Sample to Insight__
45114
QIAGEN GmbH
QIAGEN Strasse 1,
R1
April 2021
artus
SARS-CoV-2 Prep&Amp
UM Kit Instructions for Use
(Handbook)
®
768
3072
Version 1
For In Vitro Diagnostic Use Only. Validation of this test has not been reviewed
by the FDA. Review under the EUA program is pending. The test is distributed in
accordance with the guidance on Policy for Coronavirus Disease-2019 Tests
During the Public Health Emergency, Section IV.C.2.
℞
For Prescription Use Only
For use on Rotor-Gene
®
Q MDx and ABI® 7500 Fast Dx instruments
40, 4511449
40724 Hilden, GERMANY

2
artus
SARS-CoV-2 Prep&Amp UM Kit Instructions for Use (Handbook) 04/2021
Contents
Intended Use .............................................................................................................. 4
Description and Principle ............................................................................................. 5
Pathogen information ........................................................................................ 5
Summary and explanation ................................................................................. 6
Materials Provided ...................................................................................................... 9
Kit contents ...................................................................................................... 9
Kit components .............................................................................................. 10
Platforms and software .................................................................................... 11
Materials Required but Not Provided .......................................................................... 12
Consumables ................................................................................................. 12
Equipment ..................................................................................................... 12
Warnings and Precautions ......................................................................................... 13
Safety information .......................................................................................... 13
Precautions .................................................................................................... 14
Reagent Storage and Handling .................................................................................. 15
Specimen Transport, Storage and Handling ................................................................. 15
Specimen collection, transport and storage ........................................................ 15
Protocol: Sample preparation and SARS-CoV-2 detection on the RGQ MDx .................... 16
Protocol: Sample Preparation and SARS-CoV-2 Detection on ABI 7500 Fast Dx ............... 21
Results ..................................................................................................................... 25
Interpretation of Results .............................................................................................. 27

artus
SARS-CoV-2 Prep&Amp UM Kit Instructions for Use (Handbook) 04/2021
3
Limitations ................................................................................................................ 29
Performance ............................................................................................................. 30
Analytical sensitivity (Limit of detection) ............................................................. 30
Analytical specificity studies (Inclusivity and exclusivity/cross-reactivity) ................ 30
Interfering substances ...................................................................................... 36
Precision ....................................................................................................... 37
Clinical performance ...................................................................................... 38
References ............................................................................................................... 40
Troubleshooting Guide .............................................................................................. 41
Symbols ................................................................................................................... 43
Contact Information .................................................................................................. 44
Ordering Information ................................................................................................ 45
Document Revision History ......................................................................................... 46

4
artus
SARS-CoV-2 Prep&Amp UM Kit Instructions for Use (Handbook) 04/2021
Intended Use
The
artus
SARS-CoV-2 Prep&Amp UM Kit is a real-time RT-PCR test intended for the qualitative
detection of nucleic acid from the SARS-CoV-2 in nasopharyngeal swabs (NPS), nasal swabs,
and oropharyngeal swabs from individuals with signs and symptoms of infection who are
suspected of COVID-19. Testing is limited to laboratories certified under the Clinical
Laboratory Improvement Amendments of 1988 (CLIA), 42 U.S.C. §263a, to perform high
complexity tests or by similarly qualified non-U.S. laboratories.
Results are for the identification of SARS-CoV-2 RNA. The SARS-CoV-2 RNA is generally
detectable in NPS, nasal swabs, and oropharyngeal swabs during the acute phase of
infection. Positive results are indicative of active infection. Laboratories within the United States
and its territories are required to report all positive results to the appropriate public health
authorities.
Negative results do not preclude SARS-CoV-2 infection and should not be used as the sole
basis for patient management decisions. Negative results must be combined with clinical
observations, patient history, and epidemiological information.
artus
The
personnel specifically instructed and trained in the techniques of real-time PCR and
diagnostic procedures. Validation of this test has not been reviewed by the FDA. Review under
the EUA program is pending. The test is distributed in accordance with the guidance on Policy
for Coronavirus Disease-2019 Tests During the Public Health Emergency, Section IV.C.2.
The
MDx System or the ABI 7500 Fast Dx as RT-PCR instruments.
SARS-CoV-2 Prep&Amp UM Kit is intended for use by trained clinical laboratory
artus
SARS-CoV-2 PCR assay test is intended to be used with the Rotor-Gene Q (RGQ)
in vitro

artus
SARS-CoV-2 Prep&Amp UM Kit Instructions for Use (Handbook) 04/2021
5
Description and Principle
Pathogen information
Coronaviruses, a genus in the family
RNA viruses that cause highly virulent disease in humans and domestic animals (1).
Coronaviruses are known to infect humans account for one-third of common cold infections
and are also a well-known cause of nosocomial upper respiratory infections in premature
infants (2).
A novel member of the coronavirus family caused an outbreak of respiratory disease in
Wuhan City in China (1, 3). First named novel coronavirus (2019-nCoV), SARS-CoV-2 differs
from the SARS-CoV (1, 3), which was responsible for the 2003 outbreak, and the MERS-CoV,
which has been circulating in the Middle East since 2012. SARS-CoV-2 is the causative agent
of COVID-19. The SARS-CoV-2 RNA is detectable during the early and acute phases of the
infection from various upper respiratory tract specimens (nasal, oropharyngeal, and
nasopharyngeal swabs) (3).
The SARS-CoV-2 Prep&Amp UM assay targets 2 viral genes (N1 and N2 genes) detected with
the same fluorescence channel. The two gene targets are not differentiated, and amplification
of either or both gene targets leads to a fluorescence signal. Positive results are indicative of
the presence of the SARS-CoV-2 virus, but do not rule out co-infection with other pathogens.
On the other hand, negative RT-PCR results do not exclude a possible infection.
Coronaviridae
, are large enveloped, positive-stranded

