Page 1

Design of Experiments
Guide
Page 2
Get the Most from JMP
Whether you are a first-time or a long-time user, there is always something to learn about JMP.
Visit JMP.com and to find the following:
• live and recorded Webcasts about how to get started with JMP
• video demos and Webcasts of new features and advanced techniques
• schedules for seminars being held in your area
• success stories showing how others use JMP
• a blog with tips, tricks, and stories from JMP staff
• a forum to discuss JMP with other users
®
http://www.jmp.com/getstarted/
Page 3

Release 9
Design of Experiments
Guide
“The real voyage of discovery consists not in seeking new
landscapes, but in having new eyes.”
Marcel Proust
JMP, A Business Unit of SAS
SAS Campus Drive
Cary, NC 27513
Page 4

The correct bibliographic citation for this manual is as follows: SAS Institute Inc. 2009. JMP® 9 Design
of Experiments Guide, Second Edition. Cary, NC: SAS Institute Inc.
®
JMP
9 Design of Experiments Guide, Second Edition
Copyright © 2010, SAS Institute Inc., Cary, NC, USA
ISBN 978-1-60764-597-9
All rights reserved. Produced in the United States of America.
For a hard-copy book: No part of this publication may be reproduced, stored in a retrieval system, or
transmitted, in any form or by any means, electronic, mechanical, photocopying, or otherwise, without
the prior written permission of the publisher, SAS Institute Inc.
For a Web download or e-book: Your use of this publication shall be governed by the terms established
by the vendor at the time you acquire this publication.
U.S. Government Restricted Rights Notice: Use, duplication, or disclosure of this software and related
documentation by the U.S. government is subject to the Agreement with SAS Institute and the
restrictions set forth in FAR 52.227-19, Commercial Computer Software-Restricted Rights (June 1987).
SAS Institute Inc., SAS Campus Drive, Cary, North Carolina 27513.
1st printing, September 2010
®
JMP
, SAS® and all other SAS Institute Inc. product or service names are registered trademarks or
trademarks of SAS Institute Inc. in the USA and other countries. ® indicates USA registration.
Other brand and product names are registered trademarks or trademarks of their respective companies.
Page 5

1 Introduction to Designing Experiments
A Beginner’s Tutorial . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 1
About Designing Experiments . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 3
My First Experiment . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 3
The Situation . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 3
Step 1: Design the Experiment . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 4
Step 2: Define Factor Constraints . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 6
Step 3: Add Interaction Terms . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 6
Step 4: Determine the Number of Runs . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 7
Step 5: Check the Design . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 8
Step 6: Gather and Enter the Data . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 9
Step 7: Analyze the Results . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 10
2 Examples Using the Custom Designer
. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 15
Creating Screening Experiments . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 17
Creating a Main-Effects-Only Screening Design . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 17
Creating a Screening Design to Fit All Two-Factor Interactions . . . . . . . . . . . . . . . . . . . . . . . . . . . 19
A Compromise Design Between Main Effects Only and All Interactions . . . . . . . . . . . . . . . . . . . . 20
Creating ‘Super’ Screening Designs . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 22
Screening Designs with Flexible Block Sizes . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 26
Checking for Curvature Using One Extra Run . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 30
Contents
Design of Experiments
Creating Response Surface Experiments . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 33
Exploring the Prediction Variance Surface . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 33
Introducing I-Optimal Designs for Response Surface Modeling . . . . . . . . . . . . . . . . . . . . . . . . . . . 36
A Three-Factor Response Surface Design . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 37
Response Surface with a Blocking Factor . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 39
Creating Mixture Experiments . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 43
Mixtures Having Nonmixture Factors . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 43
Experiments that are Mixtures of Mixtures . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 47
Page 6

ii
Special-Purpose Uses of the Custom Designer . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 50
Designing Experiments with Fixed Covariate Factors . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 50
Creating a Design with Two Hard-to-Change Factors: Split Plot . . . . . . . . . . . . . . . . . . . . . . . . . . . 54
Technical Discussion . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 59
3 Building Custom Designs
The Basic Steps . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 63
Creating a Custom Design . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 65
Enter Responses and Factors into the Custom Designer . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 65
Describe the Model . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 69
Specifying Alias Terms . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 70
Select the Number of Runs . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 71
Understanding Design Evaluation . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 72
Specify Output Options . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 78
Make the JMP Design Table . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 79
Creating Random Block Designs . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 80
Creating Split Plot Designs . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 80
Creating Split-Split Plot Designs . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 81
Creating Strip Plot Designs . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 82
Special Custom Design Commands . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 83
Save Responses and Save Factors . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 84
Load Responses and Load Factors . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 85
Save Constraints and Load Constraints . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 85
Set Random Seed: Setting the Number Generator . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 85
Simulate Responses . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 86
Save X Matrix: Viewing the Number of Rows in the Moments Matrix and the Design Matrix (X) in the
Log . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 87
Optimality Criterion . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 88
Number of Starts: Changing the Number of Random Starts . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 88
Sphere Radius: Constraining a Design to a Hypersphere . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 90
Disallowed Combinations: Accounting for Factor Level Restrictions . . . . . . . . . . . . . . . . . . . . . . . 90
Advanced Options for the Custom Designer . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 92
Save Script to Script Window . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 93
Assigning Column Properties . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 93
Define Low and High Values (DOE Coding) for Columns . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 94
Set Columns as Factors for Mixture Experiments . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 95
Define Response Column Values . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 96
Page 7

Assign Columns a Design Role . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 97
Identify Factor Changes Column Property . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 98
How Custom Designs Work: Behind the Scenes . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 99
4 Screening Designs
. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 101
Screening Design Examples . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 103
Using Two Continuous Factors and One Categorical Factor . . . . . . . . . . . . . . . . . . . . . . . . . . . . 103
Using Five Continuous Factors . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 105
Creating a Screening Design . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 109
Enter Responses . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 109
Enter Factors . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 110
Choose a Design . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 111
Display and Modify a Design . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 115
Specify Output Options . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 119
View the Design Table . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 120
Create a Plackett-Burman design . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 120
iii
Analysis of Screening Data . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 122
Using the Screening Analysis Platform . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 123
Using the Fit Model Platform . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 124
5 Response Surface Designs
. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 127
A Box-Behnken Design: The Tennis Ball Example . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 129
The Prediction Profiler . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 132
A Response Surface Plot (Contour Profiler) . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 134
Geometry of a Box-Behnken Design . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 136
Creating a Response Surface Design . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 136
Enter Responses and Factors . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 137
Choose a Design . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 137
Specify Output Options . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 139
View the Design Table . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 140
6 Full Factorial Designs
. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 143
The Five-Factor Reactor Example . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 145
Analyze the Reactor Data . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 146
Page 8

iv
Creating a Factorial Design . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 151
Enter Responses and Factors . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 151
Select Output Options . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 152
Make the Table . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 152
7 Mixture Designs
. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 155
Mixture Design Types . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 157
The Optimal Mixture Design . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 157
The Simplex Centroid Design . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 158
Creating the Design . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 159
Simplex Centroid Design Examples . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 160
The Simplex Lattice Design . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 161
The Extreme Vertices Design . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 163
Creating the Design . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 164
An Extreme Vertices Example with Range Constraints . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 165
An Extreme Vertices Example with Linear Constraints . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 167
Extreme Vertices Method: How It Works . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 168
The ABCD Design . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 168
Creating Ternary Plots . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 169
Fitting Mixture Designs . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 170
Whole Model Tests and Analysis of Variance Reports . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 171
Understanding Response Surface Reports . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 171
A Chemical Mixture Example . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 171
Create the Design . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 172
Analyze the Mixture Model . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 174
The Prediction Profiler . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 175
The Mixture Profiler . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 177
A Ternary Plot of the Mixture Response Surface . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 178
8 Discrete Choice Designs
. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 179
Introduction . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 181
Create an Example Choice Experiment . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 183
Analyze the Example Choice Experiment . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 186
Design a Choice Experiment Using Prior Information . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 189
Page 9

Administer the Survey and Analyze Results . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 191
Initial Choice Platform Analysis . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 191
Find Unit Cost and Trade Off Costs with the Profiler . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 192
9 Space-Filling Designs
. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 195
Introduction to Space-Filling Designs . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 197
Sphere-Packing Designs . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 197
Creating a Sphere-Packing Design . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 197
Visualizing the Sphere-Packing Design . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 199
Latin Hypercube Designs . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 200
Creating a Latin Hypercube Design . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 200
Visualizing the Latin Hypercube Design . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 202
Uniform Designs . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 204
Comparing Sphere-Packing, Latin Hypercube, and Uniform Methods . . . . . . . . . . . . . . . . . . . . . . . . 206
Minimum Potential Designs . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 207
Maximum Entropy Designs . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 209
v
Gaussian Process IMSE Optimal Designs . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 211
Borehole Model: A Sphere-Packing Example . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 212
Create the Sphere-Packing Design for the Borehole Data . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 212
Guidelines for the Analysis of Deterministic Data . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 214
Results of the Borehole Experiment . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 214
10 Accelerated Life Test Designs
Designing Experiments for Accelerated Life Tests . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 219
Overview of Accelerated Life Test Designs . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 221
Using the ALT Design Platform . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 221
Platform Options . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 226
Example . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 226
11 Nonlinear Designs
. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 231
Examples of Nonlinear Designs . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 233
Using Nonlinear Fit to Find Prior Parameter Estimates . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 233
Creating a Nonlinear Design with No Prior Data . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 239
Page 10

vi
Creating a Nonlinear Design . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 243
Identify the Response and Factor Column with Formula . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 243
Set Up Factors and Parameters in the Nonlinear Design Dialog . . . . . . . . . . . . . . . . . . . . . . . . . . 244
Enter the Number of Runs and Preview the Design . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 245
Make Table or Augment the Table . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 246
Advanced Options for the Nonlinear Designer . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 247
12 Taguchi Designs
. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 249
The Taguchi Design Approach . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 251
Taguchi Design Example . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 251
Analyze the Data . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 254
Creating a Taguchi Design . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 256
Detail the Response and Add Factors . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 256
Choose Inner and Outer Array Designs . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 257
Display Coded Design . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 258
Make the Design Table . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 259
13 Augmented Designs
. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 261
A D-Optimal Augmentation of the Reactor Example . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 263
Analyze the Augmented Design . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 266
Creating an Augmented Design . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 274
Replicate a Design . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 274
Add Center Points . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 277
Creating a Foldover Design . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 278
Adding Axial Points . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 279
Adding New Runs and Terms . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 280
Special Augment Design Commands . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 283
Save the Design (X) Matrix . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 284
Modify the Design Criterion (D- or I- Optimality) . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 284
Select the Number of Random Starts . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 285
Specify the Sphere Radius Value . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 285
Disallow Factor Combinations . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 285
Page 11

14 Prospective Sample Size and Power
. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 287
Launching the Sample Size and Power Platform . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 289
One-Sample and Two-Sample Means . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 289
Single-Sample Mean . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 291
Sample Size and Power Animation for One Mean . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 294
Two-Sample Means . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 295
k-Sample Means . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 296
One Sample Standard Deviation . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 298
One Sample Standard Deviation Example . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 299
One-Sample and Two-Sample Proportions . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 300
One Sample Proportion . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 300
Two Sample Proportions . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 302
Counts per Unit . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 305
Counts per Unit Example . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 306
Sigma Quality Level . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 307
Sigma Quality Level Example . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 307
Number of Defects Computation Example . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 308
vii
Reliability Test Plan and Demonstration . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 308
Reliability Test Plan . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 308
Reliability Demonstration . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 311
Index
Design of Experiments . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 319
Page 12

viii
Page 13

Credits and Acknowledgments
Origin
JMP was developed by SAS Institute Inc., Cary, NC. JMP is not a part of the SAS System, though portions
of JMP were adapted from routines in the SAS System, particularly for linear algebra and probability
calculations. Version 1 of JMP went into production in October 1989.
Credits
JMP was conceived and started by John Sall. Design and development were done by John Sall, Chung-Wei
Ng, Michael Hecht, Richard Potter, Brian Corcoran, Annie Dudley Zangi, Bradley Jones, Craige Hales,
Chris Gotwalt, Paul Nelson, Xan Gregg, Jianfeng Ding, Eric Hill, John Schroedl, Laura Lancaster, Scott
McQuiggan, Melinda Thielbar, Clay Barker, Peng Liu, Dave Barbour, Jeff Polzin, John Ponte, and Steve
Amerige.
In the SAS Institute Technical Support division, Duane Hayes, Wendy Murphrey, Rosemary Lucas, Win
LeDinh, Bobby Riggs, Glen Grimme, Sue Walsh, Mike Stockstill, Kathleen Kiernan, and Liz Edwards
provide technical support.
Nicole Jones, Kyoko Keener, Hui Di, Joseph Morgan, Wenjun Bao, Fang Chen, Susan Shao, Yusuke Ono,
Michael Crotty, Jong-Seok Lee, Tonya Mauldin, Audrey Ventura, Ani Eloyan, Bo Meng, and Sequola
McNeill provide ongoing quality assurance. Additional testing and technical support are provided by Noriki
Inoue, Kyoko Takenaka, and Masakazu Okada from SAS Japan.
Bob Hickey and Jim Borek are the release engineers.
The JMP books were written by Ann Lehman, Lee Creighton, John Sall, Bradley Jones, Erin Vang, Melanie
Drake, Meredith Blackwelder, Diane Perhac, Jonathan Gatlin, Susan Conaghan, and Sheila Loring, with
contributions from Annie Dudley Zangi and Brian Corcoran. Creative services and production was done by
SAS Publications. Melanie Drake implemented the Help system.
Jon Weisz and Jeff Perkinson provided project management. Also thanks to Lou Valente, Ian Cox, Mark
Bailey, and Malcolm Moore for technical advice.
Thanks also to Georges Guirguis, Warren Sarle, Gordon Johnston, Duane Hayes, Russell Wolfinger,
Randall Tobias, Robert N. Rodriguez, Ying So, Warren Kuhfeld, George MacKensie, Bob Lucas, Warren
Kuhfeld, Mike Leonard, and Padraic Neville for statistical R&D support. Thanks are also due to Doug
Melzer, Bryan Wolfe, Vincent DelGobbo, Biff Beers, Russell Gonsalves, Mitchel Soltys, Dave Mackie, and
Stephanie Smith, who helped us get started with SAS Foundation Services from JMP.
Acknowledgments
We owe special gratitude to the people that encouraged us to start JMP, to the alpha and beta testers of
JMP, and to the reviewers of the documentation. In particular we thank Michael Benson, Howard
Page 14

x
Yetter (d), Andy Mauromoustakos, Al Best, Stan Young, Robert Muenchen, Lenore Herzenberg, Ramon
Leon, Tom Lange, Homer Hegedus, Skip Weed, Michael Emptage, Pat Spagan, Paul Wenz, Mike Bowen,
Lori Gates, Georgia Morgan, David Tanaka, Zoe Jewell, Sky Alibhai, David Coleman, Linda Blazek,
Michael Friendly, Joe Hockman, Frank Shen, J.H. Goodman, David Iklé, Barry Hembree, Dan Obermiller,
Jeff Sweeney, Lynn Vanatta, and Kris Ghosh.
Also, we thank Dick DeVeaux, Gray McQuarrie, Robert Stine, George Fraction, Avigdor Cahaner, José
Ramirez, Gudmunder Axelsson, Al Fulmer, Cary Tuckfield, Ron Thisted, Nancy McDermott, Veronica
Czitrom, Tom Johnson, Cy Wegman, Paul Dwyer, DaRon Huffaker, Kevin Norwood, Mike Thompson,
Jack Reese, Francois Mainville, and John Wass.
We also thank the following individuals for expert advice in their statistical specialties: R. Hocking and P.
Spector for advice on effective hypotheses; Robert Mee for screening design generators; Roselinde Kessels
for advice on choice experiments; Greg Piepel, Peter Goos, J. Stuart Hunter, Dennis Lin, Doug
Montgomery, and Chris Nachtsheim for advice on design of experiments; Jason Hsu for advice on multiple
comparisons methods (not all of which we were able to incorporate in JMP); Ralph O’Brien for advice on
homogeneity of variance tests; Ralph O’Brien and S. Paul Wright for advice on statistical power; Keith
Muller for advice in multivariate methods, Harry Martz, Wayne Nelson, Ramon Leon, Dave Trindade, Paul
Tobias, and William Q. Meeker for advice on reliability plots; Lijian Yang and J.S. Marron for bivariate
smoothing design; George Milliken and Yurii Bulavski for development of mixed models; Will Potts and
Cathy Maahs-Fladung for data mining; Clay Thompson for advice on contour plotting algorithms; and
Tom Little, Damon Stoddard, Blanton Godfrey, Tim Clapp, and Joe Ficalora for advice in the area of Six
Sigma; and Josef Schmee and Alan Bowman for advice on simulation and tolerance design.
For sample data, thanks to Patrice Strahle for Pareto examples, the Texas air control board for the pollution
data, and David Coleman for the pollen (eureka) data.
Translations
Trish O'Grady coordinates localization. Special thanks to Noriki Inoue, Kyoko Takenaka, Masakazu Okada,
Naohiro Masukawa and Yusuke Ono (SAS Japan); and Professor Toshiro Haga (retired, Tokyo University of
Science) and Professor Hirohiko Asano (Tokyo Metropolitan University) for reviewing our Japanese
translation; Professors Fengshan Bai, Xuan Lu, and Jianguo Li at Tsinghua University in Beijing, and their
assistants Rui Guo, Shan Jiang, Zhicheng Wan, and Qiang Zhao; and William Zhou (SAS China) and
Zhongguo Zheng, professor at Peking University, for reviewing the Simplified Chinese translation; Jacques
Goupy (consultant, ReConFor) and Olivier Nuñez (professor, Universidad Carlos III de Madrid) for
reviewing the French translation; Dr. Byung Chun Kim (professor, Korea Advanced Institute of Science and
Technology) and Duk-Hyun Ko (SAS Korea) for reviewing the Korean translation; Bertram Schäfer and
David Meintrup (consultants, StatCon) for reviewing the German translation; Patrizia Omodei, Maria
Scaccabarozzi, and Letizia Bazzani (SAS Italy) for reviewing the Italian translation. Finally, thanks to all the
members of our outstanding translation teams.
Past Support
Many people were important in the evolution of JMP. Special thanks to David DeLong, Mary Cole, Kristin
Nauta, Aaron Walker, Ike Walker, Eric Gjertsen, Dave Tilley, Ruth Lee, Annette Sanders, Tim Christensen,
Eric Wasserman, Charles Soper, Wenjie Bao, and Junji Kishimoto. Thanks to SAS Institute quality
assurance by Jeanne Martin, Fouad Younan, and Frank Lassiter. Additional testing for Versions 3 and 4 was
done by Li Yang, Brenda Sun, Katrina Hauser, and Andrea Ritter.
Page 15

