciferase activity (as much as 50%).
Note If this passive lysis method does not yield satisfactory results,
refer to the instructions for an active lysis method in
Troubleshooting.
§
to the wells and swirl the
PERFORMING THE LUCIFERASE ACTIVITY ASSAY
1. Mix 5–20 μl of cell extract equilibrated to room temperature with
100 μl of room temperature 1× luciferase assay reagent
BD Falcon polystyrene round-bottom tube.
2. Measure the light emitted from the reaction with a luminometer using
an integration time of 10–30 seconds.
3. Luciferase activity may be expressed in relative light units (RLU) as
detected by the luminometer from the sample. The activity may also be
expressed as RLU/well, RLU/number of cells, or RLU/mg of total
cellular protein.
§
See Preparation of Media and Reagents.
§
in a 5-ml
10 PathDetect Reporter Cell Lines

TROUBLESHOOTING
Observation Suggestions
Cells do not survive resuspension in
growth medium following storage in
liquid nitrogen
Cells do not survive passaging
The background luciferase activity is
too low to calculate
Results vary among triplicate samples Variations are occurring in pipetting, growth conditions, extraction efficiency of
All samples exhibit very low or no
luciferase activity
Cells are not stored properly. Store the cryovials of cells in liquid nitrogen
immediately on receipt
Cells are not thawed properly. Thaw the cells quickly and immediately dilute the
cells in the growth medium
Cells are over-diluted during passaging. When passaging, inoculate a fresh
tissue culture flask with 1 × 10
Cells are over-tyrpsinized during passaging. Incubate the cells with trypsin–EDTA
for the minimum time necessary for the cells to release from the flask
Split the cells when the monolayer of cells is at most 90% confluent. Cells grown
to 100% confluence are starved for growth medium
The mammalian transcription activators are binding to the GAL4 UAS inefficiently
causing the expression of the luciferase gene to be low. Increase the
concentration of cell lysate used in the assay
Plot and compare the absolute luciferase activity rather than the activation
fold increase
luciferase, etc. Use the same DNA–transfection reagent mixture for the three
wells. Take care when washing the cells to avoid removing the cells from the
wells
Passive cell lysis is not effective. Perform the following active lysis. Scrape all
surfaces of the tissue culture dish, pipet the cell lysate to microcentrifuge tube
and place on ice. Lyse the cells by brief sonication with the microtip set at the
lowest setting or freeze the cells at –80°C for 20 minutes and then thaw in a
37°C water bath and vortex 10–15 seconds. Spin the tubes in a microcentrifuge
at high speed for 2 minutes. Use the supernatant for the luciferase activity assay
Transfection is not successful. Optimize the transfection procedure with a reporter
plasmid such as pCMV-βGAL
4
–1 × 106 cells
PathDetect Reporter Cell Lines 11

APPENDIX I: PLASMID INFORMATION
r
Luciferase Reporter Cassette
5x GAL4
binding element
TATATA
Luciferase
Sequence of GAL4 Binding Element in the pFR-Luc Plasmid
AT CTTATCATGTCTGGATC CA AGCTTGCAT GCCTGCAG
GT AG CGGAGTACTGTC CTCCG CGGAGTACTGTC CTCCG
AG AG CGGAGTACTGTC CTCCG CGGAGTACTGTC CTCCG
AG AG CGGAGACTC TAGAGGGCGGAGTACTGTC CTCCG
TATATAATGGATCCC CGGGT AC CGAGCTCGAAT TC...
...CAGCTTGGCATTCCGGTACTGTTGGTAAAA TG
Fusion -Activator Cassettes
pFA2-Elk1
pFA2-CREB
pFA-CHOP
trans
CMV Promoter GAL4 DNA BD (1–147) Elk1 (307–427)
CMV Promoter CREB (1–280)
CMV Promoter CHOP (1–101)
GAL4 DNA BD (1–147)
GAL4 DNA BD (1–147)
Luciferase
5× GAL4
Binding Element
The pFA trans-Activator Plasmids
pUC ori
TK pA
neo/kan
pFA trans-Activato
Plasmids
P SV40
12 PathDetect Reporter Cell Lines
P CMV
GAL4-BD
trans-activator
f1 ori
P bla
PREPARATION OF MEDIA AND REAGENTS
Cell Lysis Buffer (5×)
40 mM tricine (pH 7.8)
50 mM NaCl
2 mM EDTA
1 mM MgSO
4
5 mM DTT
®
1% Triton
X-100
Growth Medium
500 ml of Dulbecco’s Modified Eagle
Medium (DMEM) (high glucose,
without
L-glutamine, without sodium
pyruvate)
5 ml of 200 mM
L-glutamine
5 ml of penicillin (5000 U/ml)–
streptomycin (5000 μg/ml) mixture
50 ml of fetal bovine serum, heat
inactivated
Freezing Medium
Dulbecco’s Modified Eagle Medium
(DMEM) (high glucose, without
glutamine, without sodium pyruvate)
20% (v/v) fetal bovine serum
10% (v/v) dimethylsulfoxide (DMSO)
L-
Phosphate-buffered Saline (1× PBS)
137 mM NaCl
2.6 mM KCl
10 mM Na
1.8 mM KH2PO
HPO
2
4
4
Adjust the pH to 7.4 with HCl
Filter sterilize
Luciferase Assay Reagent (1×)
40.0 mM tricine (pH 7.8)
0.5 mM ATP
10 mM MgSO
4
0.5 mM EDTA
10.0 mM DTT
0.5 mM coenzyme A
0.5 mM luciferin
Trypsin–EDTA
0.53 mM tetrasodium
ethylenediaminetetraacetic acid
0.05% trypsin
PathDetect Reporter Cell Lines 13