6
artus
SARS-CoV-2 Prep&Amp UM Kit Instructions for Use (Handbook) 04/2021
Summary and explanation
The
artus
SARS-CoV-2 Prep&Amp UM Kit constitutes a ready-to-use system with a simple sample
preparation step followed by detection of the SARS-CoV-2 RNA using RT-PCR on either the
RGQ MDx system or on ABI 7500 Fast Dx platform (Figure 1). The SARS-CoV-2 UM Amp Buffer
contains reagents and enzymes for the specific amplification of a 72 base pair (bp) and a 67 bp
regions of the SARS-CoV-2 RNA genome and for their direct detection in the “Green” fluorescence
channel of the RGQ MDx instruments and with the fluorescent filter A/1 of the ABI 7500 Fast Dx.
The Primers and Probes mix of the
artus
SARS-CoV-2 Prep&Amp UM Kit also contains the
oligonucleotides required for the RNAse P amplifications. When detected in the “Yellow”
fluorescence channel of the RGQ MDx instrument or with the fluorescent filter B/2 of the ABI 7500
Fast Dx, those amplicons assure that enough biological sample has been collected on the swab.
This control is critical to ensure the presence of biological samples in SARS-CoV-2 negative samples.
An amplification should always be detectable; otherwise, it questions the sample quality.
The
artus
SARS-CoV-2 Prep&Amp UM Kit also contains a third heterologous amplification system
to reveal possible RT-PCR inhibition. This is detected as an internal RNA control (IC) in the “Red”
fluorescence channel of the RGQ MDx instruments and with the fluorescence filter E/5 of the
ABI 7500 Fast Dx. Because the IC is included in the SARS--CoV--2 Amp Primers Mix, its
amplification should be constant, unless an RT-PCR inhibitor is present in the sample or in the RT-PCR
reaction, which delays or prevents amplification.
External positive and negative controls (SARS-CoV-2 Positive Control and nuclease-free water used
as NTC, respectively) are supplied in the
artus
SARS-CoV-2 Prep&Amp UM Kit to attest of the
performance of the PCR step. A no extraction control (SARS-CoV-2 UM Prep Buffer used as NEC)
is strongly recommended to verify the absence of RT-PCR inhibitors in the preparation buffer.
Taken together, the efficiency of the reverse transcription and the PCR steps are monitored by these
controls.
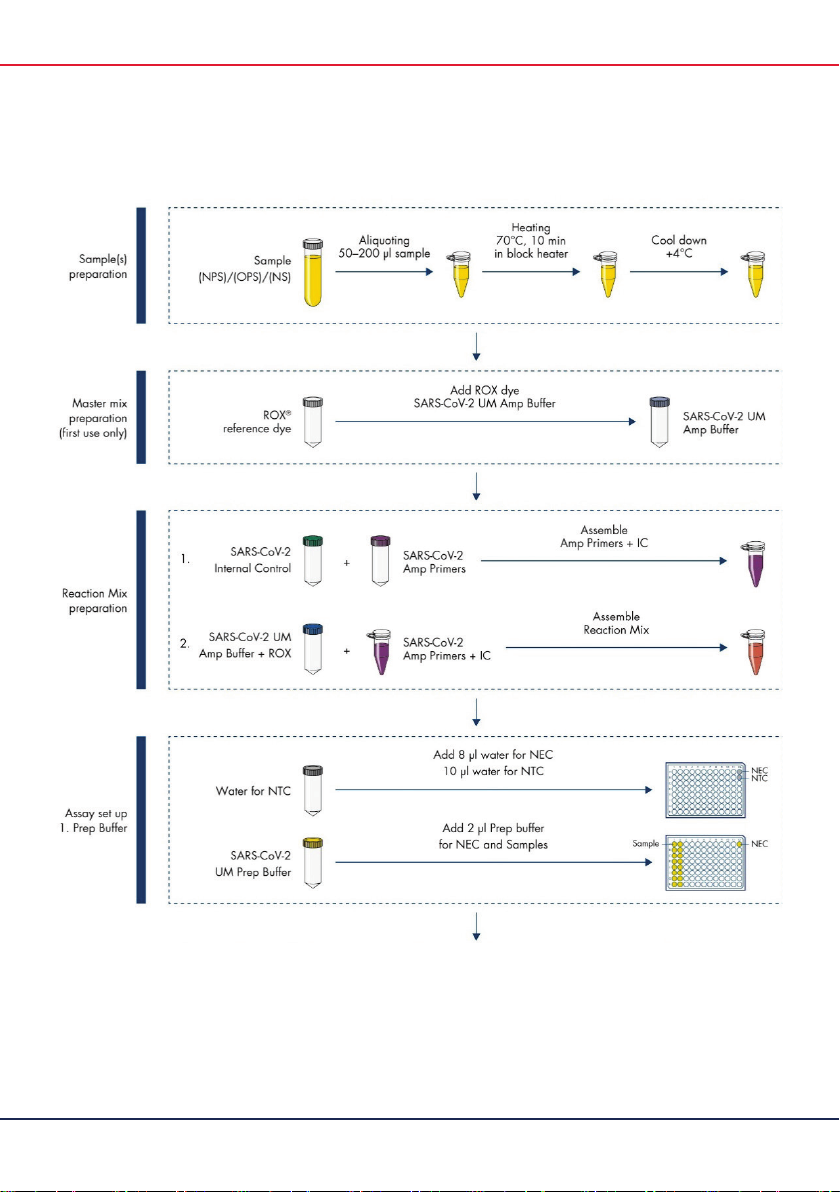
artus
SARS-CoV-2 Prep&Amp UM Kit Instructions for Use (Handbook) 04/2021
7
artus
SARS-CoV-2 Prep&Amp UM Kit Workflow
(Continued on next page)
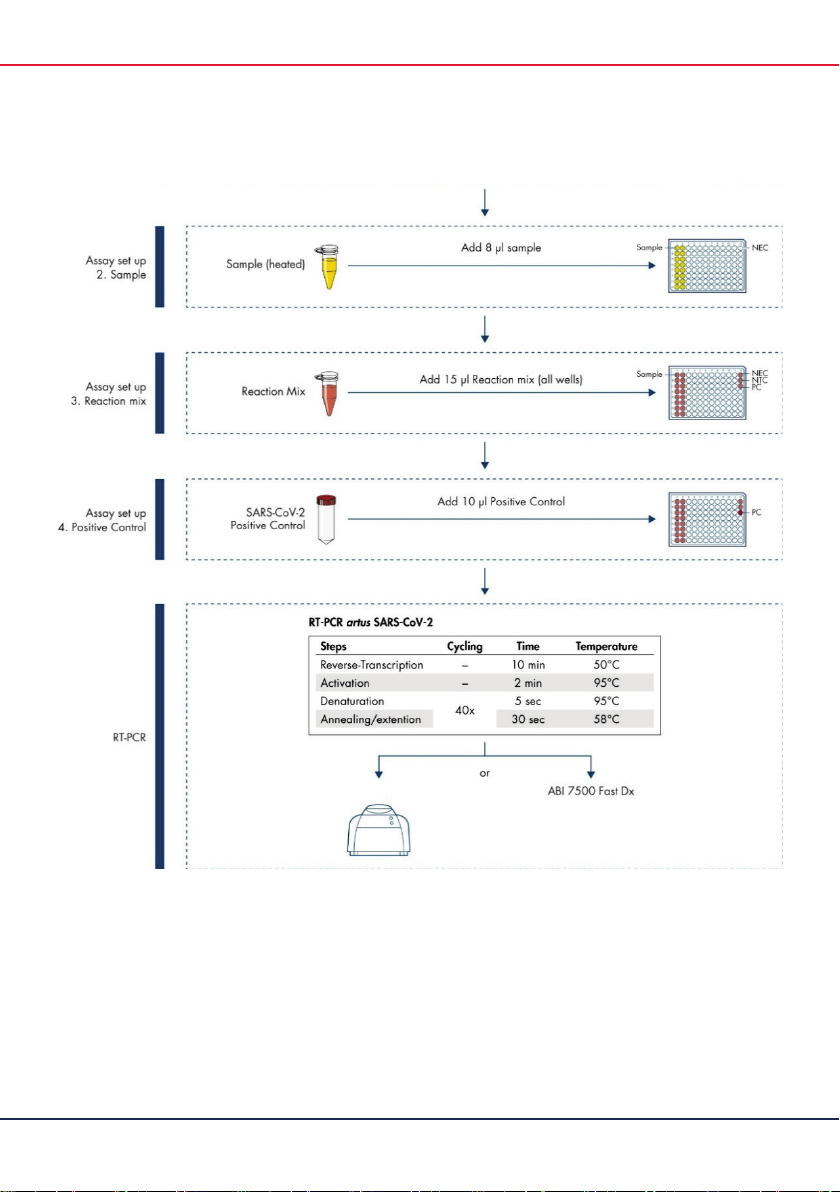
8
artus
SARS-CoV-2 Prep&Amp UM Kit Instructions for Use (Handbook) 04/2021
RGQ MDx
(Continued from previous page)
Figure 1.
artus
SARS-CoV-2 Prep&Amp UM Kit workflow.