Also thanks to Jenny Kendall, John Hansen, Eddie Routten, David Schlotzhauer, and James Mulherin.
Thanks to Steve Shack, Greg Weier, and Maura Stokes for testing JMP Version 1.
Thanks for support from Charles Shipp, Harold Gugel (d), Jim Winters, Matthew Lay, Tim Rey, Rubin
Gabriel, Brian Ruff, William Lisowski, David Morganstein, Tom Esposito, Susan West, Chris Fehily, Dan
Chilko, Jim Shook, Ken Bodner, Rick Blahunka, Dana C. Aultman, and William Fehlner.
Technology License Notices
xi
Scintilla is Copyright 1998-2003 by Neil Hodgson <neilh@scintilla.org>.
WARRANTIES WITH REGARD TO THIS SOFTWARE, INCLUDING ALL IMPLIED WARRANTIES OF
MERCHANTABILITY AND FITNESS, IN NO EVENT SHALL NEIL HODGSON BE LIABLE FOR ANY SPECIAL,
INDIRECT OR CONSEQUENTIAL DAMAGES OR ANY DAMAGES WHATSOEVER RESULTING FROM LOSS OF
USE, DATA OR PROFITS, WHETHER IN AN ACTION OF CONTRACT, NEGLIGENCE OR OTHER TORTIOUS
ACTION, ARISING OUT OF OR IN CONNECTION WITH THE USE OR PERFORMANCE OF THIS SOFTWARE.
XRender is Copyright © 2002 Keith Packard.
TO THIS SOFTWARE, INCLUDING ALL IMPLIED WARRANTIES OF MERCHANTABILITY AND FITNESS, IN NO
EVENT SHALL KEITH PACKARD BE LIABLE FOR ANY SPECIAL, INDIRECT OR CONSEQUENTIAL DAMAGES OR
ANY DAMAGES WHATSOEVER RESULTING FROM LOSS OF USE, DATA OR PROFITS, WHETHER IN AN ACTION
OF CONTRACT, NEGLIGENCE OR OTHER TORTIOUS ACTION, ARISING OUT OF OR IN CONNECTION WITH
THE USE OR PERFORMANCE OF THIS SOFTWARE.
KEITH PACKARD DISCLAIMS ALL WARRANTIES WITH REGARD
NEIL HODGSON DISCLAIMS ALL
ImageMagick software is Copyright © 1999-2010 ImageMagick Studio LLC, a non-profit organization
dedicated to making software imaging solutions freely available.
bzlib software is Copyright © 1991-2009, Thomas G. Lane, Guido Vollbeding. All Rights Reserved.
FreeType software is Copyright © 1996-2002 The FreeType Project (www.freetype.org). All rights reserved.
Page 16

xii
Page 17

Chapter 1
Introduction to Designing Experiments
A Beginner’s Tutorial
This tutorial chapter introduces you to the design of experiments (DOE) using JMP’s custom designer. It
gives a general understanding of how to design an experiment using JMP. Refer to subsequent chapters in
this book for more examples and procedures on how to design an experiment for your specific project.
Page 18

Contents
About Designing Experiments. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 3
My First Experiment . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 3
The Situation . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 3
Step 1: Design the Experiment . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 4
Step 2: Define Factor Constraints . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 6
Step 3: Add Interaction Terms . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 6
Step 4: Determine the Number of Runs . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 7
Step 5: Check the Design . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 8
Step 6: Gather and Enter the Data . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 9
Step 7: Analyze the Results . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .10
Page 19

Chapter 1 Introduction to Designing Experiments 3
About Designing Experiments
About Designing Experiments
Increasing productivity and improving quality are important goals in any business. The methods for
determining how to increase productivity and improve quality are evolving. They have changed from costly
and time-consuming trial-and-error searches to the powerful, elegant, and cost-effective statistical methods
that JMP provides.
Designing experiments in JMP is centered around factors, responses, a model, and runs. JMP helps you
determine if and how a factor affects a response.
My First Experiment
If you have never used JMP to design an experiment, this section shows you how to design the experiment
and how to understand JMP’s output.
Tip: The recommended way to create an experiment is to use the custom designer. JMP also provides
classical designs for use in textbook situations.
The Situation
Your goal is to find the best way to microwave a bag of popcorn. Because you have some experience with
this, it is easy to decide on reasonable ranges for the important factors:
• how long to cook the popcorn (between 3 and 5 minutes)
• what level of power to use on the microwave oven (between settings 5 and 10)
• which brand of popcorn to use (Top Secret or Wilbur)
When a bag of popcorn is popped, most of the kernels pop, but some remain unpopped. You prefer to have
all (or nearly all) of the kernels popped and no (or very few) unpopped kernels. Therefore, you define “the
best popped bag” based on the ratio of popped kernels to the total number of kernels.
A good way to improve any procedure is to conduct an experiment. For each experimental run, JMP’s
custom designer determines which brand to use, how long to cook each bag in the microwave and what
power setting to use. Each run involves popping one bag of corn. After popping a bag, enter the total
number of kernels and the number of popped kernels into the appropriate row of a JMP data table. After
doing all the experimental runs, use JMP’s model fitting capabilities to do the data analysis. Then, you can
use JMP’s profiling tools to determine the optimal settings of popping time, power level, and brand.
Page 20

4 Introduction to Designing Experiments Chapter 1
My First Experiment
Step 1: Design the Experiment
The first step is to select DOE > Custom Design. Then, define the responses and factors.
Define the Responses: Popped Kernels and Total Kernels
There are two responses in this experiment:
• the number of popped kernels
• the total number of kernels in the bag. After popping the bag add the number of unpopped kernels to
the number of popped kernels to get the total number of kernels in the bag.
By default, the custom designer contains one response labeled
Figure 1.1 Custom Design Responses Panel
Y (Figure 1.1).
You want to add a second response to the Responses panel and change the names to be more descriptive:
1. To rename the
increase the number of popped kernels, leave the goal at
2. To add the second response (total number of kernels), click
menu that appears. JMP labels this response
3. Double-click
Y response, double-click the name and type “Number Popped.” Since you want to
Maximize.
Add Response and choose None from the
Y2 by default.
Y2 and type “Total Kernels” to rename it.
The completed Responses panel looks like Figure 1.2.
Figure 1.2 Renamed Responses with Specified Goals
Page 21

Chapter 1 Introduction to Designing Experiments 5
My First Experiment
Define the Factors: Time, Power, and Brand
In this experiment, the factors are:
• brand of popcorn (Top Secret or Wilbur)
• cooking time for the popcorn (3 or 5 minutes)
• microwave oven power level (setting 5 or 10)
In the Factors panel, add
1. Click
Add Factor and select Categorical > 2 Level.
Brand as a two-level categorical factor:
2. To change the name of the factor (currently named
3. To rename the default levels (
Add
Time as a two-level continuous factor:
4. Click
Add Factor and select Continuous.
5. Change the default name of the factor (
6. Likewise, to rename the default levels (
L1 and L2), click the level names and type Top S ec r e t and Wilbur.
X2) by double-clicking it and typing Time.
–1 and 1) as 3 and 5, click the current level name and type in the
new value.
Add
Power as a two-level continuous factor:
7. Click
8. Change the name of the factor (currently named
9. Rename the default levels (currently named
Add Factor and select Continuous.
X3) by double-clicking it and typing Power.
-1 and 1) as 5 and 10 by clicking the current name and
typing. The completed Factors panel looks like Figure 1.3.
Figure 1.3 Renamed Factors with Specified Values
X1), double-click on its name and type Brand.
10. Click Continue.
Page 22

6 Introduction to Designing Experiments Chapter 1
My First Experiment
Step 2: Define Factor Constraints
The popping time for this experiment is either 3 or 5 minutes, and the power settings on the microwave are
5 and 10. From experience, you know that
• popping corn for a long time on a high setting tends to scorch kernels.
• not many kernels pop when the popping time is brief and the power setting is low.
You want to constrain the combined popping time and power settings to be less than or equal to 13, but
greater than or equal to 10. To define these limits:
1. Open the Constraints panel by clicking the disclosure button beside the
Define Factor Constraints title
bar (see Figure 1.4).
2. Click the
Add Constraint button twice, once for each of the known constraints.
3. Complete the information, as shown to the right in Figure 1.4. These constraints tell the Custom
Designer to avoid combinations of
to change
<= to >= in the second constraint.
Power and Time that sum to less than 10 and more than 13. Be sure
The area inside the parallelogram, illustrated on the left in Figure 1.4, is the allowable region for the runs.
You can see that popping for 5 minutes at a power of 10 is not allowed and neither is popping for 3 minutes
at a power of 5.
Figure 1.4 Defining Factor Constraints
Step 3: Add Interaction Terms
You are interested in the possibility that the effect of any factor on the proportion of popped kernels may
depend on the value of some other factor. For example, the effect of a change in popping time for the
Wilbur popcorn brand could be larger than the same change in time for the Top Secret brand. This kind of
synergistic effect of factors acting in concert is called a two-factor interaction. You can examine all possible
two-factor interactions in your a priori model of the popcorn popping process.
1. Click
Interactions in the Model panel and select 2nd. JMP adds two-factor interactions to the model as
shown to the left in Figure 1.5.
Page 23

Chapter 1 Introduction to Designing Experiments 7
My First Experiment
In addition, you suspect the graph of the relationship between any factor and any response might be curved.
You can see whether this kind of curvature exists with a quadratic model formed by adding the second order
powers of effects to the model, as follows.
2. Click
Powers and select 2nd to add quadratic effects of the continuous factors, Power and Time.
The completed Model should look like the one to the right in Figure 1.5.
Figure 1.5 Add Interaction and Power Terms to the Model
Step 4: Determine the Number of Runs
The Design Generation panel in Figure 1.6 shows the minimum number of runs needed to perform the
experiment with the effects you’ve added to the model. You can use that minimum or the default number of
runs, or you can specify your own number of runs as long as that number is more than the minimum. JMP
has no restrictions on the number of runs you request. For this example, use the default number of runs, 16.
Click
Make Design to continue.
Figure 1.6 Model and Design Generation Panels
Page 24