REFERENCES
1. Xu, L., Chau, F., Sanchez, T., Buchanan, M. and Zheng, C.-F. (2001) Strategies
14(1):17–19.
2. Xu, L., Sanchez, T., Buchanan, M. and Zheng, C.-F. (1998) Strategies 11(3):94–97.
3. Marais, R., Wynne, J. and Treisman, R. (1993) Cell 73(2):381-93.
4. Price, M. A., Rogers, A. E. and Treisman, R. (1995) Embo J 14(11):2589-601.
5. Rao, V. N., Huebner, K., Isobe, M., ar-Rushdi, A., Croce, C. M. et al. (1989) Science
244(4900):66-70.
6. Xu, L., Sanchez, T. and Zheng, C.-F. (1997) Strategies 10(1):1–3.
7. Gonzalez, G. A., Yamamoto, K. K., Fischer, W. H., Karr, D., Menzel, P. et al. (1989)
Nature 337(6209):749-52.
8. Wang, X. Z. and Ron, D. (1996) Science 272(5266):1347-9.
9. Laughon, A. and Gesteland, R. F. (1984) Mol Cell Biol 4(2):260-7.
10.Sadowski, I. and Ptashne, M. (1989) Nucleic Acids Res 17(18):7539.
ENDNOTES
Triton® is a registered trademark of Union Carbide Chemicals and Plastics Co., Inc.
MSDS INFORMATION
The Material Safety Data Sheet (MSDS) information for Stratagene products is provided on the web at
http://www.stratagene.com/MSDS/. Simply enter the catalog number to retrieve any associated MSDS’s
in a print-ready format. MSDS documents are not included with product shipments.
14 PathDetect Reporter Cell Lines

PathDetect Cell Lines
Catalog #800050, #800055, #800060, and #800065
QUICK-REFERENCE PROTOCOL
Starting the Cells in Culture
♦ Transfer 10 ml of growth medium to a 15-ml conical tube
♦ Thaw the frozen cells quickly in a 37°C water bath
♦ Transfer the cell suspension to the conical tube containing the growth medium
♦ Centrifuge the cells at 200 × g for 5 minutes at room temperature and remove the
growth medium by aspiration
a. For the HLR cell line, add 10 ml of growth medium containing hygromycin
(100 μg/ml) to a 75-cm
b. For HLR trans-reporting cell lines, add 10 ml of growth medium containing
G418 (250
μg/ml) and hygromycin (100 μg/ml) to a 75-cm
♦ Resuspend the cells in the conical tube in 5 ml of growth medium containing the
appropriate selective agent(s), and transfer the cell suspension to the tissue culture flask
2
tissue culture flask
2
tissue culture flask
♦ Place the cells in a 37°C incubator with 5% CO
2
Passaging the Cells
♦ Remove the growth medium, rinse the cells with 1× PBS, and remove the PBS by
aspiration
♦ Add 1.5 ml of trypsin–EDTA to each 75-cm
room temperature for ~2–5 minutes, and tap the flask to release the adherent cells
♦ Dilute the cells with the growth medium containing hygromycin (100mg /ml) and, for
double stables, G418 (250
μg/ml) and transfer the cells to the desired number of
tissue culture flasks
♦ Place the cells in a 37°C incubator with 5% CO
2
tissue culture flask, incubate the cells at
2

Freezing the Cells for Long-term Storage
♦ Remove the growth medium, rinse the cells with 1× PBS, and remove the PBS by
aspiration
♦ Add 1.5 ml of trypsin–EDTA to each 75-cm
2
tissue culture flask containing cells,
incubate the cells at room temperature for ~2–5 minutes, and tap the flask to release
the adherent cells
♦ Dilute the cells with 8.5 ml of growth medium
♦ Transfer the cell suspension to a 15-ml conical tube, centrifuge at 200 × g for
5 minutes at room temperature, and remove the growth medium by aspiration
♦ Resuspend the cells in 1–2 ml of freezing medium
♦ Transfer the cells to cryovials, place the cryovials in a chilled controlled-freezing
container, and incubate the cryovials at –80°C overnight
♦ The following day, transfer the cryovials to liquid nitrogen for long-term storage
16
 Loading...
Loading...