artus
SARS-CoV-2 Prep&Amp UM Kit Instructions for Use (Handbook) 04/2021
9
artus
SARS-CoV-2 Prep&Amp UM Kit
Materials Provided
Kit contents
Catalog no.
Number of reactions
Tube color Lid Color Identity Tube ID Volume (µl) Volume (µl)
Clear
Clear
Clear
Clear
Clear Red SARS-CoV-2 Positive Control Positive Control 1 x 220
Clear
Clear
Yellow
Blue
Purple
Green
Clear
Clear
SARS-CoV-2 UM Prep Buffer Preparation Buffer 2 x 930
SARS-CoV-2 UM Amp Buffer
SARS-CoV-2 Amp Primers
SARS-CoV-2 Internal Control
Water for NTC Water (NTC) 1 x 1900
ROX Reference Dye ROX Dye 1 x 210
Master Mix
Primers and Probes
Internal Control (IC)
4511440
768
4 x 1440
4 x 1680
1 x 1390
4511449
3072
8 x 930
16 x 1440
16 x 1680
4 x 1390
4 x 220
4 x 1900
4 x 210

10
artus
SARS-CoV-2 Prep&Amp UM Kit Instructions for Use (Handbook) 04/2021
Kit components
Reagents
In each tube, the reagent volumes have been optimized for 8 batches of 96 samples (for the
768 reactions kit) or 32 batches of 96 reactions (for the 3072 reactions kit), including a
positive control (PC), a no template control (NTC), and a no extraction control (NEC).
Fewer or a greater number of samples may be run, but there will be sub-optimal reagent usage.
It is recommended to avoid multiple freeze−thaw cycles. Reagents may be aliquoted to avoid
multiple freeze−thaw cycles.
Primers and probes
Primers and probes targeting the SARS-CoV-2 sequences are based on the primers and probes
designed by the Centers for Disease Control and Prevention (CDC).
Controls and calibrators
The assay contains 5 controls to monitor the RT-PCR efficiency.
Internal control (IC): The internal control is a single-strand IVT RNA that verifies the presence of
contaminants that could inhibit the reverse transcription. The internal control also monitors the
reverse transcription efficiency in the no template control (NTC) and no extraction control (NEC).
No template control (NTC): The no template control is composed of nuclease-free water. It is
added to the PCR plate to verify introduction of contaminants during the PCR plate preparation
that could lead to misinterpretation of the SARS-CoV-2 targets.
Positive control (PC): The positive control is a double-strand DNA amplified with the
SARS-CoV-2 Primers and Probes (P&P mix). Its detection verifies the efficiency of the reagent
involved in the PCR amplification step.

artus
SARS-CoV-2 Prep&Amp UM Kit Instructions for Use (Handbook) 04/2021
11
No extraction step (NEC): The no extraction control is composed of the SARS-CoV-2 UM Prep
Buffer. It is processed in parallel with the clinical samples to verify introduction of contaminants
during the sample preparation that could lead to misinterpretation of the SARS-CoV-2 targets.
Sampling Control: The Sampling Control detects the RNAse P gene and is critical to ensure
the presence of biological samples in SARS-CoV-2 negative samples. Amplification of the
sampling control should always be detectable; otherwise, it questions the sample quality.
Platforms and software
Prior to use, ensure that instruments have been maintained and calibrated according to the
manufacturer’s recommendations. This kit can be used in two workflows that require the use of the
Rotor-Gene Q MDx or of the ABI 7500 Fast Dx instruments and their appropriate software:
Rotor-Gene Q MDx: Rotor-Gene Q software version 2.3.1 or higher
ABI 7500 Fast Dx: SDS software version 1.4.1 or higher

12
artus
SARS-CoV-2 Prep&Amp UM Kit Instructions for Use (Handbook) 04/2021
Materials Required but Not Provided
Consumables
Disposable powder-free gloves
Sterile and nuclease-free pipette tips with filters
1.5 ml or 2 ml PCR-free tubes
0.1 ml PCR tubes for use with the Rotor-Gene Q MDx (Strip Tubes and Caps, 0.1 ml,
cat. no. 981103)
96-Well MicroAmp™ for use with the ABI 7500 Fast Dx qPCR platform
(Applied Biosystems 96-well plate, cat. no. N8010560)
MicroAmp Optical Adhesive film for use with the ABI 7500 Fast Dx qPCR platform
(Applied Biosystems, cat. no. 4360954)
Equipment*
Desktop centrifuge with rotor for 2 ml reaction tubes
Pipettes (adjustable)
Vortex mixer
Block heater
Rotor-Gene Q MDx (cat. no. 9002035 or 9002036) with Rotor-Gene Q software
version 2.3.1 or higher
Rotor-Disc 72 Rotor (cat. no. 9018899)
Rotor-Disc 72 Locking Ring (cat.no. 9018900)
72-well Loading Block (loading block 72 x 0.1 ml tubes, cat. no 9018901)
Alternatively: ABI 7500 Fast Dx qPCR platform (Thermo Fisher Scientific,
cat. no 4406985) with software version 1.4.1 or higher and a 96-well plate centrifuge
* Prior to use and when applicable, ensure that instruments have been checked and calibrated according to the
manufacturer’s recommendations.

artus
SARS-CoV-2 Prep&Amp UM Kit Instructions for Use (Handbook) 04/2021
13
Warnings and Precautions
Please be aware that you may be required to consult your local regulations for reporting
serious incidents that have occurred to the device to the manufacturer and the regulatory
authority in which the user and/or the patient is established.
Safety information
When working with chemicals, always wear a suitable lab coat, disposable gloves, and
protective goggles. For more information, please consult the appropriate safety data sheets
(SDSs). These are available online in convenient and compact PDF format at
www.qiagen.com/safety, where you can find, view, and print the SDS for each QIAGEN kit
and kit component.
Always wear appropriate personal protective equipment, including but not limited to
disposable powder-free gloves, a lab coat, and protective eyewear. Protect skin, eyes, and
mucus membranes. Change gloves often when handling samples.
All samples should be treated as potentially hazardous. Always observe safety precautions as
outlined in relevant guidelines, such as the Clinical and Laboratory Standards Institute
Protection of Laboratory Workers from Occupationally Acquired Infections; Approved
Guideline
(M29), or other appropriate documents.
Specimens and samples are potentially infectious. Discard sample and assay waste
according to your local safety procedures.
®
(CLSI)