8 Introduction to Designing Experiments Chapter 1
My First Experiment
Step 5: Check the Design
When you click Make Design, JMP generates and displays a design, as shown on the left in Figure 1.7.
Note that because JMP uses a random seed to generate custom designs and there is no unique optimal
design for this problem, your table may be different than the one shown here. You can see in the table that
the custom design requires 8 runs using each brand of popcorn.
Scroll to the bottom of the Custom Design window and look at the Output Options area (shown to the
right in Figure 1.7. The
data table when it is created. Keep the selection at
in a random order.
Run Order option lets you designate the order you want the runs to appear in the
Randomize so the rows (runs) in the output table appear
Now click
Figure 1.7 Design and Output Options Section of Custom Designer
Make Table in the Output Options section.
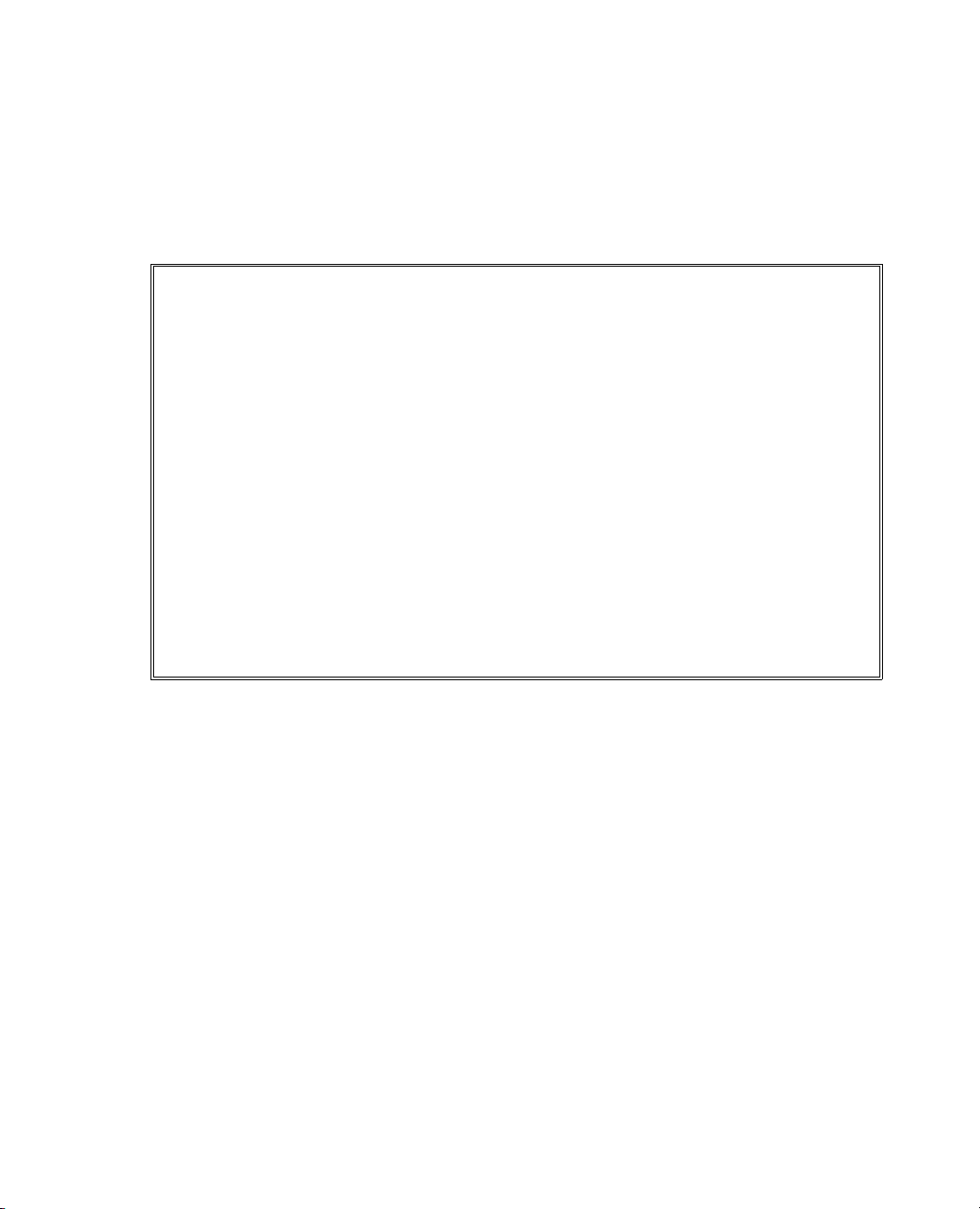
The resulting data table (Figure 1.8) shows the order in which you should do the experimental runs and
provides columns for you to enter the number of popped and total kernels.
You do not have fractional control over the power and time settings on a microwave oven, so you should
round the power and time settings, as shown in the data table. Although this altered design is slightly less
optimal than the one the custom designer suggested, the difference is negligible.
Tip: Note that optionally, before clicking
Right
in the Run Order menu to have JMP present the results in the data table according to the brand. We
have conducted this experiment for you and placed the results, called
Sample Data folder installed with JMP. These results have the columns sorted from left to right.
Make Table in the Output Options, you could select Sort Left to
Popcorn DOE Results.jmp, in the
Page 25
Chapter 1 Introduction to Designing Experiments 9
results from
experiment
scripts to
analyze data
My First Experiment
Figure 1.8 JMP Data Table of Design Runs Generated by Custom Designer
Step 6: Gather and Enter the Data
Pop the popcorn according to the design JMP provided. Then, count the number of popped and unpopped
kernels left in each bag. Finally, enter the numbers shown below into the appropriate columns of the data
table.
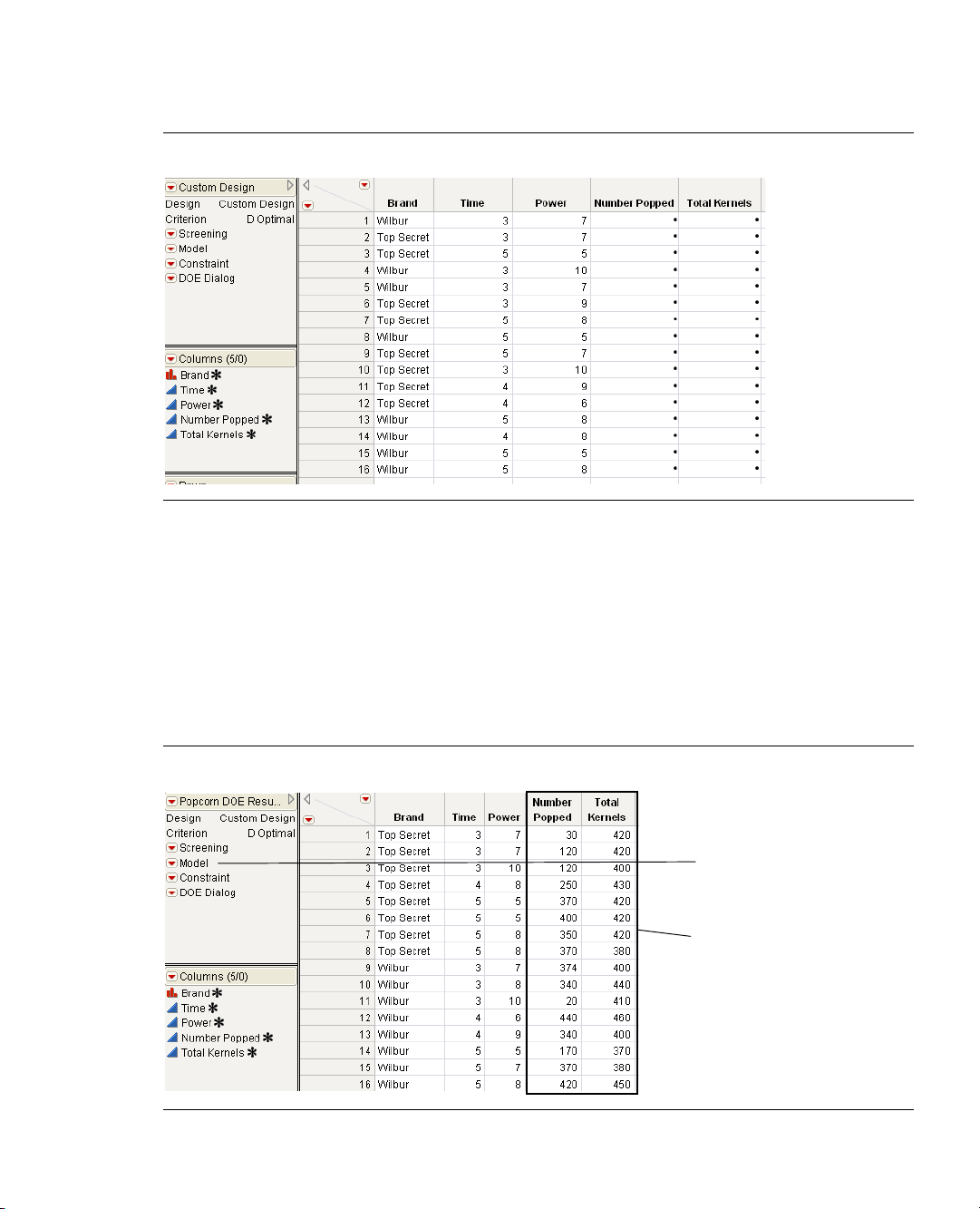
We have conducted this experiment for you and placed the results in the
JMP. To see the results, open
data.
The data table is shown in Figure 1.9.
Figure 1.9 Results of the Popcorn DOE Experiment
Popcorn DOE Results.jmp from the Design Experiment folder in the sample
Sample Data folder installed with
Page 26
10 Introduction to Designing Experiments Chapter 1
My First Experiment
Step 7: Analyze the Results
After the experiment is finished and the number of popped kernels and total kernels have been entered into
the data table, it is time to analyze the data. The design data table has a script, labeled
the top left panel of the table. When you created the design, a standard least squares analysis was stored in
the
Model script with the data table.
Model, that shows in
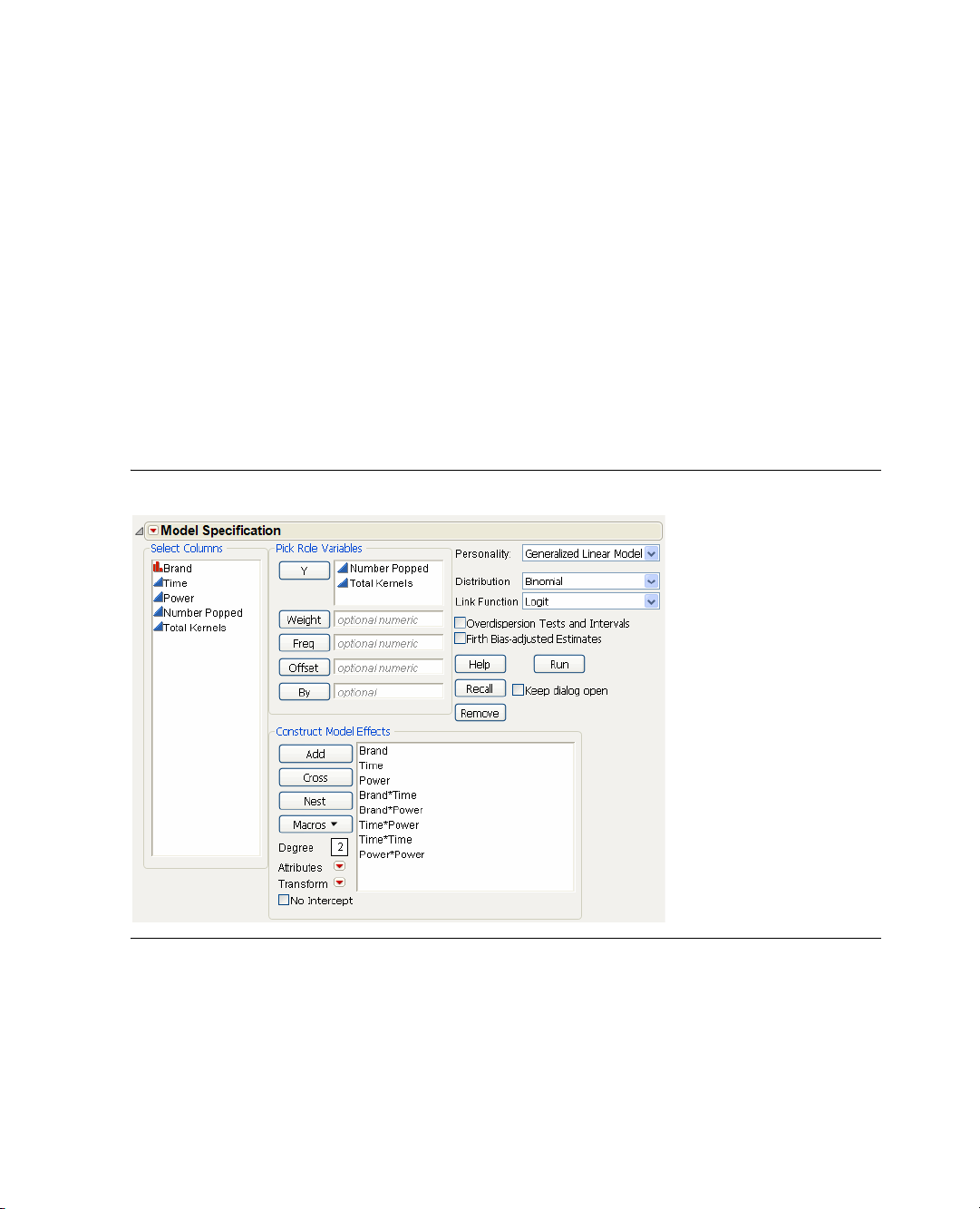
1. Click the red triangle for
The default fitting personality in the model dialog is
Model and select Run Script.
Standard Least Squares. One assumption of
standard least squares is that your responses are normally distributed. But because you are modeling the
proportion of popped kernels it is more appropriate to assume that your responses come from a binomial
distribution. You can use this assumption by changing to a generalized linear model.
2. Change the Personality to
Logit, as shown in Figure 1.10.
Figure 1.10 Fitting the Model
Generalized Linear Model, Distribution to Binomial, and Link Function to
3. Click Run.
4. Scroll down to view the Effect Tests table (Figure 1.11) and look in the column labeled Prob>Chisq.
This column lists p-values. A low p-value (a value less than 0.05) indicates that results are statistically
significant. There are asterisks that identify the low p-values. You can therefore conclude that, in this
experiment, all the model effects except for
there is a strong relationship between popping time (
(
Brand), and the proportion of popped kernels.
Time*Time are highly significant. You have confirmed that
Time), microwave setting (Power), popcorn brand
Page 27
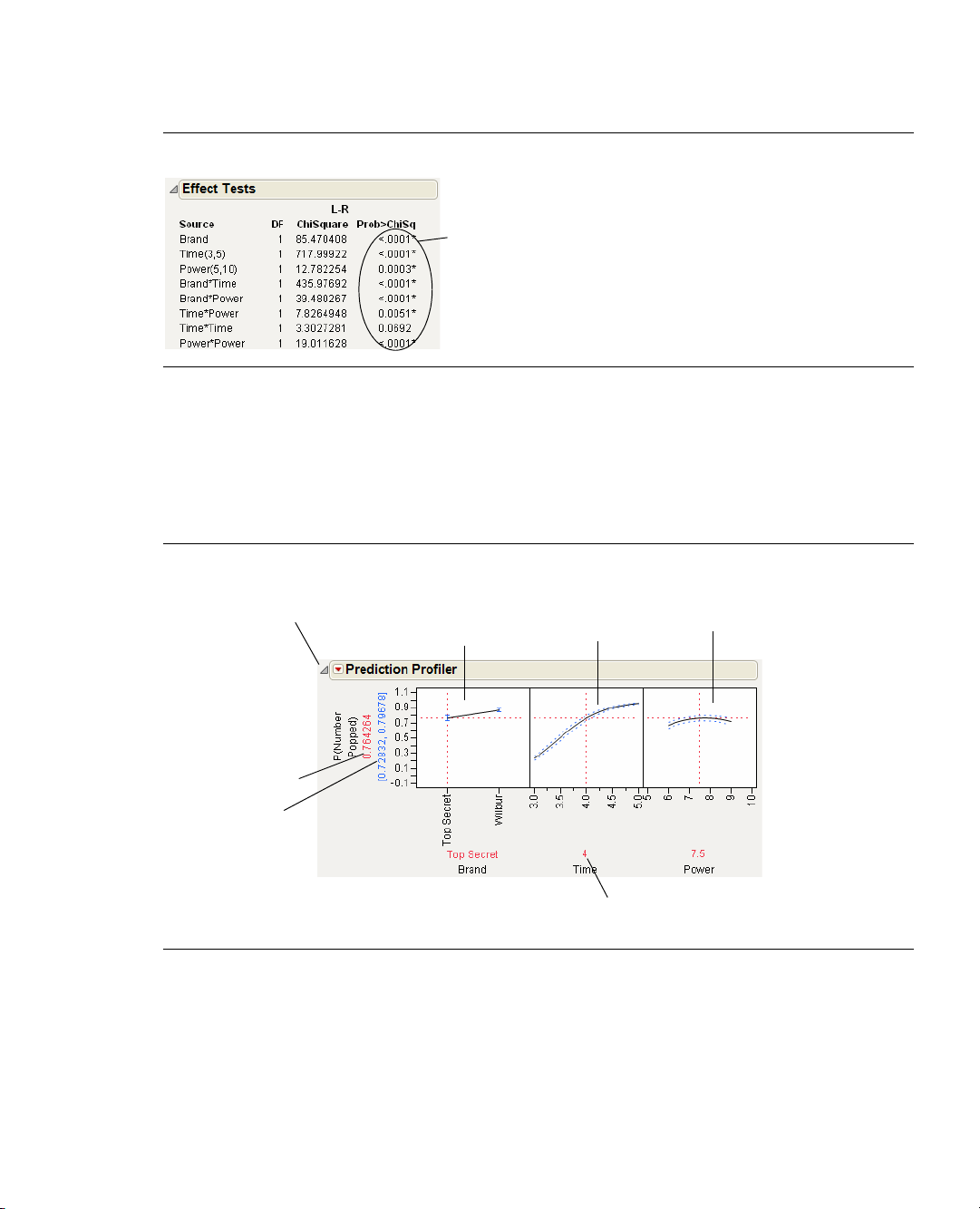
Chapter 1 Introduction to Designing Experiments 11
p-values indicate significance.
Values with * beside them are
p-values that indicate the results
are statistically significant.
Prediction trace
for
Brand
predicted value
of the response
95% confidence
interval on the mean
response
Factor values (here, time = 4)
Prediction trace
for
Time
Prediction trace
for
Power
Disclosure icon to
open or close the
Prediction Profiler
My First Experiment
Figure 1.11 Investigating p-Values
To further investigate, use the Prediction Profiler to see how changes in the factor settings affect the
numbers of popped and unpopped kernels:
1. Choose
Profilers > Profiler from the red triangle menu on the Generalized Linear Model Fit title bar.
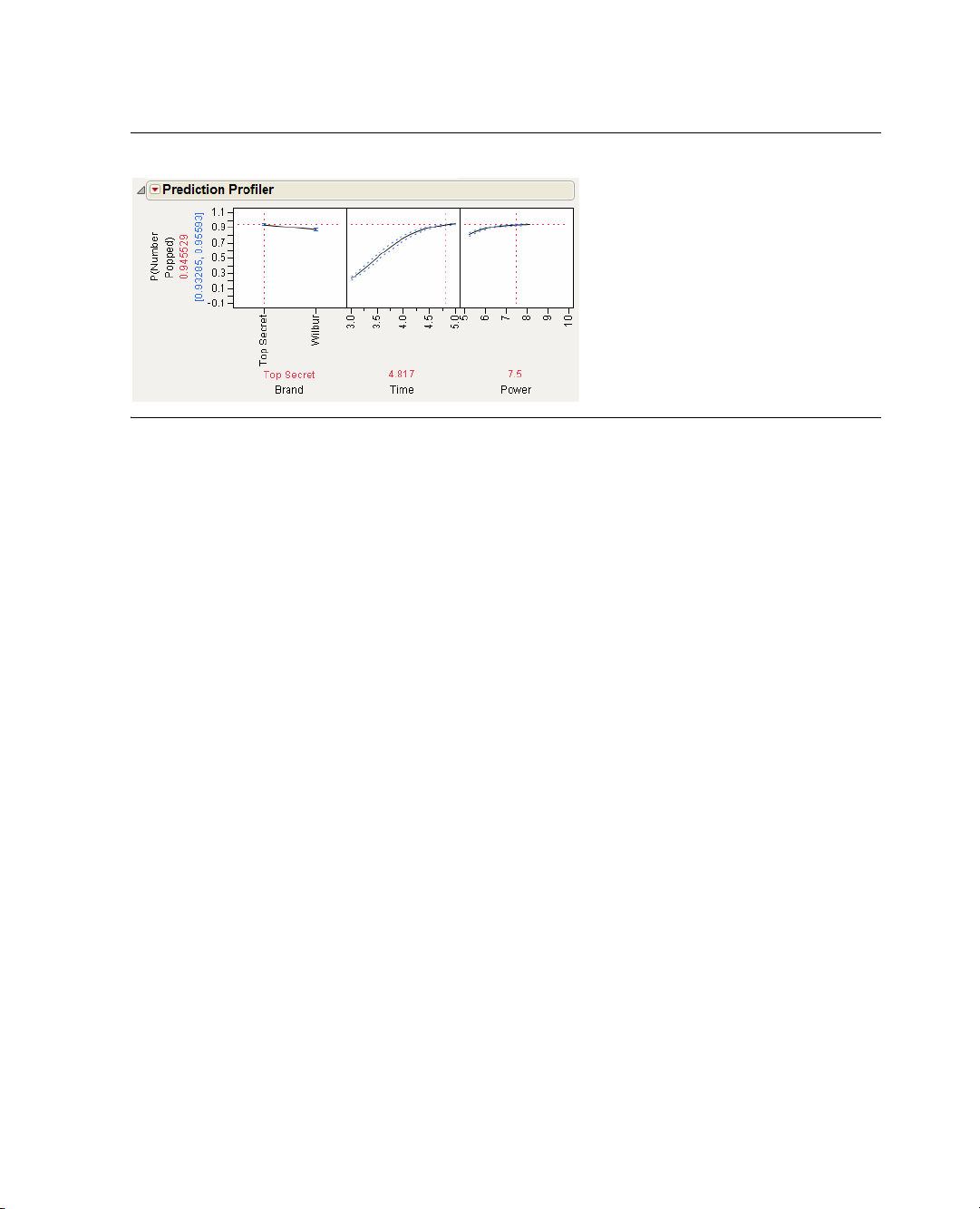
The Prediction Profiler is shown at the bottom of the report. Figure 1.12 shows the Prediction Profiler
for the popcorn experiment. Prediction traces are displayed for each factor.
Figure 1.12 The Prediction Profiler
2. Move the vertical red dotted lines to see the effect that changing a factor value has on the response. For
example, drag the red line in the
Time graph to the right and left (Figure 1.13).
Page 28
12 Introduction to Designing Experiments Chapter 1
My First Experiment
Figure 1.13 Moving the Time Value from 4 to Near 5
As Time increases and decreases, the curved Time and Power prediction traces shift their slope and
maximum/minimum values. The substantial slope shift tells you there is an interaction (synergistic effect)
involving
Time and Power.
Furthermore, the steepness of a prediction trace reveals a factor’s importance. Because the prediction trace
for
Time is steeper than that for Brand or Power (see Figure 1.13), you can see that cooking time is more
important than the brand of popcorn or the microwave power setting.
Now for the final steps.
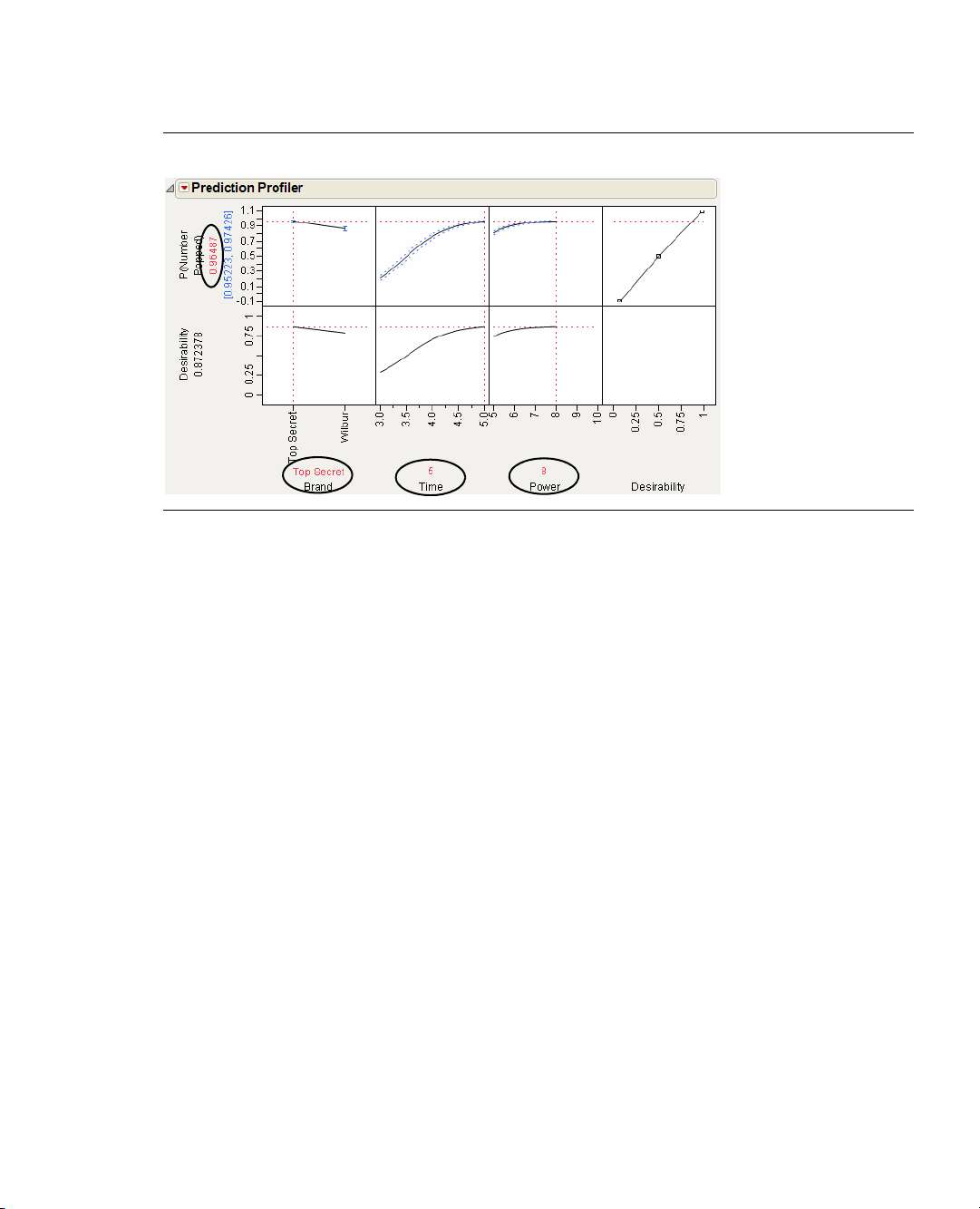
3. Click the red triangle icon in the Prediction Profiler title bar and select
4. Click the red triangle icon in the Prediction Profiler title bar and select
Desirability Functions.
Maximize Desirability. JMP
automatically adjusts the graph to display the optimal settings at which the most kernels will be popped
(Figure 1.14).
Our experiment found how to cook the bag of popcorn with the greatest proportion of popped kernels: use
Top Secret, cook for five minutes, and use a power level of 8. The experiment predicts that cooking at these
settings will yield greater than 96.5% popped kernels.
Page 29
Chapter 1 Introduction to Designing Experiments 13
My First Experiment
Figure 1.14 The Most Desirable Settings
The best settings are the Top Secret brand, cooking time at 5, and power set at 8.
Page 30

14 Introduction to Designing Experiments Chapter 1
My First Experiment
Page 31

Chapter 2
Describe Design Collect Fit Predict
Key mathematical steps: appropriate
computer-based tools are empowering.
Key engineering steps: process knowledge and
engineering judgement are important.
Identify factors
and responses.
Compute design
for maximum
information from
runs.
Use design to set
factors: measure
response for each
run.
Compute best fit
of mathematical
model to data
from test runs.
Use model to find
best factor settings
for on-target
responses and
minimum variability.
Examples Using the Custom Designer
The use of statistical methods in industry is increasing. Arguably, the most cost-beneficial of these methods
for quality and productivity improvement is statistical design of experiments. A trial-and -error search for
the vital few factors that most affect quality is costly and time-consuming. The purpose of experimental
design is to characterize, predict, and then improve the behavior of any system or process. Designed
experiments are a cost-effective way to accomplish these goals.
JMP’s custom designer is the recommended way to describe your process and create a design that works for
your situation. To use the custom designer, you first enter the process variables and constraints, then JMP
tailors a design to suit your unique case. This approach is more general and requires less experience and
expertise than previous tools supporting the statistical design of experiments.
Custom designs accommodate any number of factors of any type. You can also control the number of
experimental runs. This makes custom design more flexible and more cost effective than alternative
approaches.
This chapter presents several examples showing the use of custom designs. It shows how to drive its interface
to build a design using this easy step-by-step approach:
Figure 2.1 Approach to Experimental Design
Page 32

Contents
Creating Screening Experiments . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .17
Creating a Main-Effects-Only Screening Design . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .17
Creating a Screening Design to Fit All Two-Factor Interactions. . . . . . . . . . . . . . . . . . . . . . . . . . . . . .19
A Compromise Design Between Main Effects Only and All Interactions. . . . . . . . . . . . . . . . . . . . . . 20
Creating ‘Super’ Screening Designs . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 22
Screening Designs with Flexible Block Sizes . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 26
Checking for Curvature Using One Extra Run . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 30
Creating Response Surface Experiments . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .33
Exploring the Prediction Variance Surface . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .33
Introducing I-Optimal Designs for Response Surface Modeling . . . . . . . . . . . . . . . . . . . . . . . . . . . . .36
A Three-Factor Response Surface Design. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .37
Response Surface with a Blocking Factor . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .39
Creating Mixture Experiments . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .43
Mixtures Having Nonmixture Factors . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .43
Experiments that are Mixtures of Mixtures . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 47
Special-Purpose Uses of the Custom Designer . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 50
Designing Experiments with Fixed Covariate Factors . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 50
Creating a Design with Two Hard-to-Change Factors: Split Plot. . . . . . . . . . . . . . . . . . . . . . . . . . . . .54
Technical Discussion . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .59
Page 33

Chapter 2 Examples Using the Custom Designer 17
Creating Screening Experiments
Creating Screening Experiments
You can use the screening designer in JMP to create screening designs, but the custom designer is more
flexible and general. The straightforward screening examples described below show that ‘custom’ does not
mean ‘exotic.’ The custom designer is a general purpose design environment that can create screening
designs.
Creating a Main-Effects-Only Screening Design
To create a main-effects-only screening design using the custom designer:
1. Select
2. Enter six continuous factors into the Factors panel (see “Step 1: Design the Experiment,” p. 4, for
3. Click
4. Using the default of eight runs, click
Note to DOE experts: The result is a resolution-three screening design. All the main effects are
estimable, but they are confounded with two factor interactions.
DOE > Custom Design.
details). Figure 2.2 shows the six factors.
Continue. The default model contains only the main effects.
Make Design. Click the disclosure button ( on Windows and
on the Macintosh) to open the
Design Evaluation outline node.
Page 34

18 Examples Using the Custom Designer Chapter 2
Creating Screening Experiments
Figure 2.2 A Main-Effects-Only Screening Design
5. Click the disclosure buttons beside Design Evaluation and then beside Alias Matrix ( on Windows
and on the Macintosh) to open the Alias Matrix. Figure 2.3 shows the Alias Matrix, which is a
table of zeros, ones, and negative ones.
The Alias Matrix shows how the coefficients of the constant and main effect terms in the model are biased
by any active two-factor interaction effects not already added to the model. The column labels identify
interactions. For example, the columns labeled X2*X6 and X3*X4 in the table have a 1 and -1 in the row
for
X1. This means that the expected value of the main effect of X1 is actually the sum of the main effect of
X1 and X2*X6, minus the effect of X3*X4. You are assuming that these interactions are negligible in size
compared to the effect of
Figure 2.3 The Alias Matrix
X1.
Page 35

Chapter 2 Examples Using the Custom Designer 19
Open
outline
nodes
Creating Screening Experiments
Note to DOE experts: The Alias matrix is a generalization of the confounding pattern in fractional
factorial designs.
Creating a Screening Design to Fit All Two-Factor Interactions
There is risk involved in designs for main effects only. The risk is that two-factor interactions, if they are
strong, can confuse the results of such experiments. To avoid this risk, you can create experiments resolving
all the two-factor interactions.
Note to DOE experts: The result in this example is a resolution-five screening design. Two-factor
interactions are estimable but are confounded with three-factor interactions.
1. Select
DOE > Custom Design.
2. Enter five continuous factors into the Factors panel (see “Step 1: Design the Experiment,” p. 4 in the
“Introduction to Designing Experiments” chapter for details).
3. Click
4. In the Model panel, select
5. In the Design Generation Panel choose
Continue.
Interactions > 2nd.
Minimum for Number of Runs and click Make Design.
Figure 2.4 shows the runs of the two-factor design with all interactions. The sample size, 16 (a power of
two) is large enough to fit all the terms in the model. The values in your table may be different from those
shown below.
Figure 2.4 All Two-Factor Interactions
Page 36

20 Examples Using the Custom Designer Chapter 2
Creating Screening Experiments
6. Click the disclosure button ( on Windows and on the Macintosh) and to open the Design
Evaluation
The columns labels identify an interaction. For example, the column labelled
interaction of the first and second effect, the column labelled
outlines, then open Alias Matrix. Figure 2.5 shows the alias matrix table of zeros and ones.
1 2 refers to the
2 3 refers to the interaction between the
second and third effect, and so forth.
Look at the column labelled
occurs in the row labelled
expected value of the two-factor interaction
the row labelled
X1*X2 contain only zeros, which means that the Intercept and main effect terms are not
1 2. There is only one value of 1 in that column. All others are 0. The 1
X1*X2. All the other rows and columns are similar. This means that the
X1*X2 is not biased by any other terms. All the rows above
biased by any two-factor interactions.
Figure 2.5 Alias Matrix Showing all Two-Factor Interactions Clear of all Main Effects
A Compromise Design Between Main Effects Only and All Interactions
In a screening situation, suppose there are six continuous factors and resources for n =16 runs. The first
example in this section showed an eight-run design that fit all the main effects. With six factors, there are 15
possible two-factor interactions. The minimum number of runs that could fit the constant, six main effects
and 15 two-factor interactions is 22. This is more than the resource budget of 16 runs. It would be good to
find a compromise between the main-effects only design and a design capable of fitting all the two-factor
interactions.
This example shows how to obtain such a design compromise using the custom designer.
1. Select
2. Define six continuous factors (X1 - X6).
3. Click
4. Click the
DOE > Custom Design.
Continue. The model includes the main effect terms by default. The default estimability of these
terms is
Necessary.
Interactions button and choose 2nd to add all the two-factor interactions.
Page 37

Chapter 2 Examples Using the Custom Designer 21
Creating Screening Experiments
5. Select all the interaction terms and click the current estimability (Necessary) to reveal a menu. Change
Necessary to If Possible, as shown in Figure 2.6.
Figure 2.6 Model for Six-Variable Design with Two-Factor Interactions Designated If Possible
6. Type 16 in the User Specified edit box in the Number of Runs section, as shown. Although the desired
number of runs (16) is less than the total number of model terms, the custom designer builds a design to
estimate as many two-factor interactions as possible.
7. Click
After the custom designer creates the design, click the disclosure button beside
Make Design.
Design Evaluation to open
the Alias Matrix (Figure 2.7). The values in your table may be different from those shown below, but with a
similar pattern.
Page 38

22 Examples Using the Custom Designer Chapter 2
Creating Screening Experiments
Figure 2.7 Alias Matrix
All the rows above the row labelled X1*X2 contain only zeros, which means that the Intercept and main
effect terms are not biased by any two-factor interactions. The row labelled
1 2 column and the same value in the 3 6 column. That means the expected value of the estimate for X1*X2
is actually the sum of
X1*X2 and any real effect due to X3*X6.
Note to DOE experts: The result in this particular example is a resolution-four screening design.
Two-factor interactions are estimable but are aliased with other two-factor interactions.
Creating ‘Super’ Screening Designs
This section shows how to use the technique shown in the previous example to create ‘super’
(supersaturated) screening designs. Supersaturated designs have fewer runs than factors, which makes them
attractive for factor screening when there are many factors and experimental runs are expensive.
In a saturated design, the number of runs equals the number of model terms. In a supersaturated design, as
the name suggests, the number of model terms exceeds the number of runs (Lin, 1993). A supersaturated
design can examine dozens of factors using fewer than half as many runs as factors.
The Need for Supersaturated Designs
7–4
The 2
and the 2
with respect to a main effects model. In the analysis of a saturated design, you can (barely) fit the model, but
there are no degrees of freedom for error or for lack of fit. Until recently, saturated designs represented the
limit of efficiency in designs for screening.
15–11
fractional factorial designs available using the screening designer are both saturated
X1*X2 has the value 0.333 in the
Page 39

Chapter 2 Examples Using the Custom Designer 23
Creating Screening Experiments
Factor screening relies on the sparsity principle. The experimenter expects that only a few of the factors in a
screening experiment are active. The problem is not knowing which are the vital few factors and which are
the trivial many. It is common for brainstorming sessions to turn up dozens of factors. Yet, in practice,
screening experiments rarely involve more than ten factors. What happens to winnow the list from dozens to
ten or so?
If the experimenter is limited to designs that have more runs than factors, then dozens of factors translate
into dozens of runs. Often, this is not economically feasible. The result is that the factor list is reduced
without the benefit of data. In a supersaturated design, the number of model terms exceeds the number of
runs, and you can examine dozens of factors using less than half as many runs.
There are drawbacks:
• If the number of active factors approaches the number of runs in the experiment, then it is likely that
these factors will be impossible to identify. A rule of thumb is that the number of runs should be at least
four times larger than the number of active factors. If you expect that there might be as many as five
active factors, you should have at least 20 runs.
• Analysis of supersaturated designs cannot yet be reduced to an automatic procedure. However, using
forward stepwise regression is reasonable and the Screening platform (
Screening) offers a more streamlined analysis.
Analyze > Modeling >
Example: Twelve Factors in Eight Runs
As an example, consider a supersaturated design with twelve factors. Using model terms designated
Possible
In the last example, two-factor interaction terms were specified
terms—including main effects—are
provides the software machinery for creating a supersaturated design.
If Possible. In a supersaturated design, all
If Possible. Note in Figure 2.8, the only primary term is the intercept.
To see an example of a supersaturated design with twelve factors in eight runs:
1. Select
2. Add 12 continuous factors and click
3. Highlight all terms except the Intercept and click the current estimability (
DOE > Custom Design.
menu. Change
Necessary to If Possible, as shown in Figure 2.8.
Continue.
Necessary) to reveal the
If
Page 40

24 Examples Using the Custom Designer Chapter 2
Creating Screening Experiments
Figure 2.8 Changing the Estimability
4. The desired number of runs is eight so type 8 in the User Specified edit box in the Number of Runs
section.
5. Click the red triangle on the Custom Design title bar and select
6. Click
Make Design, then click Make Table. A window named Simulate Responses and a design table
appear, similar to the one in Figure 2.9. The
Y column values are controlled by the coefficients of the
Simulate Responses.
model in the Simulate Responses window. The values in your table may be different from those shown
below.
Figure 2.9 Simulated Responses and Design Table
Page 41

Chapter 2 Examples Using the Custom Designer 25
Creating Screening Experiments
7. Change the default settings of the coefficients in the Simulate Responses dialog to match those in
Figure 2.10 and click
Apply. The numbers in the Y column change. Because you have set X2 and X10 as
active factors in the simulation, the analysis should be able to identify the same two factors.
Note that random noise is added to the
Y column formula, so the numbers you see might not necessarily
match those in the figure. The values in your table may be different from those shown below.
Figure 2.10 Give Values to Two Main Effects and Specify the Standard Error as 0.5
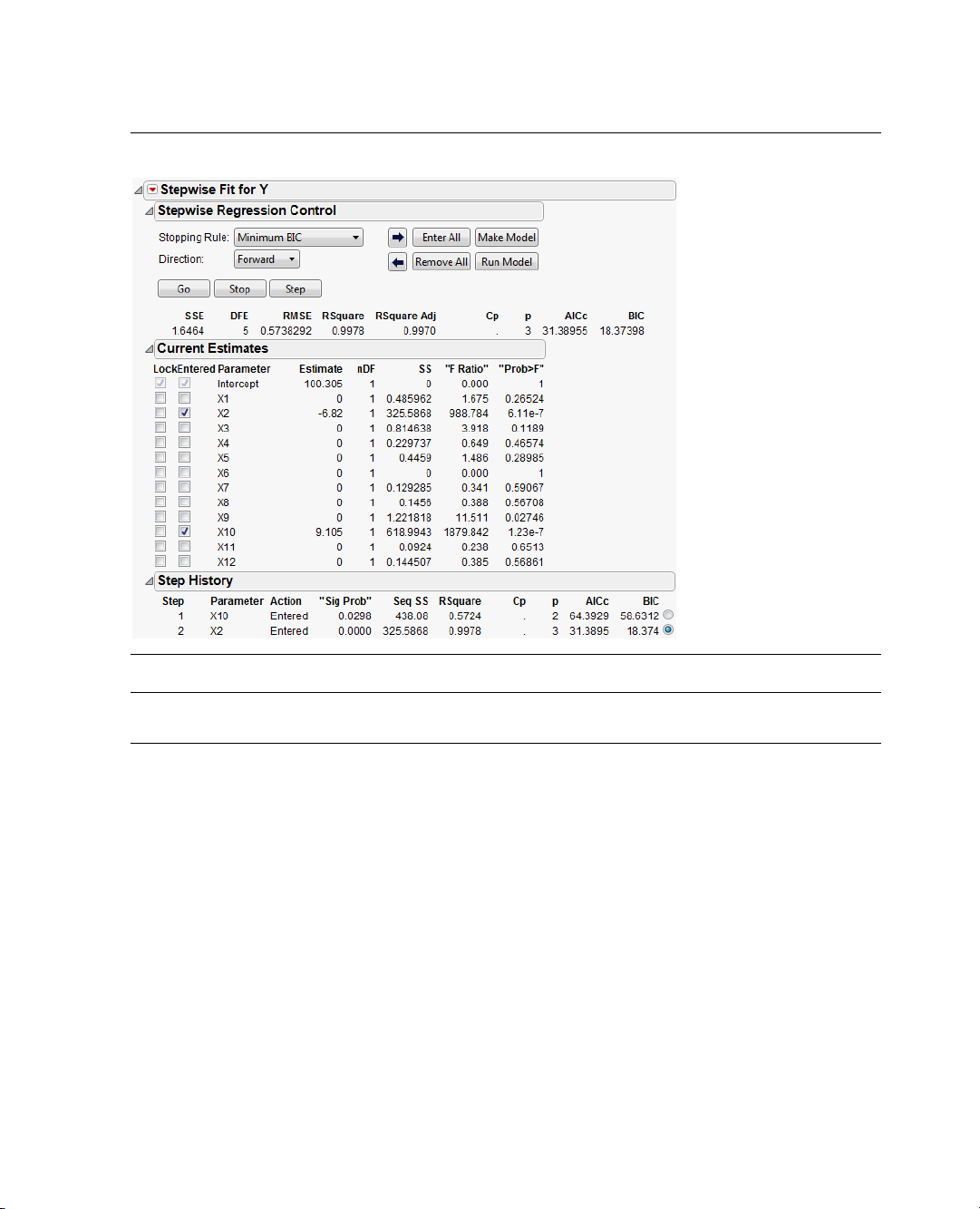
To identify active factors using stepwise regression:
1. To run the
2. Change the
Stepwise.
3. Click
4. In the resulting display click the
Model script in the design table, click the red triangle beside Model and select Run Script.
Personality in the Model Specification window from Standard Least Squares to
Run on the Fit Model dialog.
Step button two times. JMP enters the factors with the largest effects.
From the report that appears, you should identify two active factors, X2 and X10, as shown in
Figure 2.11. The step history appears in the bottom part of the report. Because random noise is added,
your estimates will be slightly different from those shown below.
Page 42
26 Examples Using the Custom Designer Chapter 2
Creating Screening Experiments
Figure 2.11 Stepwise Regression Identifies Active Factors
Note: This example defines two large main effects and sets the rest to zero. In real-world situations, it may
be less likely to have such clearly differentiated effects.
Screening Designs with Flexible Block Sizes
When you create a design using the Screening designer (DOE > Screening), the available block sizes for the
listed designs are a power of two. However, custom designs in JMP can have blocks of any size. The
blocking example shown in this section is flexible because it is using three runs per block, instead of a power
of two.
After you select
Values section of the Factors panel because the sample size is unknown at this point. After you complete the
design, JMP shows the appropriate number of blocks, which is calculated as the sample size divided by the
number of runs per block.
DOE > Custom Design and enter factors, the blocking factor shows only one level in the
Page 43