14
artus
SARS-CoV-2 Prep&Amp UM Kit Instructions for Use (Handbook) 04/2021
Precautions
Observe standard laboratory procedures for keeping the working area clean and
contamination-free. Dedicate an area with specific equipment to manipulate RNA.
Follow good laboratory practices to minimize cross-contamination.
Pay attention to avoid contamination with RNAse during the experiment and use
RNAse-free plasticware.
Make sure to have a good traceability with records, especially for sample identification.

artus
SARS-CoV-2 Prep&Amp UM Kit Instructions for Use (Handbook) 04/2021
15
Reagent Storage and Handling
Attention should be paid to expiration dates and storage conditions printed on the box and
all components’ labels. Do not use expired or incorrectly stored components.
artus
The
until expiry date.
SARS-CoV-2 Prep&Amp UM Kit can be kept at −30°C to −15°C for 6 months, or
Specimen Transport, Storage and Handling
The
artus
SARS-CoV-2 Prep&Amp UM Kit is for use with nasopharyngeal, nasal, and
oropharyngeal swabs. All samples should be treated as potentially hazardous.
The Centers for Disease Control and Prevention (CDC) and Public Health England have
provided guidelines for sample collection, handling, and testing clinical specimens. Refer to
these guidelines or to other relevant national reference laboratory protocols for additional
information.
Specimen collection, transport and storage
For swab specimen collection, storage, and transport, please refer to the supplier’s
recommendations. Swabs must be fully immersed in transport media to maintain specimen
integrity.

16
artus
SARS-CoV-2 Prep&Amp UM Kit Instructions for Use (Handbook) 04/2021
Protocol: Sample preparation and SARS-CoV-2 detection on the RGQ MDx
This protocol describes the sample and the RT-PCR preparation to detect the SARS-CoV-2
targets in human nasal, nasopharyngeal, or oropharyngeal swabs stored in transport media
on the RGQ MDx.
Important points before starting
Verify that the expiration dates and storage conditions printed on the box and all
component labels are followed. Do not use expired or incorrectly stored components.
Use well-maintained and calibrated equipment.
Pay attention to avoid contamination with RNAses during the experiment and use
nuclease-free plasticware.
Things to do before starting
Samples may be kept at room temperature during preparation steps and reaction setup,
but it is recommended to keep them on ice or at 4°C on a cooling rack.
Before use, let the SARS-CoV-2 UM Prep Buffer, SARS-CoV-2 UM Amp Buffer, SARS--CoV--2
Amp Primers, SARS-CoV-2 IC, Water for NTC, and SARS-CoV-2 Positive Control
completely thaw at room temperature (15–25°C). Keep tubes at room temperature and
protected from light until use.
Before use, homogenize the SARS-CoV-2 UM Prep Buffer and the SARS-CoV-2 UM Amp
Buffer by inverting them 2-3 times (do not vortex), followed by a quick spin. All the other
individual reagents can be homogenized by pulse vortexing for 3-5 seconds or by inverting
2-3 times, followed by a quick spin.
The SARS-CoV-2 UM Prep Buffer inhibits RNAses present in the clinical samples for the
detection step, but it is not a virus-inactivating solution. All samples should be treated as
potentially hazardous.
Verify that the cycling conditions of the qPCR platform are as specified in this protocol.

artus
SARS-CoV-2 Prep&Amp UM Kit Instructions for Use (Handbook) 04/2021
17
*
Reagents may be aliquoted to avoid multiple freeze–thaw cycles.
Freshly prepare the reaction mix (<2 h to the RT-PCR plate launch).
To minimize contamination, the sample and the RT-PCR preparations should be done in
distinct zones.
Procedure
1. Sample preparation
1a. Vortex the swab containing the sample vigorously.
1b. Aliquot 50-200 µl of the sample into 1.5mL PCR-free tubes
1c. Perform heating step at 70°C for 10 min on a block heater. Cool down the samples
on ice for at least 5 min, then, keep the samples on ice or at 4°C.
2. At first use, complete the SARS-CoV-2 UM Amp Buffer with the ROX reference dye.
2a. Add 32.8 µl of the ROX dye to 1 tube of SARS-CoV-2 UM Amp Buffer.
2b. Close the lid containing the SARS-CoV-2 UM Amp Buffer and the ROX dye and
invert the tube 3 times.
2c. Spin down the SARS-CoV-2 UM Amp Buffer containing ROX dye at the bottom of the tube.
3. For a full RGQ MDx plate (72 wells), prepare an aliquot mix of the SARS-CoV-2 Amp
Primers with the SARS-CoV-2 Internal Control.
3a. Transfer the required volumes of the SARS-CoV-2 Amp Primers and the SARS-CoV-2
Internal Control according to Table 1 into a new 1.5 mL PCR-free tube.
3b. Close the lid and invert the tube 3 times or pulse vortex the tube for 3-5 s.
3c. Spin down the SARS-CoV-2 Amp Primers containing the IC at the bottom of the tube.
Table 1. SARS-CoV-2 Amp Primers + IC mix setup
SARS-CoV-2 Amp Primers + IC mix
Reagents
SARS-CoV-2 Amp Primers
SARS-CoV-2 Internal Control 166.67 cp/µl 10 cp/µl 1.5 132
Total SARS-CoV-2 Amp Primers + IC mix 8.75 770
Stock
concentration
3.45x
Final
concentration
1x 7.25 638
1 rxn
(+22% extra volume
Number of reactions
Volume (µl)
72 rxns
)
* Note: Adjust the volumes of SARS-CoV-2 Amp Primers and SARS-CoV-2 Internal Control according to the number of
samples to be tested. Consider extra volume to compensate for the dead volume.

18
artus
SARS-CoV-2 Prep&Amp UM Kit Instructions for Use (Handbook) 04/2021
*
4. Prepare a reaction mix according to Table 2 and mix thoroughly.
Table 2. Reaction mix setup
RT-PCR reaction mix Number of reactions
Reagents
SARS-CoV-2 UM Amp buffer†
SARS-CoV-2 Amp Primers‡
Total reaction volume – 15.00 1296
* Note: Adjust the volumes of SARS-CoV-2 UM Amp buffer, SARS-CoV-2 Amp Primers according to the number of
samples to be tested. Consider extra volume to compensate for the dead volume.
†
SARS-CoV-2 UM Amp buffer completed with the ROX Reference Dye
‡
SARS-CoV-2 Amp Primers completed with the SARS-CoV-2 Internal Control
Stock
concentration
4x 1x 6.25 540
2.9x
Final
concentration
1x 8.75 756
1 rxn
Volume (µl)
72 rxns
(+20% extra volume
)
5. Dispense 8 µl of nuclease-free water to the PCR tube assigned to the NEC.
6. Load 10 µl of nuclease-free water into the PCR tube assigned to the NTC.
7. Dispense 2 μl of SARS-CoV-2 UM Prep Buffer into each PCR tube assigned to the NEC and
the prepared samples.
8. Add 8 μl of the prepared sample to a PCR tube containing the SARS-CoV-2 UM Prep Buffer.
Mix by pipetting up and down 5 times.
9. Add 15 μl of the reaction mix prepared in Step 4 to the tubes dedicated to samples and
controls (Figure 2 provided as an example). Mix by pipetting up and down 5 times, then
close the PCR tube lids, except for the one reserved as the SARS-CoV-2 Positive Control.
Note: Verify that tubes are well closed to prevent cross-contamination.
10. Load 10 µl of the SARS-CoV-2 Positive Control into the appropriate PCR tube. Mix by
pipetting up and down 5 times.
11. Set the RT-PCR program of the RGQ MDx according to specifications in Table 3.
Note: Data acquisition should be performed during the annealing/extension step.
12. Place tubes in the real-time cycler (an example of tube layout is represented in Figure 2),
and start the cycling program as described in Table 3.
Note: Be careful to follow the same tube position and order between the assay set-up and
the real-time cycler steps.