Chapter 2 Examples Using the Custom Designer 27
Creating Screening Experiments
For example, Figure 2.12 shows that when you enter three continuous factors and one blocking factor with
three runs per block, only one block appears in the Factors panel.
Figure 2.12 One Block Appears in the Factors Panel
The default sample size of nine requires three blocks. After you click Continue, there are three blocks in the
Factors panel (Figure 2.13). This is because the default sample size is nine, which requires three blocks with
three runs each.
Figure 2.13 Three Blocks in the Factors Panel
Page 44

28 Examples Using the Custom Designer Chapter 2
Creating Screening Experiments
If you enter 24 runs in the User Specified box of the Number of Runs section, the Factors panel changes
and now contains 8 blocks (Figure 2.14).
Figure 2.14 Number of Runs is 24 Gives Eight Blocks
If you add all the two-factor interactions and change the number of runs to 15, three runs per block
produces five blocks (as shown in Figure 2.15), so the Factors panel displays five blocks in the Values
section.
Page 45
Chapter 2 Examples Using the Custom Designer 29
Creating Screening Experiments
Figure 2.15 Changing the Runs to 15
Click Make Design, then click the disclosure button ( on Windows and on the Macintosh) to
open the
Design Evaluation outline node. Then, click the disclosure button to open the Relative Variance
of Coefficients report. Figure 2.16 shows the variance of each coefficient in the model relative to the
unknown error variance.
The values in your table may be slightly different from those shown below. Notice that the variance of each
coefficient is about one-tenth the error variance and that all the variances are roughly the same size. The
error variance is assumed to be 1.
Page 46

30 Examples Using the Custom Designer Chapter 2
Creating Screening Experiments
Figure 2.16 Table of Relative Variance of the Model Coefficients
The main question here is whether the relative size of the coefficient variance is acceptably small. If not,
adding more runs (18 or more) will lower the variance of each coefficient.
For more details, see “The Relative Variance of Coefficients and Power Table,” p. 75.
Note to DOE experts: There are four rows associated with X4 (the block factor). That is because X4 has
5 blocks and, therefore, 4 degrees of freedom. Each degree of freedom is associated with one unknown
coefficient in the model.
Checking for Curvature Using One Extra Run
In screening designs, experimenters often add center points and other check points to a design to help
determine whether the assumed model is adequate. Although this is good practice, it is also ad hoc. The
custom designer provides a way to improve on this ad hoc practice while supplying a theoretical foundation
and an easy-to-use interface for choosing a design robust to the modeling assumptions.
The purpose of check points in a design is to provide a detection mechanism for higher-order effects that are
contained in the assumed model. These higher-order terms are called potential terms. (Let q denote the
potential terms, designated
denote the primary terms designated
To take advantage of the benefits of the approach using
larger than the number of
Possible
(potential) terms. That is, p < n < p+q. The formal name of the approach using If Possible model
terms is Bayesian D-Optimal design. This type of design allows the precise estimation of all of the
terms while providing omnibus detectability (and some estimability) for the
For a two-factor design having a model with an intercept, two main effects, and an interaction, there are
p = 4 primary terms. When you enter this model in the custom designer, the default minimum runs value is
a four-run design with the factor settings shown in Figure 2.17.
If Possible in JMP.) The assumed model consists of the primary terms. (Let p
Necessary in JMP.)
Necessary (primary) terms but smaller than the sum of the Necessary and If
If Possible model terms, the sample size should be
Necessary
If Possible terms.
Page 47

Chapter 2 Examples Using the Custom Designer 31
Creating Screening Experiments
Figure 2.17 Two Continuous Factors with Interaction
Now suppose you can afford an extra run (n = 5). You would like to use this point as a check point for
curvature. If you leave the model the same and increase the sample size, the custom designer replicates one
of the four vertices. Replicating any run is the optimal choice for improving the estimates of the terms in the
model, but it provides no way to check for lack of fit.
Adding the two quadratic terms to the model makes a total of six terms. This is a way to model curvature
directly. However, to do this the custom designer requires two additional runs (at a minimum), which
exceeds your budget of five runs.
The Bayesian D-Optimal design provides a way to check for curvature while adding only one extra run. To
create this design:
1. Select
2. Define two continuous factors (
3. Click
4. Choose
DOE > Custom Design.
X1 and X2).
Continue.
2nd from the Interactions menu in the Model panel. The results appear as shown in
Figure 2.18.
Figure 2.18 Second-Level Interactions
Page 48

32 Examples Using the Custom Designer Chapter 2
Creating Screening Experiments
5. Choose 2nd from the Powers button in the Model panel. This adds two quadratic terms.
6. Select the two quadratic terms (
the menu and change
Figure 2.19 Changing the Estimability
Necessary to If Possible, as shown in Figure 2.19.
X1*X1 and X2*X2) and click the current estimability (Necessary) to see
Now, the p = 4 primary terms (the intercept, two main effects, and the interaction) are designated as
Necessary while the q = 2 potential terms (the two quadratic terms) are designated as If Possible. The
desired number of runs, five, is between p = 4 and p + q = 6.
7. Enter 5 into the User Specified edit box in the Number of Runs section of the Design Generation panel.
8. Click
Make Design. The resulting factor settings appear in Figure 2.20. The values in your design may
be different from those shown below.
Figure 2.20 Five-Run Bayesian D-Optimal Design
9. Click Make Table to create a JMP data table of the runs.
10. Create the overlay plot in Figure 2.21 with
Graph > Overlay Plot, and assign X1 as Y and X2 as X. The
overlay plot illustrates how the design incorporates the single extra run. In this example the design places
the factor settings at the center of the design instead of at one of the corners.
Page 49

Chapter 2 Examples Using the Custom Designer 33
Creating Response Surface Experiments
Figure 2.21 Overlay Plot of Five-run Bayesian D-Optimal Design
Creating Response Surface Experiments
Response surface experiments traditionally involve a small number (generally 2 to 8) of continuous factors.
The a priori model for a response surface experiment is usually quadratic.
In contrast to screening experiments, researchers use response surface experiments when they already know
which factors are important. The main goal of response surface experiments is to create a predictive model
of the relationship between the factors and the response. Using this predictive model allows the
experimenter to find better operating settings for the process.
In screening experiments one measure of the quality of the design is the size of the relative variance of the
coefficients. In response surface experiments, the prediction variance over the range of the factors is more
important than the variance of the coefficients. One way to visualize the prediction variance is JMP’s
prediction variance profile plot. This plot is a powerful diagnostic tool for evaluating and comparing
response surface designs.
Exploring the Prediction Variance Surface
The purpose of the example below is to generate and interpret a simple Prediction Variance Profile Plot.
Follow the steps below to create a design for a quadratic model with a single continuous factor.
1. Select
2. Add one continuous factor by selecting
3. In the Model panel, select Powers > 2nd to create a quadratic term (Figure 2.22).
DOE > Custom Design.
Add Factor > Continuous (Figure 2.22), and click Continue.
Page 50

34 Examples Using the Custom Designer Chapter 2
Creating Response Surface Experiments
Figure 2.22 Adding a Factor and a Quadratic Term
4. In the Design Generation panel, use the default number of runs (six) and click Make Design
(Figure 2.23). The number of runs is inversely proportional to the size of variance of the predicted
response. As the number of runs increases, the prediction variances decrease.
Figure 2.23 Using the Default Number of Runs
5. Click the disclosure button ( on Windows and on the Macintosh) to open the Design
Evaluation
outline node, and then the Prediction Variance Profile, as shown in Figure 2.24.
For continuous factors, the initial setting is at the mid-range of the factor values. For categorical factors, the
initial setting is the first level. If the design model is quadratic, then the prediction variance function is
quartic. The y-axis is the relative variance of prediction of the expected value of the response.
In this design, the three design points are –1, 0, and 1. The prediction variance profile shows that the
variance is a maximum at each of these points on the interval –1 to 1.
Figure 2.24 Prediction Profile for Single Factor Quadratic Model
Page 51

Chapter 2 Examples Using the Custom Designer 35
Creating Response Surface Experiments
The prediction variance is relative to the error variance. When the relative prediction variance is one, the
absolute variance is equal to the error variance of the regression model. More detail on the Prediction
Variance Profiler is in “Understanding Design Evaluation,” p. 72.
6. To compare profile plots, click the
Back button and choose Minimum in the Design Generation panel,
which gives a sample size of three.
7. Click
Make Design and then open the Prediction Variance Profile again.
Now you see a curve that has the same shape as the previous plot, but the maxima are at one instead of 0.5.
Figure 2.25 compares plots for a sample size of six and sample size of three for this quadratic model. You can
see the prediction variance increase as the sample size decreases. Since the prediction variance is inversely
proportional to the sample size, doubling the number of runs halves the prediction variance. These profiles
show settings for the maximum variance and minimum variance, for sample sizes six (top charts) and sample
size three (bottom charts). The axes on the bottom plots are adjusted to match the axes on the top plot.
Figure 2.25 Comparison of Prediction Variance Profiles
Tip: Click on the factor to set a factor level precisely.
8. To create an unbalanced design, click the
text edit box in the Design Generation panel, then click
Back button and enter a sample size of 7 in the User Specified
Make Design. The results are shown in
Figure 2.26.
You can see that the variance of prediction at –1 is lower than the other sample points (its value is 0.33
instead of 0.5). The symmetry of the plot is related to the balance of the factor settings. When the design is
balanced, the plot is symmetric, as shown in Figure 2.25. When the design is unbalanced, the prediction
plot might not be symmetric, as shown in Figure 2.26.
Page 52

36 Examples Using the Custom Designer Chapter 2
Prediction variance at X1= 0
Prediction variance at X1= –1
Creating Response Surface Experiments
Figure 2.26 Sample Size of Seven for the One-Factor Quadratic Model
Introducing I-Optimal Designs for Response Surface Modeling
The custom designer generates designs using a mathematical optimality criterion. All the designs in this
chapter so far have been D-Optimal designs. D-Optimal designs are most appropriate for screening
experiments because the optimality criterion focuses on precise estimates of the coefficients. If an
experimenter has precise estimates of the factor effects, then it is easy to tell which factors’ effects are
important and which are negligible. However, D-Optimal designs are not as appropriate for designing
experiments where the primary goal is prediction.
I-Optimal designs minimize the average prediction variance inside the region of the factors. This makes
I-Optimal designs more appropriate for prediction. As a result I-Optimality is the recommended criterion
for JMP response surface designs.
An I-Optimal design tends to place fewer runs at the extremes of the design space than does a D-Optimal
design. As an example, consider a one-factor design for a quadratic model using n = 12 experimental runs.
The D-Optimal design for this model puts four runs at each end of the range of interest and four runs in the
middle. The I-Optimal design puts three runs at each end point and six runs in the middle. In this case, the
D-Optimal design places two-thirds of its runs at the extremes versus one-half for the I-Optimal design.
Figure 2.27 compares prediction variance profiles of the one-factor I- and D-Optimal designs for a
quadratic model with (n = 12) runs. The variance function for the I-Optimal design is less than the
corresponding function for the D-Optimal design in the center of the design space; the converse is true at
the edges.
Page 53

Chapter 2 Examples Using the Custom Designer 37
Creating Response Surface Experiments
Figure 2.27 Prediction Variance Profiles for 12-Run I-Optimal (left) and D-Optimal (right) Designs
At the center of the design space, the average variance (relative to the error variance) for the I-Optimal
design is 0.1667 compared to the D-Optimal design, which is 0.25. This means that confidence intervals
for prediction will be nearly 10% shorter on average for the I-Optimal design.
To compare the two design criteria, create a one-factor design with a quadratic model that uses the
I-Optimality criterion, and another one that uses D-Optimality:
1. Select
2. Add one continuous factor:
3. Click
4. Click the
DOE > Custom Design.
X1.
Continue.
RSM button in the Model panel to make the design I-Optimal.
5. Change the number of runs to 12.
6. Click
Make Design.
7. Click the disclosure button ( on Windows and on the Macintosh) to open the
Evaluation
outline node.
8. Click the disclosure button ( on Windows and on the Macintosh) to open the
Variance Profile
. (The Prediction Variance Profile is shown on the left in Figure 2.27.)
9. Repeat the same steps to create a D-Optimal design, but select
Design
from the red triangle menu on the custom design title bar. The results in the Prediction Variance
Profile should look the same as those on the right in Figure 2.27.
A Three-Factor Response Surface Design
In higher dimensions, the I-Optimal design continues to place more emphasis on the center of the region of
the factors. The D-Optimal and I-Optimal designs for fitting a full quadratic model in three factors using
16 runs are shown in Figure 2.28.
To compare the two design criteria, create a three-factor design that uses the I-Optimality criterion, and
another one that uses D-Optimality:
Design
Prediction
Optimality Criterion > Make D-Optimal
1. Select
DOE > Custom Design.
2. Add three continuous factors:
3. Click
Continue.
X1, X2, and X3.
Page 54
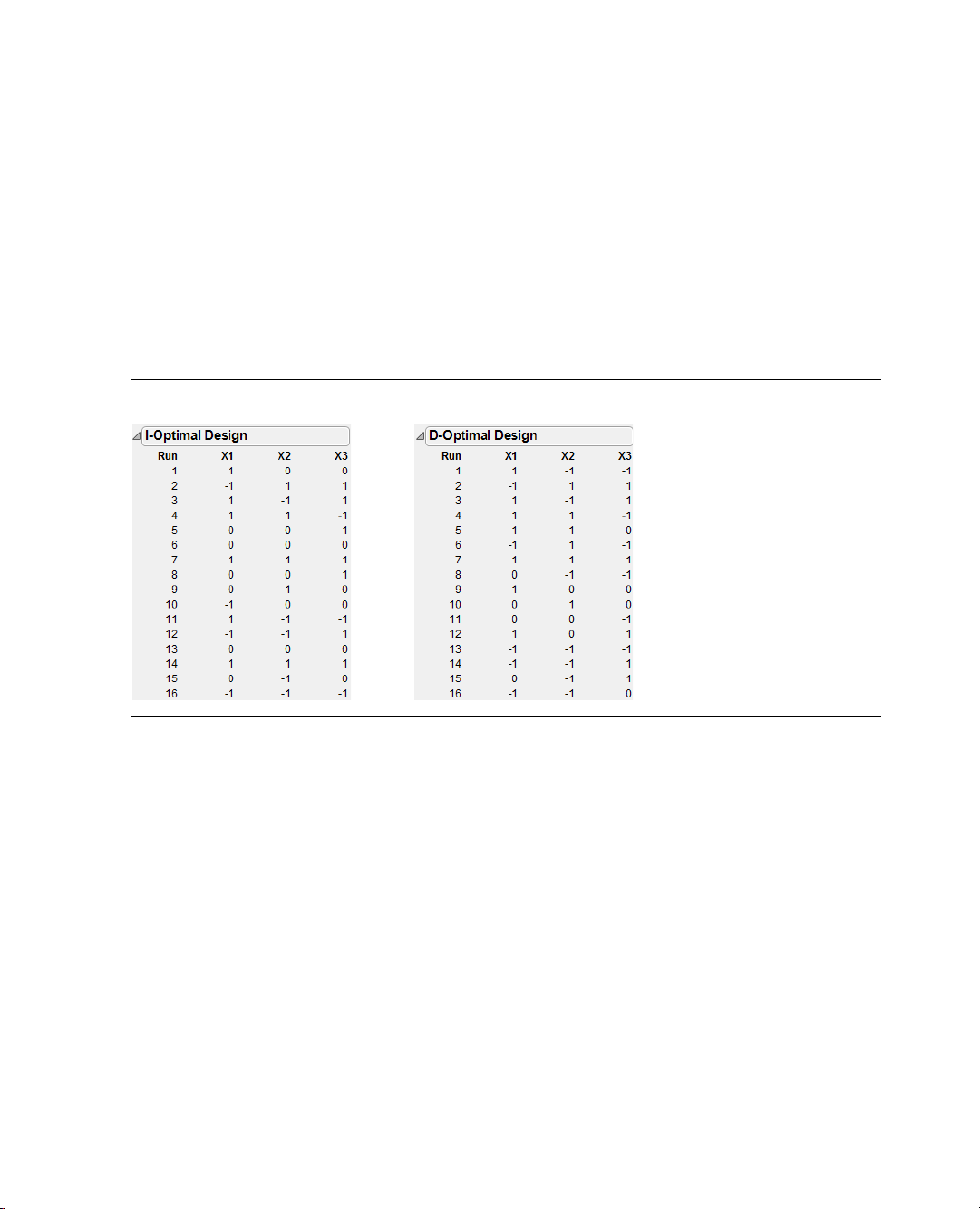
38 Examples Using the Custom Designer Chapter 2
Creating Response Surface Experiments
4. Click the RSM button in the Model panel to add interaction and quadratic terms to the model and to
change the default optimality criterion to I-Optimal.
5. Use the default of 16 runs.
6. Click
Make Design.
The design is shown in the Design panel (the left in Figure 2.28).
7. If you want to create a D-Optimal design for comparison, repeat the same steps but select
Criterion > Make D-Optimal Design
from the red triangle menu on the custom design title bar. The
Optimality
design should look similar to those on the right in Figure 2.28. The values in your design may be
different from those shown below.
Figure 2.28 16-run I-Optimal and D-Optimal designs for RSM Model
Profile plots of the variance function are displayed in Figure 2.29. These plots show slices of the variance
function as a function of each factor, with all other factors fixed at zero. The I-Optimal design has the lowest
prediction variance at the center. Note that there are two center points in this design.
The D-Optimal design has no center points and its prediction variance at the center of the factor space is
almost three times the variance of the I-Optimal design. The variance at the vertices of the D-Optimal
design is not shown. However, note that the D-Optimal design predicts better than the I-Optimal design
near the vertices.
Page 55

Chapter 2 Examples Using the Custom Designer 39
I-Optimal RSM Design
with 16 runs
D-Optimal RSM Design
with 16 runs
Creating Response Surface Experiments
Figure 2.29 Variance Profile Plots for 16 run I-Optimal and D-Optimal RSM Designs
Response Surface with a Blocking Factor
It is not unusual for a process to depend on both qualitative and quantitative factors. For example, in the
chemical industry, the yield of a process might depend not only on the quantitative factors temperature and
pressure, but also on such qualitative factors as the batch of raw material and the type of reactor. Likewise,
an antibiotic might be given orally or by injection, a qualitative factor with two levels. The composition and
dosage of the antibiotic could be quantitative factors (Atkinson and Donev, 1992).
The response surface designer (described in “Response Surface Designs,” p. 127) only deals with
quantitative factors. You could use the response surface designer to produce a Response Surface Model
(RSM) design with a qualitative factor by replicating the design over each level of the factor. But, this is
unnecessarily time-consuming and expensive. Using custom designer is simpler and more cost-effective
because fewer runs are required. The following steps show how to accommodate a blocking factor in a
response surface design using the custom designer:
1. First, define two continuous factors (
2. Now, click
Add Factor and select Blocking > 4 runs per block to create a blocking factor(X3). The
blocking factor appears with one level, as shown in Figure 2.30, but the number of levels adjusts later to
accommodate the number of runs specified for the design.
X1 and X2).
Page 56

40 Examples Using the Custom Designer Chapter 2
Creating Response Surface Experiments
Figure 2.30 Add Two Continuous Factors and a Blocking Factor
3. Click Continue, and then click RSM in the Model panel to add the quadratic terms to the model
(Figure 2.31). This automatically changes the recommended optimality criterion from D-Optimal to
I-Optimal. Note that when you click RSM, a message reminds you that nominal factors (such as the
blocking factor) cannot have quadratic effects.
Figure 2.31 Add Response Surface Terms
4. Enter 12 in the User Specified text edit box in the Design Generation panel, and note that the Factors
panel now shows the Blocking factor,
X3, with three levels (Figure 2.32). Twelve runs defines three
blocks with four runs per block.
Page 57

Chapter 2 Examples Using the Custom Designer 41
Creating Response Surface Experiments
Figure 2.32 Blocking Factor Now Shows Three Levels
5. Click Make Design.
6. In the Output Options, select
7. Click
Make Table to see an I-Optimal table similar to the one on the left in Figure 2.33.
Sort Right to Left from the Run Order list.
Figure 2.33 compares the results of a 12-run I-Optimal design and a 12-run D-Optimal Design.
To see the D-Optimal design:
1. Click the
2. Click the red triangle icon on the Custom Design title bar and select
D-Optimal Design
Back button.
Optimality Criterion > Make
.
3. Click Make Design, then click Make Table.
Figure 2.33 JMP Design Tables for 12-Run I-Optimal and D-Optimal Designs
Page 58

42 Examples Using the Custom Designer Chapter 2
Creating Response Surface Experiments
Figure 2.34 gives a graphical view of the designs generated by this example. These plots were generated for
the runs in each JMP table by choosing
factor (
X3) as the Grouping variable.
Graph > Overlay Plot from the main menu andusing the blocking
Note that there is a center point in each block of the I-Optimal design. The D-Optimal design has only one
center point. The values in your graph may be different from those shown in Figure 2.34.
Figure 2.34 Plots of I-Optimal (left) and D-Optimal (right) Design Points by Block.
Either of the designs in Figure 2.34 supports fitting the specified model. The D-Optimal design does a
slightly better job of estimating the model coefficients. The diagnostics (Figure 2.35) for the designs show
beneath the design tables. In this example, the D-efficiency of the I-Optimal design is about 51%, and is
55% for the D-Optimal design.
The I-Optimal design is preferable for predicting the response inside the design region. Using the formulas
given in “Technical Discussion,” p. 59, you can compute the relative average variance for these designs. The
average variance (relative to the error variance) for the I-Optimal design is 0.5 compared to 0.59 for the
D-Optimal design (See Figure 2.35). This means confidence intervals for prediction will be almost 20%
longer on average for D-Optimal designs.
Page 59

Chapter 2 Examples Using the Custom Designer 43
Creating Mixture Experiments
Figure 2.35 Design Diagnostics for I-Optimal and D-Optimal Designs
Creating Mixture Experiments
If you have factors that are ingredients in a mixture, you can use either the custom designer or the
specialized mixture designer. However, the mixture designer is limited because it requires all factors to be
mixture components and you might want to vary the process settings along with the percentages of the
mixture ingredients. The optimal formulation could change depending on the operating environment. The
custom designer can handle mixture ingredients and process variables in the same study. You are not forced
to modify your problem to conform to the restrictions of a special-purpose design approach.
Mixtures Having Nonmixture Factors
The following example from Atkinson and Donev (1992) shows how to create designs for experiments with
mixtures where one or more factors are not ingredients in the mixture. In this example:
• The response is the electromagnetic damping of an acrylonitrile powder.
• The three mixture ingredients are copper sulphate, sodium thiosulphate, and glyoxal.
• The nonmixture environmental factor of interest is the wavelength of light.
Though
wavelength is a continuous variable, the researchers were only interested in predictions at three
discrete wavelengths. As a result, they treated it as a categorical factor with three levels. To create this custom
design:
1. Select
2. Create
3. In the Factors panel, add the three mixture ingredients and the categorical factor,
DOE > Custom Design.
Damping as the response. The authors do not mention how much damping is desirable, so
right-click the goal and create
Damping’s response goal to be None.
Wavelength. The
mixture ingredients have range constraints that arise from the mechanism of the chemical reaction.
Rather than entering them by hand, load them from the Sample Data folder that was installed with
JMP: click the red triangle icon on the Custom Design title bar and select
Mixture Factors.jmp
, from the Design Experiment sample data folder. The custom design panels should
Load Factors. Open Donev
now look like those shown in Figure 2.36.
Page 60

44 Examples Using the Custom Designer Chapter 2
Creating Mixture Experiments
Figure 2.36 Mixture Experiment Response Panel and Factors Panel
The model, shown in Figure 2.37 is a response surface model in the mixture ingredients along with the
additive effect of the wavelength. To create this model:
1. Click
Interactions, and choose 2nd. A warning dialog appears telling you that JMP removes the main
effect terms for non-mixture factors that interact with all the mixture factors. Click
2. In the Design Generation panel, type 18 in the
User Specified text edit box (Figure 2.37), which results
in six runs each for the three levels of the wavelength factor.
OK.
Page 61

Chapter 2 Examples Using the Custom Designer 45
Creating Mixture Experiments
Figure 2.37 Mixture Experiment Design Generation Panel
3. Click Make Design, and then click Make Table.
The resulting data table is shown in Figure 2.38. The values in your table may be different from those
shown below.
Page 62

46 Examples Using the Custom Designer Chapter 2
Creating Mixture Experiments
Figure 2.38 Mixture Experiment Design Table
Atkinson and Donev also discuss the design where the number of runs is limited to 10. In that case, it is not
possible to run a complete mixture response surface design for every wavelength.
To v i e w t h is :
1. Click the
2. Remove all the effects by highlighting them and clicking
3. Add the main effects by clicking the
Back button.
Remove Term.
Main Effects button.
4. In the Design Generation panel, change the number of runs to 10 (Figure 2.39) and click
Design
. The Design table to the right in Figure 2.39 shows the factor settings for 10 runs.
Figure 2.39 Ten-Run Mixture Response Surface Design
Make
Page 63

Chapter 2 Examples Using the Custom Designer 47
Creating Mixture Experiments
Note that there are necessarily unequal numbers of runs for each wavelength. Because of this lack of balance
it is a good idea to look at the prediction variance plot (top plot in Figure 2.40).
5. Open the
Design Evaluation outline node, then open the Prediction Variance Profile.
The prediction variance is almost constant across the three wavelengths, which is a good indication that the
lack of balance is not a problem.
The values of the first three ingredients sum to one because they are mixture ingredients. If you vary one of
the values, the others adjust to keep the sum constant.
6. Select
Maximize Desirability from red triangle menu on the Prediction Variance Profile title bar, as
shown in the bottom profiler in Figure 2.40.
The most desirable wavelength is
percentage is zero, and
Figure 2.40 Prediction Variance Plots for Ten-Run Design
Na2S2O3 is 0.8, which maintains the mixture.
L3, with the CuSO4 percentage decreasing from about 0.4 to 0.2, Glyoxal
Experiments that are Mixtures of Mixtures
As a way to illustrate the idea of a ‘mixture of mixtures’ situation, imagine the ingredients that go into
baking a cake and assume the following:
• dry ingredients composed of flour, sugar, and cocoa
• wet (or non-dry) ingredients consisting of milk, melted butter, and eggs.
These two components (wet and dry) of the cake are two mixtures that are first mixed separately and then
blended together.
Page 64

48 Examples Using the Custom Designer Chapter 2
Creating Mixture Experiments
The dessert chef knows that the dry component (the mixture of flour, sugar, and cocoa) contributes 45% of
the combined mixture and the wet component (butter, milk, and eggs) contributes 55%.
The objective of such an experiment might be to identify proportions within the two components that
maximize some measure of taste or consistency.
This is a main effects model except that you must leave out one of the factors in order to avoid singularity.
The choice of which factor to leave out of the model is arbitrary.
For now, consider these upper and lower levels of the various factors:
Within the dry mixture:
• cocoa must be greater than 10% but less than 20%
• sugar must be greater than 0% but less than 15%
• flour must be greater than 20% but less than 30%
Within the wet mixture:
• melted butter must be greater than 10% but less than 20%
• milk must be greater than 25% and less than 35%
• eggs constitute more than 5% but less than 20%
You want to bake cakes and measure taste on a scale from 1 to 10
Use the Custom Designer to set up this example, as follows:
1. In the Response Panel, enter one response and call it
2. Give Taste a
Lower Limit of 1 and an Upper Limit of 10. (You are assuming a taste test where the
Ta st e .
respondents reply on a scale of 1 to 10.)
3. In the Factors Panel, enter the six cake factors described above.
4. Enter the given percentage values of the factors as proportions in the
Values section of the Factors panel.
The completed Response and Factors panels should look like those shown in Figure 2.41.
Page 65

Chapter 2 Examples Using the Custom Designer 49
Creating Mixture Experiments
Figure 2.41 Completed Responses and Factors Panel for the Cake Example
5. Next, click Continue.
6. Open the Define Factor Constraints pane and click Add Constraint twice.
7. Enter the constraints as shown in Figure 2.42. For the second constraint setting, click on the less than or
equal to button and select the greater than or equal to direction.
By confining the dry factors to exactly 45% in this way, the mixture role of all the factors ensures that the
wet factors constitute the remaining 55%.
8. Open the Model dialog and note that it lists all 6 effects. Because these are mixture factors, including all
effects would render the model singular. Highlight any one of the terms in the model and click
Te rm
, as shown.
Figure 2.42 Constraints to Define the Double Mixture Experiment
Remove
Page 66

50 Examples Using the Custom Designer Chapter 2
Each run sums to 0.55 (55%)
Each run sums to 0.45 (45%)
Special-Purpose Uses of the Custom Designer
9. To see a completed example, choose Simulate Responses from the menu on the Custom Design title
bar.
10. In the Design Generation panel, enter 10 as the number of runs for the example. That is, you would
bake cakes with 10 different sets of ingredient proportions.
11. Click
The table inFigure 2.43 shows that the two sets of cake ingredients (dry and wet) adhere to the proportions
45% and 55% as defined by the entered constraints. In addition, the amount of each ingredient in each
cake recipe (run) conforms to the upper and lower limits given in the factors dialog.
Figure 2.43 Cake Experiment Conforming to a Mixture of Mixture Design
Make Design in the Design Generation panel, and then click Make Table.
Note: As a word of caution, keep in mind that it is easy to define constraints in such a way that it is
impossible to construct a design that fits the model. In such a case, you will get a message saying “Could not
find a valid starting design. Please check your constraints for consistency.”
Special-Purpose Uses of the Custom Designer
While some of the designs discussed in previous sections can be created using other designers in JMP or by
looking them up in a textbook containing tables of designs, the designs presented in this section cannot be
created without using the custom designer.
Designing Experiments with Fixed Covariate Factors
Pre-tabulated designs rely on the assumption that the experimenter controls all the factors. Sometimes you
have quantitative measurements (a covariate) on the experimental units before the experiment begins. If this
variable affects the experimental response, the covariate should be a design factor. The pre-defined design
that allows only a few discrete values is too restrictive. The custom designer supplies a reasonable design
option.
Page 67

Chapter 2 Examples Using the Custom Designer 51
Special-Purpose Uses of the Custom Designer
For this example, suppose there are a group of students participating in a study. A physical education
researcher has proposed an experiment where you vary the number of hours of sleep and the calories for
breakfast and ask each student to run 1/4 mile. The weight of the student is known and it seems important
to include this information in the experimental design.
To follow along with this example that shows column properties, open
Big Class.jmp from the Sample Data
folder that was installed when you installed JMP.
Build the custom design as follows:
1. Select
DOE > Custom Design.
2. Add two continuous variables to the models by entering 2 beside Add N Factors, clicking
and selecting
3. Click
Continuous, naming them calories and sleep.
Add Factor and select Covariate, as shown in Figure 2.44. The Covariate selection displays a list
of the variables in the current data table.
Figure 2.44 Add a Covariate Factor
4. Select weight from the variable list (Figure 2.45) and click OK.
Add Factor
Figure 2.45 Design with Fixed Covariate
5. Click Continue.
6. Add the interaction to the model by selecting
panel, and then clicking the
Cross button (Figure 2.46).
calories in the Factors panel, selecting sleep in the Model
Page 68

52 Examples Using the Custom Designer Chapter 2
Special-Purpose Uses of the Custom Designer
Figure 2.46 Design With Fixed Covariate Factor
7. Click Make Design, then click Make Table. The data table in Figure 2.47 shows the design table. Your
runs might not look the same because the order of the runs has been randomized.
Figure 2.47 Design Table for Covariate Example
Note: Covariate factors cannot have missing values.
Remember that
custom designer has calculated settings for
correlations between
designer did by fitting a model of
weight is the covariate factor, measured for each student, but it is not controlled. The
calories and sleep for each student. It would be desirable if the
calories, sleep and weight were as small as possible. You can see how well the custom
weight as a function of calories and sleep. If that fit has a small model
sum of squares, that means the custom designer has successfully separated the effect of weight from the
effects of calories and sleep.
Page 69

Chapter 2 Examples Using the Custom Designer 53
Special-Purpose Uses of the Custom Designer
8. Click the red triangle icon beside Model in the data table and select Run Script, as shown on the left in
Figure 2.48.
Figure 2.48 Model Script
9. Rearrange the dialog so weight is Y and calories, sleep, and calories*sleep are the model effects, as
shown to the right in Figure 2.48. Click
Run.
The leverage plots are nearly horizontal, and the analysis of variance table shows that the model sum of
squares is near zero compared to the residuals (Figure 2.49). Therefore,
and
sleep. The values in your analysis may be a little different from those shown below.
weight is independent of calories
Figure 2.49 Analysis to Check That Weight is Independent of Calories and Sleep
Page 70

54 Examples Using the Custom Designer Chapter 2
Special-Purpose Uses of the Custom Designer
Creating a Design with Two Hard-to-Change Factors: Split Plot
While there is substantial research literature covering the analysis of split plot designs, it has only been
possible in the last few years to create optimal split plot designs (Goos 2002). The split plot design
capability accessible in the JMP custom designer is the first commercially available tool for generating
optimal split plot designs.
The split plot design originated in agriculture, but is commonplace in manufacturing and engineering
studies. In split plot experiments, hard-to-change factors only change between one whole plot and the next.
The whole plot is divided into subplots, and the levels of the easy-to-change factors are randomly assigned
to each subplot.
The example in this section is adapted from Kowalski, Cornell, and Vining (2002). The experiment studies
the effect of five factors on the thickness of vinyl used to make automobile seat covers. The response and
factors in the experiment are described below:
• Three of the factors are ingredients in a mixture. They are plasticizers whose proportions,
m3, sum to one. Additionally, the mixture components are the subplot factors of the experiment.
• Two of the factors are process variables. They are the rate of extrusion (
temperature (
temperature) of drying. These process variables are the whole plot factors of the
extrusion rate) and the
m1, m2, and
experiment. They are hard to change.
• The response in the experiment is the thickness of the vinyl used for automobile seat covers. The
response of interest (
thickness) depends both on the proportions of the mixtures and on the effects of
the process variables.
To create this design in JMP:
1. Select
2. By default, there is one response,
DOE > Custom Design.
default goal,
Maximize (Figure 2.50).
Y, showing. Double-click the name and change it to thickness. Use the
3. Enter the lower limit of 10.
4. To add three mixture factors, type 3 in the box beside
Add N Factors, and click Add Factor > Mixture.
5. Name the three mixture factors m1, m2, and m3. Use the default levels 0 and 1 for those three factors.
6. Add two continuous factors by typing 2 in the box beside
Continuous
. Name these factors extrusion rate and temperature.
Add N Factors, and click Add Factor >
7. Ensure that you are using the default levels, –1 and 1, in the Values area corresponding to these two
factors.
8. To make
9. To make
extrusion rate a whole plot factor, click Easy and select Hard.
temperature a whole plot factor, click Easy and select Hard. Your dialog should look like the
one in Figure 2.50.
Page 71

Chapter 2 Examples Using the Custom Designer 55
Special-Purpose Uses of the Custom Designer
Figure 2.50 Entering Responses and Factors
10. Click Continue.
11. Next, add interaction terms to the model by selecting
Interactions > 2nd (Figure 2.51). This causes a
warning that JMP removes the main effect terms for non-mixture factors that interact with all the
mixture factors. Click
OK.
Figure 2.51 Adding Interaction Terms
12. In the Design Generation panel, type 7 in the Number of Whole Plots text edit box.
13. For
Number of Runs, type 28 in the User Specified text edit box (Figure 2.52).
Page 72

56 Examples Using the Custom Designer Chapter 2
X′V1–X
Special-Purpose Uses of the Custom Designer
Figure 2.52 Assigning the Number of Whole Plots and Number of Runs
Note: If you enter a missing value in the Number of Whole Plots edit box, then JMP considers many
different numbers of whole plots and chooses the number that maximizes the information about the
coefficients in the model. It maximizes the determinant of where V
-
1
is the inverse of the
variance matrix of the responses. The matrix, V, is a function of how many whole plots there are, so
changing the number of whole plots changes V, which can make a difference in the amount of information
a design contains.
14. Click
Figure 2.53 Partial Listing of the Final Design Structure
Make Design. The result is shown in Figure 2.53.
15. Click Make Table.
16. From the Sample Data folder that was installed with JMP, open
Experiment
folder, which contains 28 runs as well as response values. The values in the table you
Vinyl Data.jmp from the Design
generated with the custom designer may be different from those from the Sample Data folder, shown in
Figure 2.54.
Page 73

Chapter 2 Examples Using the Custom Designer 57
Special-Purpose Uses of the Custom Designer
Figure 2.54 The Vinyl Data Design Table
17. Click the red triangle icon next to the Model script and select Run Script. The dialog in Figure 2.55
appears.
The models for split plots have a random effect associated with the whole plots’ effect.
As shown in the dialog in Figure 2.55, JMP designates the error term by appending &Random to the name
of the effect. REML will be used for the analysis, as indicated in the menu beside
Method in Figure 2.55.
For more information about REML models, see Modeling and Multivariate Methods.
Page 74

58 Examples Using the Custom Designer Chapter 2
Special-Purpose Uses of the Custom Designer
Figure 2.55 Define the Model in the Fit Model Dialog
18. Click Run to run the analysis. The results are shown in Figure 2.56.
Page 75