artus
SARS-CoV-2 Prep&Amp UM Kit Instructions for Use (Handbook) 04/2021
19
1: PC
Table 3. SARS-CoV-2 Prep&Amp UM program
Steps Time Temperature (°C) Number of cycles Acquisition
Reverse transcription 10 min 50 1 No
PCR initial heat activation 2 min 95 1 No
2-step cycling
Denaturation
Annealing/Extension
5 s
30 s
95
58
40
Green (FAM),
Yellow (HEX),
and Red (Atto)
2: NTC
3: NEC
4:Sample 1
5. Sample 2
6. Sample 3
7: …
No
Figure 2. Example of tube layout on the RGQ MDx platform
13. Click Gain optimization in the “New Run Wizard” and open Auto-gain Optimization Setup.
14. Verify that the acquisition channels are set as described in Table 4.

20
artus
SARS-CoV-2 Prep&Amp UM Kit Instructions for Use (Handbook) 04/2021
Name
PC tube position
Min reading (Fl)
Max reading (Fl)
Min gain
Max gain
Table 4. RGQ MDx configuration
Green 1* 5 10 –10 10
Yellow 1* 5 10 –10 10
Red 1* 5 10 –10 10
* Note: This needs to be changed according to the SARS-CoV-2 Positive Control tube position.
15. Select Perform optimization before the first acquisition.
16. Start the run.
17. At the end of the run, analyze the results (see the Results section).

artus
SARS-CoV-2 Prep&Amp UM Kit Instructions for Use (Handbook) 04/2021
21
Protocol: Sample Preparation and SARS-CoV-2 Detection on ABI 7500 Fast Dx
This protocol is for preparing and detecting SARS-CoV-2 targets in human nasal, nasopharyngeal,
or oropharyngeal swabs stored in transport media on the ABI 7500 Fast Dx qPCR instrument.
Important points before starting
Verify that the expiration dates and storage conditions printed on the box and all
component labels are followed. Do not use expired or incorrectly stored components.
Use well-maintained and calibrated equipment.
Pay attention to avoid contamination with RNAses during the experiment, and use
nuclease-free plasticware.
When using ABI 7500 Fast Dx, ROX Dye must be added to the master mix tube before first use.
Things to do before starting
Samples may be kept at room temperature during preparation steps and reaction setup,
but it is recommended to keep them on ice or at 4°C on a cooling rack.
The ROX dye is required when using the ABI 7500 Fast Dx.
Data must be acquired with the ROX passive dye setting.
Before use, let the SARS-CoV-2 UM Prep Buffer, SARS-CoV-2 UM Amp Buffer, SARS--CoV--2
Amp Primers, SARS-CoV-2 IC, Water for NTC, and SARS-CoV-2 Positive Control completely
thaw at (15–25°C). Keep tubes at room temperature and protected from light until use.
Before use, homogenize the SARS-CoV-2 UM Prep Buffer and the SARS-CoV-2 UM Amp
Buffer by inverting them 2-3 times (do not vortex), followed by a quick spin. All the other
individual reagents can be homogenized by pulse vortexing for 3-5 seconds or by inverting
2-3 times, followed by a quick spin.
The SARS-CoV-2 UM Prep Buffer inhibits RNAses present in the clinical samples for the
detection step, but is not a virus-inactivating solution. All samples should be treated as
potentially hazardous.
Verify that the cycling conditions of the qPCR platform are as specified in this protocol.

22
artus
SARS-CoV-2 Prep&Amp UM Kit Instructions for Use (Handbook) 04/2021
*
Reagents may be aliquoted to avoid multiple freeze–thaw cycles.
Freshly prepare the reaction mix (<2 h to the RT-PCR plate launch).
To minimize contamination, the sample and the RT-PCR preparations should be done in
distinct zones.
Procedure
1. Sample preparation
1a. Vortex the swab containing sample vigorously.
1b. Aliquot 50-200 µl of sample into 1.5mL PCR-free tubes.
1c. Perform heating step at 70°C for 10 min on a block heater. Cool down samples on
ice for at least 5 min, then keep the samples on ice or at 4°C.
2. At first use, complete the SARS-CoV-2 UM Amp Buffer with the ROX Reference Dye.
2a. Add 32.8 µl of the ROX dye to a tube of SARS-CoV-2 UM Amp Buffer.
2b. Close the lid containing the SARS-CoV-2 UM Amp Buffer and the ROX Dye and invert
the tube 3 times.
2c. Spin down the SARS-CoV-2 UM Amp Buffer containing ROX Dye at the bottom of the tube.
3. For a full ABI 7500 Fast Dx plate (96 wells), prepare an aliquot mix of the SARS-CoV-2
Amp Primers with the SARS-CoV-2 Internal Control.
3a. Transfer the required volume of the SARS-CoV-2 Amp Primers and the SARS-CoV-2
Internal Control according to Table 5 into a new 1.5 mL PCR-free tube.
3b. Close the lid and invert the tube 3 times or pulse vortex the tube for 3-5 s.
3c. Spin down the SARS-CoV-2 Amp Primers containing the IC to bring the solution to the
bottom of the tube.
Table 5. SARS-CoV-2 Amp Primers + IC mix setup
SARS-CoV-2 Amp Primers + IC mix
Reagents
SARS-CoV-2 Amp Primers
SARS-CoV-2 Internal Control 166.67 cp/µl 10 cp/µl 1.5 174
Total SARS-CoV-2 Amp Primers + IC mix 8.75 1015
Stock
concentration
3.45x
Final
concentration
1x 7.25 841
1 rxn
Number of reactions
(+ 21% extra volume
Volume (µl)
96 rxns
)
* Note: Adjust the volumes of SARS-CoV-2 UM Amp Primers and SARS-CoV-2 Internal Control according to the number
of samples to test. Consider extra volume to compensate for the dead volume.