Chapter 2 Examples Using the Custom Designer 59
Technical Discussion
Figure 2.56 Split Plot Analysis Results
Technical Discussion
This section provides information about I-, D-, Bayesian I-, Bayesian D-, and Alias-Optimal designs.
D-Optimality:
• is the default design type produced by the custom designer except when the RSM button has been
clicked to create a full quadratic model.
• minimizes the variance of the model coefficient estimates. This is appropriate for first-order models and
in screening situations, because the experimental goal in such situations is often to identify the active
factors; parameter estimation is key.
• is dependent on a pre-stated model. This is a limitation because in most real situations, the form of the
pre-stated model is not known in advance.
• has runs whose purpose is to lower the variability of the coefficients of this pre-stated model. By focusing
on minimizing the standard errors of coefficients, a D-Optimal design may not allow for checking that
the model is correct. It will not include center points when investigating a first-order model. In the
extreme, a D-Optimal design may have just p distinct runs with no degrees of freedom for lack of fit.
Page 76

60 Examples Using the Custom Designer Chapter 2
D det X′X[]=
D det X′V1–X[]=
If
R
′ x()X′X()=
1–
fx()dx
Trace X′X()1–M[]=
Technical Discussion
• maximizes D when
D-optimal split plot designs maximize D when
-1
where V
is the block diagonal variance matrix of the responses (Goos 2002).
Bayesian D-Optimality:
• is a modification of the D-Optimality criterion that effectively estimates the coefficients in a model, and
at the same time has the ability to detect and estimate some higher-order terms. If there are interactions
or curvature, the Bayesian D-Optimality criterion is advantageous.
• works best when the sample size is larger than the number of
of the
Necessary and If Possible terms. That is, p + q > n > p. The Bayesian D-Optimal design is an
approach that allows the precise estimation of all of the
detectability (and some estimability) for the
• uses the
containing only the
If Possible terms to force in runs that allow for detecting any inadequacy in the model
Necessary terms. Let K be the (p + q) by (p + q) diagonal matrix whose first p
If Possible terms.
Necessary terms but smaller than the sum
Necessary terms while providing omnibus
diagonal elements are equal to 0 and whose last q diagonal elements are the constant, k. If there are
2-factor interactions then k = 4. Otherwise k = 1. The Bayesian D-Optimal design maximizes the
determinant of (X'X + K). The difference between the criterion for D-Optimality and Bayesian
D-Optimality is this constant added to the diagonal elements corresponding to the
If Possible terms in
the X'X matrix.
I-Optimality:
• minimizes the average variance of prediction over the region of the data.
• is more appropriate than D-Optimality if your goal is to predict the response rather than the
coefficients, such as in response surface design problems. Using the I-Optimality criterion is more
appropriate because you can predict the response anywhere inside the region of data and therefore find
the factor settings that produce the most desirable response value. It is more appropriate when your
objective is to determine optimum operating conditions, and also is appropriate to determine regions in
the design space where the response falls within an acceptable range. Precise estimation of the response
therefore takes precedence over precise estimation of the parameters.
• maximizes this criterion: If f '(x) denotes a row of the X matrix corresponding to factor combinations x,
then
where
Page 77

Chapter 2 Examples Using the Custom Designer 61
M f
R
x()fx()′dx=
var
Y
ˆ
x
0
x0′
X′XK+
1–
x
0
=
Trace X′XK+()
1–
M[]=
X1′X
1
()1–X1′X
2
tr AA'()
AA'
Technical Discussion
is a moment matrix that is independent of the design and can be computed in advance.
Bayesian I-Optimality:
Bayesian I-Optimality has a different objective function to optimize than the Bayesian D-optimal design, so
the designs that result are different. The variance matrix of the coefficients for Bayesian I-optimality is X'X
+ K where K is a matrix having zeros for the
If Possible model terms.
Necessary model terms and some constant value for the
The variance of the predicted value at a point x
is:
0
The Bayesian I-Optimal design minimizes the average prediction variance over the design region:
where M is defined as before.
Alias Optimality:
• seeks to minimize the aliasing between model effects and alias effects.
Specifically, let X
be the design matrix corresponding to the model effects, and let X2 be the matrix of alias
1
effects, and let
A =
be the alias matrix.
Then, alias optimality seeks to minimize the , subject to the D-Efficiency being greater than some
lower bound. In other words, it seeks to minimize the sum of the squared diagonal elements of .
Page 78

62 Examples Using the Custom Designer Chapter 2
Technical Discussion
Page 79

Chapter 3
Building Custom Designs
The Basic Steps
JMP can build a custom design that both matches the description of your engineering problem and remains
within your budget for time and material. Custom designs are general, flexible, and good for routine factor
screening or response optimization. To create these tailor-made designs, use the
found on the
This chapter introduces you to the steps you need to complete to build a custom design.
DOE menu or the Custom Design button found on the DOE panel of the JMP Starter.
Custom Design command
Page 80

Contents
Creating a Custom Design . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .65
Enter Responses and Factors into the Custom Designer . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .65
Describe the Model . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 69
Specifying Alias Terms. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 70
Select the Number of Runs . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .71
Understanding Design Evaluation . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 72
Specify Output Options . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 78
Make the JMP Design Table . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 79
Creating Random Block Designs. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 80
Creating Split Plot Designs . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 80
Creating Split-Split Plot Designs . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .81
Creating Strip Plot Designs . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .82
Special Custom Design Commands. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .83
Save Responses and Save Factors . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 84
Load Responses and Load Factors . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .85
Save Constraints and Load Constraints . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .85
Set Random Seed: Setting the Number Generator. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .85
Simulate Responses . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 86
Save X Matrix: Viewing the Number of Rows in the Moments Matrix and the Design Matrix (X) in the
Log . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 87
Optimality Criterion . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .88
Number of Starts: Changing the Number of Random Starts . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .88
Sphere Radius: Constraining a Design to a Hypersphere . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 90
Disallowed Combinations: Accounting for Factor Level Restrictions. . . . . . . . . . . . . . . . . . . . . . . . . 90
Advanced Options for the Custom Designer . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 92
Assigning Column Properties . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .93
Define Low and High Values (DOE Coding) for Columns . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 94
Set Columns as Factors for Mixture Experiments . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .95
Define Response Column Values . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 96
Assign Columns a Design Role . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 97
Identify Factor Changes Column Property . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 98
How Custom Designs Work: Behind the Scenes . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 99
Page 81

Chapter 3 Building Custom Designs 65
2
3
4
1
Creating a Custom Design
Creating a Custom Design
To begin, select DOE > Custom Design, or click the Custom Design button on the JMP Starter DOE page.
Then, follow the steps below.
• Enter responses and factors into the custom designer.
• Describe the model.
• Select the number of runs.
• Check the design diagnostics, if desired.
• Specify output options.
• Make the JMP design table.
The following sections describe each of these steps.
Enter Responses and Factors into the Custom Designer
How to Enter Responses
To enter responses, follow the steps in Figure 3.1.
1. To enter one response at a time, click Add Response, and then select a goal type. Possible goal types are
Maximize, Match Target, Minimize, or None.
2. (Optional) Double-click to edit the response name.
3. (Optional) Click to change the response goal.
4. Click to enter lower and upper limits and importance weights.
Figure 3.1 Entering Responses
Tip: To quickly enter multiple responses, click Number of Responses and enter the number of responses
you want.
Page 82

66 Building Custom Designs Chapter 3
Creating a Custom Design
Specifying Response Goal Types and Lower and Upper Limits
When entering responses, you can tell JMP that your goal is to obtain the maximum or minimum value
possible, to match a specific value, or that there is no response goal.
The following description explains the relationship between the goal type (step 3 in Figure 3.1) and the
lower and upper limits (step 4 in Figure 3.1):
• For responses such as strength or yield, the best value is usually the largest possible. A goal of
supports this objective.
•The
Minimize goal supports an objective of having the smallest value, such as when the response is
impurity or defects.
•The
Match Target goal supports the objective when the best value for a response is a specific target
value, such as a dimension for a manufactured part. The default target value is assumed to be midway
between the given lower and upper limits.
Note: If your target response is not equidistant from the lower and upper acceptable bounds, you can alter
the default target after you make a table from the design. In the data table, open the Column Info dialog for
the response column (Cols > Column Info) and enter the desired target value.
Understanding Response Importance Weights
To compute and maximize overall desirability, JMP uses the value you enter as the importance weight (step
4 in Figure 3.1) of each response. If there is only one response, then importance weight is unnecessary. With
two responses you can give greater weight to one response by assigning it a higher importance value.
Adding Simulated Responses, If Desired
If you do not have values for specific responses, you might want to add simulated responses to see a
prospective analysis in advance of real data collection.
1. Create the design.
2. Before you click
3. Click
Make Table to create the design table. The Y column contains values for simulated responses.
Make Table, click the red triangle icon in the title bar and select Simulate Responses.
4. For custom and augment designs, a window (Figure 3.2) appears along with the design data table. In
this window, enter values you want to apply to the
numbers you enter represent the coefficients in an equation. An example of such an equation, as shown
in Figure 3.2, would be, y =28+4X1+5X2+random noise, where the random noise is distributed with
mean zero and standard deviation one.
Maximize
Y column in the data table and click Apply. The
Page 83

Chapter 3 Building Custom Designs 67
1
2
3
4
5
6
Creating a Custom Design
Figure 3.2 In Custom and Augment Designs, Specify Values for Simulated Responses
How to Enter Factors
To enter factors, follow the steps in Figure 3.3.
1. To add one factor, click
Categorical, Blocking, Covariate, Mixture, Constant, or Uncontrolled. See “Types of Factors,” p. 67.
2. Click a factor and select
Add Factor and select a factor type. Possible factor types are Continuous,
Add Level to increase the number of levels.
3. Double-click a factor to edit the factor name.
4. Click to indicate that changing a factor’s setting from run to run is
Easy or Hard. Changing to Hard
will cause the resulting design to be a split plot design.
5. Click to enter or change factor values. To remove a level, click it, press the delete key on the keyboard,
then press the Return or Enter key on the keyboard.
6. To add multiple factors, type the number of factors in the
Add N Factors box, click the Add Factor
button, and select the factor type.
Figure 3.3 Entering Factors in a Custom Design
Types of Factors
When adding factors, click the
Continuous Continuous factors are numeric data types only. In theory, you can set a continuous
factor to any value between the lower and upper limits you supply.
Add Factor button and choose the type of factor.
Page 84

68 Building Custom Designs Chapter 3
Creating a Custom Design
Categorical
Either numeric or character data types. Categorical data types have no implied order. If
the values are numbers, the order is the numeric magnitude. If the values are character, the order is
the sorting sequence. The settings of a categorical factor are discrete and have no intrinsic order.
Examples of categorical factors are
Blocking Either numeric or character data types. Blocking factors are a special kind of categorical
machine, operator, and gender.
factor. Blocking factors differ from other categorical factors in that there is a limit to the number of
runs that you can perform within one level of a blocking factor.
Covariate Either numeric or character data types. Covariate factors are not controllable, but their
values are known in advance of an experiment.
Mixture Mixture factors are continuous factors that are ingredients in a mixture. Factor settings for a
run are the proportion of that factor in a mixture and vary between zero and one.
Constant Either numeric or character data types. Constant factors are factors whose values are fixed
during an experiment.
Uncontrolled Either numeric or character data types. Uncontrolled factors have values that cannot be
controlled during an experiment, but they are factors you want to include in the model.
Factors that are Easy, Hard, or Very Hard, to Change: Creating Optimal Split-Plot and Split-Split-Plot Designs
Split plot experiments are performed in groups of runs where one or more factors are held constant within a
group but vary between groups. In industrial experimentation this structure is desirable because certain
factors may be difficult and expensive to change from one run to the next. It is convenient to make several
runs while keeping such factors constant. Until now, commercial software has not supplied a general
capability for the design and analysis of these experiments.
To indicate the difficulty level of changing a factor’s setting, click in
for a given factor and select
results in a split-plot design and
Easy, Hard, or Very Hard from the menu that appears. Changing to Hard
Very Ha rd results in a split-split-plot design.
See “Creating Random Block Designs,” p. 80, for more details.
Defining Factor Constraints, If Necessary
Sometimes it is impossible to vary factors simultaneously over their entire experimental range. For example,
if you are studying the affect of cooking time and microwave power level on the number of kernels popped
in a microwave popcorn bag, the study cannot simultaneously set high power and long time without
burning all the kernels. Therefore, you have factors whose levels are constrained.
To define the constraints:
1. After you add factors and click
Continue, click the disclosure button ( on Windows and on
the Macintosh) to open the Define Factor Constraints panel.
2. Click the
Add Constraint button. Note that this feature is disabled if you have already controlled the
design region by entering disallowed combinations or chosen a sphere radius.
Changes column of the Factors panel
Page 85

Chapter 3 Building Custom Designs 69
Creating a Custom Design
Figure 3.4 Add Constraint
3. Specify the coefficients and their limiting value in the boxes provided, as shown to the right. When you
need to change the direction of the constraint, click on the default less than or equal button and select
the greater than or equal to direction.
4. To add another constraint, click the
Add Constraint button again and repeat the above steps.
Describe the Model
Initially, the Model panel lists only the main effects corresponding to the factors you entered, as shown in
Figure 3.5. However, you can add factor interactions or powers of continuous factors to the model. For
example, to add all the two-factor interactions and quadratic effects at once, click the
RSM button.
Figure 3.5 Add Terms
Table 3.1 summarizes the ways to add specific factor types to the model.
Ta bl e 3 .1 How to Add Terms to a Model
Action Instructions
Add interaction terms involving selected
factors. If none are selected, JMP adds
all of the interactions to the specified
order.
Click the
or
you click
Interactions button and select 2nd, 3rd, 4th,
5th. For example, if the factors are X1 and X2 and
Interactions > 2nd, X1*X2 is added to the list
of model terms.
Page 86

70 Building Custom Designs Chapter 3
Creating a Custom Design
Ta bl e 3 .1 How to Add Terms to a Model (Continued)
Action Instructions
Add all second-order effects, including
two-factor interactions and quadratic
effects
Add selected cross product terms 1. Highlight the factor names.
Add powers of continuous factors to the
model effects
Specifying Alias Terms
You can investigate the aliasing between the model terms and terms you specify in the Alias Terms panel.
For example, suppose you specify a design with three main effects in six runs, and you want to see how those
main effects are aliased by the two-way interactions and the three-way interaction. In the Alias Terms panel,
specify the interactions as shown in Figure 3.6. Also, specify six runs in the Design Generation panel.
Figure 3.6 Alias Terms
Click the RSM button. The design now uses
I-Optimality criterion rather than D-Optimality
criterion.
2. Highlight term(s) in the model list.
3. Click the
Click the
5th.
Cross button.
Powers button and select 2nd, 3rd, 4th, or
After you click the Make Design button at the bottom of the Custom Design panel, open the Alias Matrix
panel in the Design Evaluation panel to see the alias matrix. See Figure 3.7.
Figure 3.7 Aliasing
Page 87

Chapter 3 Building Custom Designs 71
Creating a Custom Design
In this example, all the main effects are partially aliased with two of the interactions. Also see “The Alias
Matrix (Confounding Pattern),” p. 76.
Select the Number of Runs
The Design Generation panel (Figure 3.8) shows the minimum number of runs needed to perform the
experiment based on the effects you’ve added to the model (two main effects in the example above). It also
shows alternate (default) numbers of runs, or lets you choose your own number of runs. Balancing the cost
of each run with the information gained by extra runs you add is a judgment call that you control.
Figure 3.8 Options for Selecting the Number of Runs
The Design Generation panel has these options for selecting the number of runs you want:
Minimum is the smallest number of terms that can create a design. When you use Minimum, the
resulting design is saturated (no degrees of freedom for error). This is an extreme and risky choice,
and is appropriate only when the cost of extra runs is prohibitive.
Default is a custom design suggestion for the number of runs. This value is based on heuristics for
creating balanced designs with a few additional runs above the minimum.
User Specified is a value that specifies the number of runs you want. Enter that value into the
Number of Runs text box.
Note: In general, the custom design suggests a number of runs that is the smallest number that can be
evenly divided by the number of levels of each of the factors and is larger than the minimum possible sample
size. For designs with factors at two levels only, the default sample size is the smallest power of two larger
than the minimum sample size.
When the Design Generation panel shows the number of runs you want, click
Make Design.
Page 88

72 Building Custom Designs Chapter 3
Creating a Custom Design
Understanding Design Evaluation
After making the design, you can preview the design and investigate details by looking at various plots and
tables that serve as design diagnostic tools.
Although different tools are available depending on the model you specify, most designs display
• the Prediction Variance Profile Plot
• the Fraction of Design Space Plot
• the Prediction Variance Surface Plot
• the Relative Variance of Coefficients and Power Table
•the Alias Matrix
•Design Diagnostic Table
These diagnostic tools are outline nodes beneath the Design Evaluation panel, as shown in Figure 3.9. JMP
always provides the Prediction Variance Profile, but the Prediction Surface Plot only appears if there are two
or more variables.
Figure 3.9 Custom Design Evaluation and Diagnostic Tools
The Prediction Variance Profile
The example in Figure 3.10 shows the prediction variance profile for a response surface model (RSM) with
2 variables and 8 runs. To see a response surface design similar to this:
1. Chose
DOE > Custom Design.
2. In the Factors panel, add 2 continuous factors.
3. Click
Continue.
4. In the Model panel, click
RSM.
Page 89

Chapter 3 Building Custom Designs 73
Creating a Custom Design
5. Click Make Design.
6. Open the Prediction Variance Profile.
Figure 3.10 A Factor Design Layout For a Response Surface Design with 2 Variables
The prediction variance for any factor setting is the product of the error variance and a quantity that
depends on the design and the factor setting. Before you collect the data the error variance is unknown, so
the prediction variance is also unknown. However, the ratio of the prediction variance to the error variance
is not a function of the error variance. This ratio, called the relative variance of prediction, depends only on
the design and the factor setting and can be calculated before acquiring the data. The prediction variance
profile plots the relative variance of prediction as a function of each factor at fixed values of the other factors
After you run the experiment, collect the data, and fit the model, you can estimate the actual variance of
prediction at any setting by multiplying the relative variance of prediction by the mean squared error (MSE)
of the least squares fit.
It is ideal for the prediction variance to be small throughout the allowable regions of the factors. Generally,
the error variance drops as the sample size increases. Comparing the prediction variance profilers for two
designs side-by-side, is one way to compare two designs. A design that has lower prediction variance on the
average is preferred.
In the profiler, drag the vertical lines in the plot to change the factor settings to different points. Dragging
the lines reveals any points that have prediction variances that are larger than you would like.
Another way to evaluate a design, or to compare designs, is to try and minimize the maximum variance. You
can use the
Maximize Desirability command on the Prediction Variance Profile title bar to identify the
maximum prediction variance for a model. Consider the Prediction Variance profile for the two-factor RSM
model shown in Figure 3.11. The plots on the left are the default plots. The plots on the right identify the
factor values where the maximum variance (or worst-case scenario) occur, which helps you evaluate the
acceptability of the model.
Page 90