artus
SARS-CoV-2 Prep&Amp UM Kit Instructions for Use (Handbook) 04/2021
23
*
4. Prepare a reaction mix according to Table 6 and mix thoroughly.
Table 6. Reaction mix setup
RT-PCR reaction mix
Reagents
SARS-CoV-2 UM Amp buffer†
SARS-CoV-2 Amp Primers‡ 2.9x 1x 8.75 1008
Total reaction volume – 15.00 1728
* Note: Adjust the volumes of SARS-CoV-2 UM Amp Buffer and SARS-CoV-2 Amp Primers according to the number of
samples to test. Consider extra volume to compensate for the dead volume.
†
SARS-CoV-2 UM Amp Buffer completed with the ROX Reference Dye
‡
SARS-CoV-2 Amp Primers completed with the SARS-CoV-2 Internal Control
Stock
concentration
4x 1x 6.25 720
Final
concentration
1 rxn
Number of reactions
Volume (µl)
96 rxns
(+20% extra volume
)
5. Dispense 8 µl of nuclease-free water to the well assigned to the NEC.
6. Load 10 µl of nuclease-free water into the well assigned to the NTC.
7. Dispense 2 μl of SARS-CoV-2 UM Prep Buffer into each well assigned to the NEC and the
prepared samples.
8. Add 8 μl of the prepared sample to a well containing the SARS-CoV-2 UM Prep Buffer.
Mix by pipetting up and down 5 times.
9. Add 15 μl of the reaction mix prepared in step 4 to the wells dedicated to samples and
controls (Figure 3 provided as an example). Mix by pipetting up and down 5 times.
10. Load 10 µl of the SARS-CoV-2 Positive Control into the appropriate well. Mix by
pipetting up and down 5 times.
11. Seal the PCR plate well to prevent cross-contamination. Make sure to apply pressure
uniformly across the entire plate to obtain a tight seal across individual wells.
12. Centrifuge the PCR plate briefly to collect liquid at the bottom of the well.
13. Set the RT-PCR program on the “Standard 7500” Run Mode of the ABI 7500 Fast Dx
according to Table 7.
Note: Data acquisition should be performed during the annealing/extension step.
Note: Please refer to the
ABI 7500 Fast Dx Instruction for Use
for more details.

24
artus
SARS-CoV-2 Prep&Amp UM Kit Instructions for Use (Handbook) 04/2021
Steps
Time
Temperature (°C)
Number of cycles
Acquisition
14. Place the plate in the real-time cycler (an example of a PCR plate layout is represented in
the Figure 3) and start the cycling program as described in Table 7.
15. Select the used wells and apply the FAM, VIC, and Cy5 reporters. Data must be acquired
with the ROX passive dye ON.
16. Verify that the Standard Curve of the ABI 7500 Fast Dx is configured to Absolute
Quantitation.
17. Start the run.
18. At the end of the run, analyze the results (see the Results section).
Table 7. SARS-CoV-2 Prep&Amp UM program
Reverse transcription 10 min 50 1 No
PCR initial heat activation 2 min 95 1 No
2-step cycling
Denaturation
Annealing/Extension
5 s
30 s
95
58
40 No
Green (FAM), Yellow
(VIC), and Red (Cy5)
Figure 3. Example of plate layout on ABI 7500 Fast Dx

artus
SARS-CoV-2 Prep&Amp UM Kit Instructions for Use (Handbook) 04/2021
25
Channels
Green
Red
Yellow
Results
On the RGQ MDx, the data are analyzed with the Rotor-Gene Q software version 2.3.1
(or higher) according to the manufacturer’s instructions (Rotor-Gene Q MDx User Manual,
Revision 6, September 2018). The following analysis parameters are needed for consistency
between different analyses (Table 8).
Table 8. Analysis parameters for the RGQ MDx
Fluorescence threshold 0.03 0.03 0.03
Slope correction Yes Yes Yes
Dynamic tube Yes Yes Yes
Take-off point No 10-20 10-20
Outlier Removal:
Reaction Efficiency Threshold
Cropped start cycles 5 5 5
Cut-off cycles Ct > 38.00 is considered
Yes
Enabled 0%
as 40.00
No No
No Ct > 35.00 is considered
as 40.00
In the RGQ software, run results are available in the quantitation results grid opened during
the analysis. Data from selected samples are summarized in the table and can be exported as
®
an Excel
file by right-clicking the mouse button in the grid and selecting Export to Excel. Make
sure that all samples are selected before exporting the results.
On the ABI, the data are analyzed with the 7500 Fast System Software version 1.4.1
(or higher) according to the manufacturer’s instructions. The following parameters are needed
for consistency between different analyses (Table 9).

26
artus
SARS-CoV-2 Prep&Amp UM Kit Instructions for Use (Handbook) 04/2021
Table 9. Analysis parameters for the ABI 7500 Fast Dx
Channels FAM* VIC/HEX* CY5/Atto*
Passive dye ROX ROX
Fluorescence threshold 0.13 0.05
Baseline set Auto Auto
Cut-off cycles
* FAM = Filter A/1 in ABI platform, VIC/HEX = Filter B/2 in ABI platform, Cy5/Atto = Filter E/5 in ABI platform
Ct > 39.00 is considered
as 40.00
No
Ct > 35.00 is considered
ROX
0.025
Auto
as 40.00
In the ABI SDS software, Ct values of a selected group of wells or the entire plate are available
in the Report sheet of the Results main section. Data can be exported in comma separated
value text (.csv) format (recommended): In the SDS Software window, select File > Export >
Results (menu item Ct can also be chosen). Select the format of the exported file as .csv.

artus
SARS-CoV-2 Prep&Amp UM Kit Instructions for Use (Handbook) 04/2021
27
Interpretation of Results
The positive control (PC), the N1, and the N2 genes are detected in the Green fluorescence
channel with the RGQ MDx (or in the fluorescent channel FAM on the ABI).
The sampling control, composed of the RNAse P, is detected in the Yellow fluorescence channel
with the RGQ MDx (or in the fluorescence channel VIC/HEX with the ABI). Every clinical sample
should display a sampling control amplification. In the PC, a yellow amplification may be seen
despite the absence of human sequences. In this case, a signal in the PC yellow channel may be
ignored because a strong fluorescence signal in the green channel may bleed in the yellow
channel.
The internal control (IC) is included in the SARS-CoV-2 Amp Primers. It is detected in the no
template control (NTC), the no extraction control (NEC), the positive control (PC), and the
clinical samples with the Red fluorescence channel with the RGQ MDx (or in the fluorescence
channel Cy5/Atto with the ABI).
To validate the RT-PCR runs, the PC, the NTC, and the NEC controls must be amplified and
detected as expected.
Table 10. Run validity criteria and result interpretation on the RGQ MDx.
Control
Positive control (PC)
No template control (NTC) or
No extraction control (NEC)
Detection in
green channel
Ct ≤ 38.00 Indifferent Indifferent Run is validated.
Ct > 38.00
or No Ct
Ct > 38.00
or No Ct
Any other combinations with
amplification in green or yellow
Detection in
Yellow channel
Indifferent Indifferent
Ct > 35.00
or No Ct
Detection in
Red channel
Yes
Indifferent
Interpretation
Run is invalidated.
Run is validated.
Run is invalidated.

28
artus
SARS-CoV-2 Prep&Amp UM Kit Instructions for Use (Handbook) 04/2021
Table 11. Run validity criteria and result interpretation on the ABI 7500 Fast Dx
Control
Positive control (PC)
No template control (NTC) or
No extraction control (NEC)
* FAM = Filter A/1 in ABI platform, VIC/HEX = Filter B/2 in ABI platform, Cy5/Atto = Filter E/5 in ABI platform
Detection in
FAM dye*
Ct ≤ 39.00 Indifferent Indifferent Run is validated.
Ct > 39.00
or No Ct
Ct > 39.00
or No Ct
Any other combinations with
amplification in FAM or VIC/HEX
Detection in
VIC/HEX dye*
Indifferent Indifferent
Ct > 35.00
or No Ct
Detection in
Cy5/Atto dye*
Yes
Indifferent Run is invalidated.
Interpretation
Run is invalidated.
Run is validated.
To validate the tested samples, the samples must be amplified and detected as expected.
Table 12. Sample validity criteria and results interpretation on the RGQ MDx.
Detection in
Green channel
Ct ≤ 38 Indifferent Indifferent Sample is positive for SARS-CoV-2 RNA.
Ct > 38
or No Ct
Ct > 38
or No Ct
Ct > 38
or No Ct
Detection in
Yellow channel
Ct ≤ 35.00 Indifferent Sample is negative, SARS-CoV-2 RNA is not detected.
Ct > 35.00
or No Ct
Ct > 35.00
or No Ct
Detection in
Red channel
Yes
No
Interpretation
Invalid sample. No or insufficient human material
detected. Re-sampling is required.
Invalid sample. RT-qPCR reaction is inhibited.
A retest is required.
Table 13. Sample validity criteria and results interpretation on the ABI 7500 Fast Dx.
Detection in
FAM dye
Ct ≤ 39 Indifferent Indifferent Sample is positive for SARS-CoV-2 RNA.
Ct > 39
or No Ct
Ct > 39
or No Ct
Ct > 39
or No Ct
* FAM = Filter A/1 in ABI platform, VIC/HEX = Filter B/2 in ABI platform, Cy5/Atto = Filter E/5 in ABI platform
Detection in
VIC/HEX dye
Ct ≤ 35.00 Indifferent
Ct > 35.00
or No Ct
Ct > 35.00
or No Ct
Detection in
Cy5/Atto dye
Yes
No
Interpretation
Sample is negative, SARS-CoV-2 RNA is not detected.
Invalid sample. No or insufficient human material
detected. Re-sampling is required.
Invalid sample. RT-qPCR reaction is inhibited.
A retest is required.

artus
SARS-CoV-2 Prep&Amp UM Kit Instructions for Use (Handbook) 04/2021
29
Limitations
For
in vitro
diagnostic use only. Validation of this test has not been reviewed by FDA.
Review under the EUA program is pending. The test is distributed in accordance with the
guidance on Policy for Coronavirus Disease-2019 Tests During the Public Health
Emergency, Section IV.C.2.
A statement such as “The test has been validated, but FDA’s independent review of this
validation is pending” should be included in test reports to healthcare providers.
Results from the
artus
SARS-CoV-2 Prep&Amp UM Kit are not intended to be used as the
sole basis for diagnosis, treatment, or other patient management decisions. Negative
results do not preclude infection with SARS-CoV-2 and should not be the sole basis of a
patient treatment decision.
The product is to be used by personnel specially instructed and trained in the
in vitro
diagnostics procedures.
Strict compliance with the qPCR platform's user manual (Rotor-Gene Q MDx or ABI 7500
Fast Dx) is required for optimal PCR results.
Attention should be paid to expiration dates printed on the box and labels of all
components. Do not use expired components.
The performance of this test has not been established for patients without signs and
symptoms of respiratory infection.

30
artus
SARS-CoV-2 Prep&Amp UM Kit Instructions for Use (Handbook) 04/2021
Performance
Analytical sensitivity (Limit of detection)
The analytical sensitivity, or the limit of detection, is defined as the lowest concentration at
which ≥95% of the tested samples generate a positive call.
The LoD was assessed by analyzing serial dilutions of negative nasopharyngeal samples
prepared with high-titer stocks of inactivated viral particles obtained from commercial suppliers
®
(ZeptoMetrix
replicates must be ≥95% (at least 19/20 replicates must generate a positive signal). The LoD
concentration was confirmed on both claimed real-time PCR platforms using two different lots
of reagents.
The claimed limit of detection for both real-time PCR platforms for the
Prep&Amp UM Kit is 950 cp/ml.
Analytical specificity studies (Inclusivity and exclusivity/cross-reactivity)
). To confirm the established LoD concentration, the detection rate of all
artus
SARS-CoV-2
Inclusivity
The inclusivity of the
analysis on sequences available in GISAID database (www.gisaid.org). A total of 722,488
sequences (available at the 23/03/202) were analyzed on COVID CG (https://covidcg.org),
alimented by GISAID metadata. Sequences were aligned to the WIV04 reference sequence (100%
identical to Wuhan-Hu-1/NC_045512.2, except for the length of the poly-A tail) and the single
nucleotide variations (SNVs) were analyzed in the genomic region targeted by the
SARS-CoV--2 Prep&Amp UM Kit Primers and Probes. The prevalence of the identified SNVs stayed
below 1%, as well as the frequency of the co-occurring mutations. There was no SNV located at
the last 1 to 3 nucleotides from the 3’ end in the respective oligonucleotides, which would be
expected to impact performance. The
detect 100% of the published sequences.
artus
SARS-CoV-2 Amp Primers and Probes has been assessed with an
artus
SARS--CoV--2 Prep&Amp UM Kit is considered able to
in silico
artus

artus
SARS-CoV-2 Prep&Amp UM Kit Instructions for Use (Handbook) 04/2021
31
Exclusivity/Cross-reactivity
In silico
analysis
The exclusivity of the
analysis on sequences stored in the NCBI databank. The
tested pathogens have more than 80% homology with one of the
probes. Among these are
Streptococcus salivarius. Pseudomonas aeruginosa
primers/probes of the SARS-CoV-2 assay. However, the
artus
SARS-CoV-2 Amp Primers and Probes has been assessed with an
Candida albicans
in silico
, SARS-CoV-1,
analysis showed that some of the
artus
SARS--CoV--2 primers or
Streptococcus pyogenes
had less than 80% homology with one of the
artus
SARS-CoV-2 Amp Primers and Probes
showed no possible amplification with the different sequences stored in the NCBI nr/nt database.
A total of 36 bacterial, viral, and fungal strains have been analyzed by
in silico
limited potential amplicon size of 500 bp. Pathogen sequences were collected from the NCBI
database. However, none of these pathogens showed amplification
Table 14. List of
Pathogens Strain/Type Taxonomy ID
Adenovirus Type 3
Adenovirus Type 4
Adenovirus Type 5
Adenovirus Type 7A
Adenovirus Type 14
Adenovirus Type 31
Bordetella pertussis
Candida albicans
Chlamydia pneumoniae
Enterovirus
in silico
tested pathogens.
Type 3 45659 No match
Type 4 28280 No match
Type 5 28285 No match
Type 7A 85755 No match
Type 14 10521 No match
Type 31 10529 No match
A639 520 No match
Z006
SC5314
CWL-029
TW-183
Type 68 42789 No match
in silico
5476 No possible amplification*†
115713 No match
.
In silico
PCR with a
PCR results
in silico
, and
* Sequence match with one of the primers/probes showed <80% homology.
†
Sequence match with one of the primers/probes shoved ≥80% homology.
(Continued on next page)