74 Building Custom Designs Chapter 3
Creating a Custom Design
Figure 3.11 Find Maximum Prediction Variance
The Fraction of Design Space Plot
The Fraction of Design Space plot is a way to see how much of the model prediction variance lies above (or
below) a given value. As a simple example, consider the Prediction Variance plot for a single factor quadratic
model, shown on the left in Figure 3.12. The Prediction Variance plot shows that 100% of the values are
smaller than 0.5. You can move the vertical trace and also see that all the values are above 0.332. The
Fraction of Design Space plot displays the same information. The X axis is the proportion of prediction
variance values, ranging from 0 to 100%, and the Y axis is the range of prediction variance values. In this
simple example, the Fraction of Design plot verifies that 100% of the values are below 0.5 and 0% of the
values are below approximately 0.3. You can use the crosshair tool and find the percentage of values for any
value of the prediction variance. The example to the right in Figure 3.12 shows that 75% of the prediction
variance values are below approximately 0.46.
The Fraction of Design space is most useful when there are multiple factors. It summarizes the prediction
variance, showing the fractional design space for all the factors taken together.
Figure 3.12 Variance Profile and Fraction of Design Space
Page 91

Chapter 3 Building Custom Designs 75
Creating a Custom Design
The Prediction Variance Surface
When there are two or more factors, the Prediction Variance Surface plots the surface of the prediction
variance for any two variables. This feature uses the
Graph > Surface Plot platform in JMP, and has all its
functionality. Drag on the plot to rotate and change the perspective. Figure 3.13 shows the Prediction
Variance Surface plot for a two-factor RSM model. The factors are on the x and y axes, and the prediction
variance is on the z axis. You can clearly see that there are high and low variance areas for both factors.
Compare this plot to the Prediction Variance Profile shown in Figure 3.11.
Figure 3.13 Prediction Variance Surface Plot for Two-Factor RSM Model
You can find complete documentation for the Surface Plot platform in Basic Analysis and Graphing.
The Relative Variance of Coefficients and Power Table
Before clicking
Make Table in the custom designer, click the disclosure button ( on Windows and
on the Macintosh) to open Design Evaluation and then again to open the Relative Variance of
Coefficients table.
The Relative Variance of Coefficients table (Figure 3.14) shows the relative variance of all the coefficients for
the example RSM custom design (see Figure 3.10). The variances are relative to the error variance, which is
unknown before the experiment, and is assumed to be one. Once you complete the experiment and have an
estimate for the error variance, you can multiply it by the relative variance to get the estimated variance of
the coefficient. The square root of this value should match the standard error of prediction for the
coefficient when you fit a model using
The
Power column shows the power of the design as specified to detect effects of a certain size. In the text
Analyze > Fit Model.
edit boxes, you can change the alpha level of the test and the magnitude of the effects compared to the error
standard deviation. The alpha level edit box is called Significance Level. The magnitude of the effects edit
box is called Signal to Noise Ratio. This is the ratio of the absolute value of the regression parameter to
sigma (the square root of the error variance).
Page 92
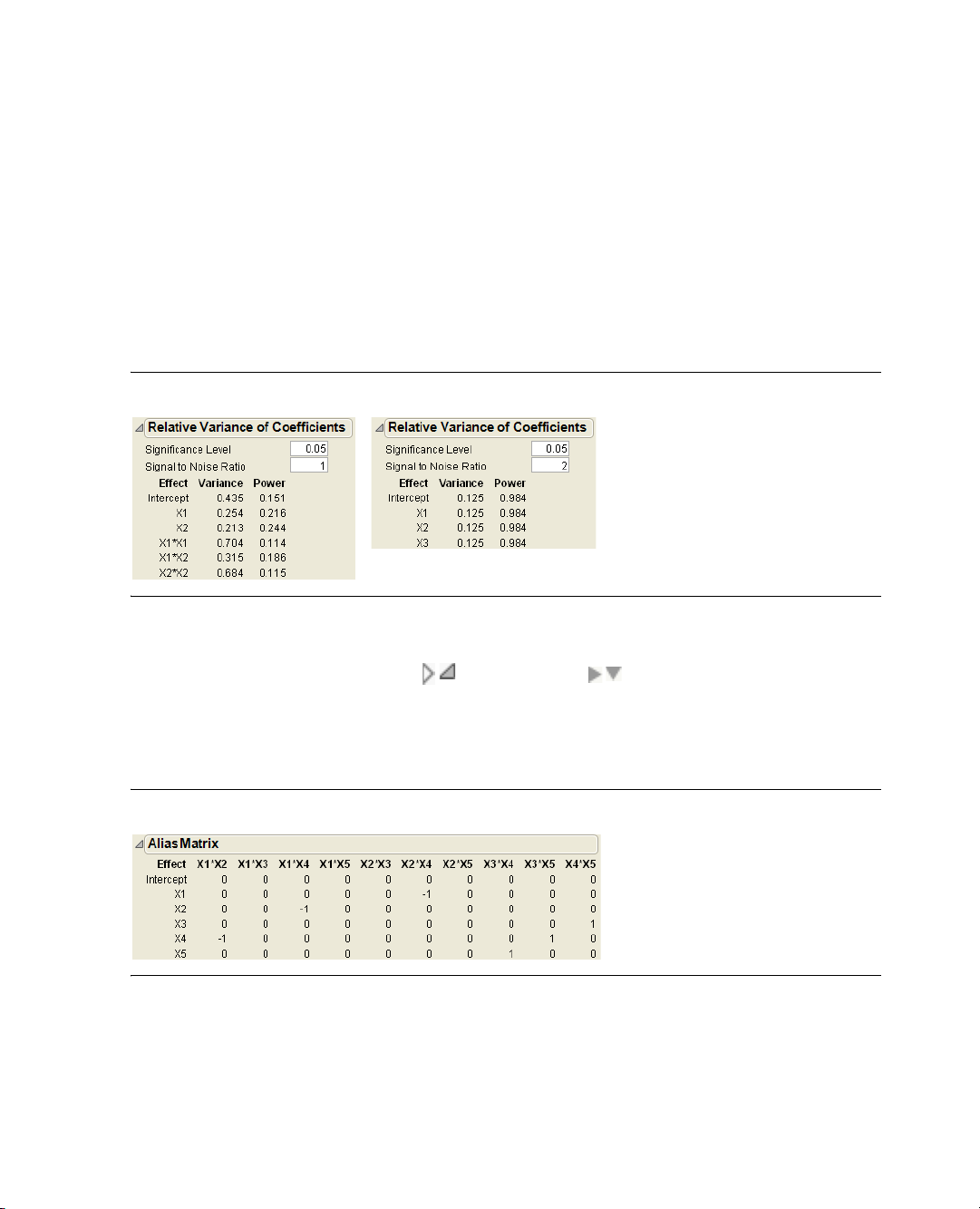
76 Building Custom Designs Chapter 3
Creating a Custom Design
If you enter a smaller alpha (requiring a more significant test), then the power falls. If you increase the
magnitude of the effect you want to detect, the power rises.
The power reported is the probability of finding a significant model parameter if the true effect is Signal to
Noise Ratio times sigma. The Relative Variance of Coefficients table on the left in Figure 3.14 shows the
results for the two-factor RSM model.
As another example, suppose you have a 3-factor 8-run experiment with a linear model and you want to
detect any regression coefficient that is twice as large as the error standard deviation, with an alpha level of
0.05. The Relative Variance of Coefficients table on the right in Figure 3.14 shows that the resulting power
is 0.984 for all the parameters.
Figure 3.14 Table of Relative Variance of Coefficients
The Alias Matrix (Confounding Pattern)
Click the Alias Matrix disclosure button ( on Windows and on the Macintosh) to open the alias
matrix (Figure 3.15).
The alias matrix shows the aliasing between the model terms and the terms you specify in the Alias Terms
panel (see “Specifying Alias Terms,” p. 70). It allows you to see the confounding patterns.
Figure 3.15 Alias Matrix
Color Map on Correlations
The Color Map On Correlations panel (see Figure 3.16) shows the correlations between all model terms
and alias terms you specify in the Alias Terms panel (see “Specifying Alias Terms,” p. 70). The colors
correspond to the absolute value of the correlations.
Page 93

Chapter 3 Building Custom Designs 77
D-efficiency =
100
1
N
D
-------
X′ X
1 p/
A-efficiency =
100
p
trace N
D
X′ X()
1–
()
-----------------------------------------------
G-efficiency =
100
p
N
D
------
σ
M
---------- -
Creating a Custom Design
Figure 3.16 Color Map of Correlations
The Design Diagnostics Table
Open the Design Diagnostics outline node to display a table with relative D-, G-, and A-efficiencies, average
variance of prediction, and length of time to create the design. The design efficiencies are computed as
follows:
where
• N
is the number of points in the design
D
• p is the number of effects in the model including the intercept
• σ
is the maximum standard error for prediction over the design points.
M
Page 94

78 Building Custom Designs Chapter 3
Creating a Custom Design
These efficiency measures are single numbers attempting to quantify one mathematical design characteristic.
While the maximum efficiency is 100 for any criterion, an efficiency of 100% is impossible for many design
problems. It is best to use these design measures to compare two competitive designs with the same model
and number of runs rather than as some absolute measure of design quality.
Figure 3.17 Custom Design Showing Diagnostics
Specify Output Options
Use the Output Options panel to specify how you want the output data table to appear.
Figure 3.18 Output Options Panel
Run Order lets you designate the order you want the runs to appear in the data table when it is created.
Choices are:
Keep the Same the rows (runs) in the output table will appear as they do in the Design panel.
Sort Left to Right the rows (runs) in the output table will appear sorted from left to right.
Randomize the rows (runs) in the output table will appear in a random order.
Sort Right to Left the rows (runs) in the output table will appear sorted from right to left.
Randomize within Blocks the rows (runs) in the output table will appear in random order within the
blocks you set up.
Add additional points using options from Make JMP Table from design plus:
Number of Center Points: Specifies additional runs placed at the center of each continuous factor’s
range.
Page 95

Chapter 3 Building Custom Designs 79
Creating a Custom Design
Number of Replicates:
centerpoints. Type the number of times you want to replicate the design in the associated text box.
One replicate doubles the number of runs.
Make the JMP Design Table
When the Design panel shows the layout you want, click Make Table. Parts of the table contain
information you might need to continue working with the table in JMP. The upper-left of the design table
can have one or more of the following scripts:
• a Screening script runs the
generated design.
• a Model script runs the
• a constraint script that shows any model constraints you entered in the Define Factor Constraints panel
of the Custom Design dialog.
•a DOE Dialog
script that recreates the dialog used to generate the design table, and regenerates the
design table.
Figure 3.19 Example Design Table
Analyze > Fit Model platform with the model appropriate for the design.
Specify the number of times to replicate the entire design, including
Analyze > Modeling > Screening platform when appropriate for the
1. This area identifies the design type that generated the table. Click Custom Design to edit.
2. Model is a script. Click the red triangle icon and select
Run Script to open the Fit Model dialog, which
is used to generate the analysis appropriate to the design.
3. DOE Dialog is a script. Click the red triangle icon and select
Run Script to recreate the DOE Custom
Dialog and generate a new design table.
Page 96

80 Building Custom Designs Chapter 3
Creating Random Block Designs
Creating Random Block Designs
It is often necessary to group the runs of an experiment into blocks. Runs within a block of runs are more
homogeneous than runs in different blocks. For example, the experiment described in Goos (2002),
describes a pastry dough mixing experiment that took several days to run. It is likely that random day-to-day
differences in environmental variables have some effect on all the runs performed on a given day. Random
block designs are useful in situations like this, where there is a non-reproducible shock to the system
between each block of runs. In Goos (2002), the purpose of the experiment was to understand how certain
properties of the dough depend on three factors: feed flow rate, initial moisture content, and rotational
screw speed. It was only possible to conduct four runs a day. Because day-to-day variation was likely, it was
important to group the runs so that this variation would not compromise the information about the three
factors. Thus, blocking the runs into groups of four was necessary. Each day's experimentation was one
block. The factor, Day, is an example of a random block factor.
To create a random block, use the custom design and enter responses and factors, and define your model as
usual. In the Design Generation panel, check the Group runs into random blocks of size check box and
enter the number of runs you want in each block. When you select or enter the sample size, the number of
runs specified are assigned to the blocks.
Figure 3.20 Assigning Runs to Blocks
In this example, the Design Generation Panel shown here designates four runs per block, and the number of
runs (16) indicates there will be four days (blocks) of 4 runs. If the number of runs is not an even multiple
of the random block size, some blocks will have a fewer runs than others.
Creating Split Plot Designs
Split plot experiments happen when it is convenient to run an experiment in groups of runs (called whole
plots) where one or more factors stay constant within each group. Usually this is because these factors are
difficult or expensive to change from run to run. JMP calls these factors
usually how split plotting arises in industrial practice.
In a completely randomized design, any factor can change its setting from one run to the next. When
certain factors are hard to change, the completely randomized design may require more changes in the
settings of hard-to-change factors than desired.
Hard to change because this is
Page 97

Chapter 3 Building Custom Designs 81
X′V1–X
Creating Split-Split Plot Designs
If you know that a factor or two are difficult to change, then you can set the Changes setting of a factor from
the default of
Easy to Hard. Before making the design, you can set the number of whole plots you are
willing to run.
For an example of creating a split plot design, see “Creating a Design with Two Hard-to-Change Factors:
Split Plot,” p. 54.
To create a split plot design using the custom designer:
1. In the factors table there is a column called
however, you click in the changes area for a factor, you can choose to make the factor
2. Once you finish defining the factors and click continue, you see an edit box for supplying the number of
whole plots. You can supply any value as long as it is above the minimum necessary to fit all the model
parameters. You can also leave this field empty. In this case, JMP chooses a number of whole plots to
minimize the omnibus uncertainty of the fixed parameters.
Note: If you enter a missing value in the Number of Whole Plots edit box, then JMP considers many
different numbers of whole plots and chooses the number that maximizes the information about the
coefficients in the model. It maximizes the determinant of where V
matrix of the responses. The matrix, V, is a function of how many whole plots there are, so changing the
number of whole plots changes V, which can make a difference in the amount of information a design
contains.
To create a split plot design every time you use a certain factor, save steps by setting up that factor to be
“hard” in all experiments. See “Identify Factor Changes Column Property,” p. 98, for details.
Creating Split-Split Plot Designs
Split-split plot designs are a three stratum extension of split plot designs. Now there are factors that are
Very-Hard-to-change, Hard-to-change, and Easy-to-change. Here, in the top stratum, the Very-Hard-tochange factors stay fixed within each whole plot. In the middle stratum the Hard-to-change factors stay
fixed within each subplot. Finally, the Easy-to-change factors may vary (and should be reset) between runs
within a subplot. This structure is natural when an experiment covers three processing steps. The factors in
the first step are Very-Hard-to-change in the sense that once the material passes through the first processing
stage, these factor settings are fixed. Now the material passes to the second stage where the factors are all
Hard-to-change. In the third stage, the factors are Easy-to-change.
Changes. By default, changes are Easy for all factors. If,
Hard to change.
-
1
is the inverse of the variance
Schoen (1999) provides an example of three-stage processing involving the production of cheese that leads
to a split-split plot design. The first processing step is milk storage. Typically milk from one storage facility
provides the raw material for several curds processing units—the second processing stage. Then the curds are
further processed to yield individual cheeses.
In a split-split plot design the material from one processing stage passes to the next stage in such a way that
nests the subplots within a whole plot. In the example above, milk from a storage facility becomes divided
into two curds processing units. Each milk storage tank provided milk to a different set of curds processors.
So, the curds processors were nested within the milk storage unit.
Page 98

82 Building Custom Designs Chapter 3
Creating Strip Plot Designs
Figure 3.21 shows an example of how factors might be defined for the cheese processing example.
Figure 3.21 Example of Split-Split Response and Factors in Custom Designer Dialog
Creating Strip Plot Designs
In a strip plot design it is possible to reorder material between processing stages. Suppose units are labelled
and go through the first stage in a particular order. If it is possible to collect all the units at the end of the
first stage and reorder them for the second stage process, then the second stage variables are not nested
within the blocks of the first stage variables. For example, in semiconductor manufacturing a boat of wafers
may go through the first processing step together. However, after this step, the wafers in a given boat may be
divided among many boats for the second stage.
To set up a strip plot design, enter responses and factors as usual, designating factors as Very Hard, Hard, or
Easy to change. Then, in the Design Generation panel, check the box that says
can vary independently of Very Hard to change factors
, as shown in Figure 3.22. Note that the Design
Hard to change factors
Generation panel specified 6 whole plots, 12 subplots, and 24 runs.
When you click
Make Design, the design table on the right in Figure 3.22 lists the run with subplots that
are not nested in the whole plots.
Page 99

Chapter 3 Building Custom Designs 83
Special Custom Design Commands
Figure 3.22 Example of Strip Split Factors and Design Generation panel in Custom Designer Dialog
Special Custom Design Commands
After you select DOE > Custom Design, click the red triangle icon on the title bar to see the list of
commands available to the Custom designer (Figure 3.23). The commands found on this menu vary,
depending on which DOE command you select. However, the commands to save and load responses and
factors, the command to set the random seed, and the command to simulate responses are available to all
designers. You should examine the red triangle menu for each designer you use to determine which
commands are available. If a designer has additional commands, they are described in the appropriate
chapter.
Page 100

84 Building Custom Designs Chapter 3
Special Custom Design Commands
Figure 3.23 Click the Red Triangle Icon to Reveal Commands
The following sections describe these menu commands and how to use them.
Save Responses and Save Factors
If you plan to do further experiments with factors and/or responses to which you have given meaningful
names and values, you can save them for later use.
To save factors or responses:
1. Select a design type from the DOE menu.
2. Enter the factors and responses into the appropriate panels (see “Enter Responses and Factors into the
Custom Designer,” p. 65, for details).
3. Click the red triangle icon on the title bar and select
Save Responses creates a data table containing a row for each response with a column called
Response Name that identifies the responses. Four additional columns identify more information
about the responses:
Save Factors creates a data table containing a column for each factor and a row for each factor level.
The columns have two column properties (noted with asterisks icons in the column panel). These
properties include:
Design Role that identifies the factor as a DOE factor and lists its type (continuous, categorical,
blocking, and so on).
Factor Changes that identifies how difficult it is to change the factor level. Factor Changes options
are
Easy, Hard, and Very H ard.
4. Save the data table.
Lower Limit, Upper Limit, Response Goal, and Importance.
Save Responses or Save Factors.
 Loading...
Loading...