32
artus
SARS-CoV-2 Prep&Amp UM Kit Instructions for Use (Handbook) 04/2021
Table 14 (Continued from previous page)
Pathogens Strain/Type Taxonomy ID
Haemophilus influenzae
Human coronavirus 229E 11137 No match
Human coronavirus NL63 277944 No match
Human coronavirus HKU-1 290028 No match
Human coronavirus OC43 OC43 31631 No match
Human Coronavirus MERS-CoV 1335626 No match
Human Metapneumovirus n/a 162145 No match
Influenza A H1N1 114727 No match
Influenza A H3N2 119210 No match
Influenza B n/a 11520 No match
Mycoplasma pneumoniae
Parainfluenza virus Type 1 12730 No match
Parainfluenza virus Type 2 2560525 No match
Parainfluenza virus Type 3 11216 No match
Parainfluenza virus Type 4 2560526 No match
Pneumocystis jirovecii
Pseudomonas aeruginosa
Respiratory syncytial virus Type A (RSV-A) 208893 No match
Respiratory syncytial virus Type B (RSV-B) 208895 No match
Rhinovirus Type A 147711 No match
Rhinovirus Type B 147712 No match
SARS-coronavirus Tor2 694009 No possible amplification†
Staphylococcus epidermidis
Streptococcus pyogenes
Streptococcus salivarius
Streptococcus pneumoniae
* Sequence match with one of the primers/probes showed <80% homology.
†
Sequence match with one of the primers/probes shoved ≥80% homology.
KW20 727 No match
M129
FH
RU7 42068 No match
PAO1 287 No possible amplification*
n/a 1282 No match
n/a 1314 No possible amplification†
ATCC® BAA-1024D-5
CCHSS3
ATCC 700669
NCTC11032
In silico
PCR results
272634 No match
1304 No possible amplification
1313 No match
†

artus
SARS-CoV-2 Prep&Amp UM Kit Instructions for Use (Handbook) 04/2021
33
In vitro
analysis
The cross-reactivity was verified
SARS-CoV-2 Amp Primers in the
in vitro
in silico
with pathogens showing ≥ 80% homology with the
analysis. Samples were prepared by spiking potential
cross-reactive organisms into nasopharyngeal swab matrix at 106 cp/ml, except for
SARS-CoV-1, which was tested undiluted according to its supplier’s recommendation. None of
these pathogens showed
The microbial interference of the
assessed
in vitro
on a panel of recommended pathogens. Samples were prepared by spiking
a maximum of 5 pathogens - at 10
in vitro
cross-reactivity.
artus
SARS-CoV-2 Prep&Amp UM Kit assay has been
5
TCID50/mL for viral targets, 106 cp/mL for bacterial and
fungal targets, or at the highest concentration possible based on the stock concentration - into
negative nasopharyngeal swabs spiked at 2.87 x LoD with inactivated SARS-CoV-2 particles
(Zeptometrix). The NATtrol™ Panels and the SARS-CoV-1 were spiked directly with inactivated
SARS-CoV-2 viral particles (Zeptometrix) at 2.87 x LoD. The results for each tested
microorganism pools and the respective concentrations are summarized below.
Table 15. List of
Pool ID /
Sample ID
Pool 1
Pool 2
in vitro
tested pathogens in microbial interference.
Microorganism Source
SARS-CoV-2
Human coronavirus 229E Zeptometrix (0810229CFHI) 1.43E+05 TCID50/ml
Human coronavirus OC43 Zeptometrix (0810024CFHI) 5.86E+04 TCID50/ml
Human coronavirus NL63 Zeptometrix (0810228CFHI) 2.84E+04 TCID50/ml
Adenovirus T3 Zeptometrix (0810016CFHI) 1.43E+05 TCID50/ml
Parainfluenza virus 1 Zeptometrix (0810014CFHI) 9.14E+06 TCID50/ml
SARS-CoV-2
Adenovirus T31 Zeptometrix (0810073CFHI) 1.67E+04 TCID50/ml
Parainfluenza virus 2 Zeptometrix (0810015CFHI) 4.29E+04 TCID50/ml
Influenza B
Florida/02/2006
Rhinovirus T 1A Zeptometrix (0810012CFNHI) 2.86E+04 TCID50/ml
Zeptometrix
(NATSARS(COV2)-ERC)
Zeptometrix
(NATSARS(COV2)-ERC)
Zeptometrix (0810037CFHI) 1.43E+05 TCID50/ml
Final
concentration
2.72E+03 cp/ml
2.72E+03 cp/ml
Unit Result
(Continued on next page)
No
interference
No
interference

34
artus
SARS-CoV-2 Prep&Amp UM Kit Instructions for Use (Handbook) 04/2021
SARS
Parainfluenza Virus T3
Haemophilus influenzae
Streptococcus pneumoniae
Candida albicans
Staphylococcus epidermi
Table 15 (Continued from previous page)
Pool ID /
Sample ID
Pool 3
Pool 4
Pool 5
Pool 6
Microorganism Source
-CoV-2
Zeptometrix (0801504DNA) 1.00E+06 CFU/ml
SARS-CoV-2
Adenovirus T7A Zeptometrix (0810021CFHI) 1.02E+06 TCID50/ml
Streptococcus pyogenes
Mycoplasma pneumoniae
Pseudomonas aeruginosa
SARS-CoV-2
Respiratory syncytial virus
RSVA
Influenza A H1N1
California
Enterovirus Type 68 Major
Group
Adenovirus T14 Zeptometrix (0810108CFHI) 2.86E+04 TCID50/ml
SARS-CoV-2
MERS-coronavirus Zeptometrix (0810575CFHI) 1.43E+04 TCID50/ml
AdenoVirus T4 Zeptometrix (0810070CFHI) 1.43E+05 TCID50/ml
Human Metapneumovirus
(hMPV) Type B
Respiratory Syncytial Virus
Type B (RSV-B)
Zeptometrix
(NATSARS(COV2)-ERC)
Zeptometrix (0810016CFHI) 1.43E+07 TCID50/ml
ATCC (51907D-5) 1.00E+06 CFU/ml
ATCC (700669DQ) 3.30E+06 CFU/ml
dis
ATCC (12228DQ) 4.60E+06 CFU/ml
Zeptometrix
(NATSARS(COV2)-ERC)
ATCC (700294DQ) 1.00E+07 CFU/ml
Zeptometrix (0801579DNA) 1.00E+08 CFU/ml
ATCC (47085DQ) 1.00E+07 CFU/ml
Zeptometrix
(NATSARS(COV2)-ERC)
Zeptometrix (0810482CFHI) 7.14E+04 TCID50/ml
Zeptometrix (0810165CFHI) 1.43E+04 TCID50/ml
Zeptometrix (0810300CFHI) 1.43E+05 TCID50/ml
Zeptometrix
(NATSARS(COV2)-ERC)
Zeptometrix (0810156CFHI) 7.14E+03 TCID50/ml
Zeptometrix (0810040CFHI) 1.43E+03 TCID50/ml
Final
concentration
2.72E+03 cp/ml
2.73E+03 cp/ml
2.72E+03 cp/ml
2.73E+03 cp/ml
Unit Result
No
interference
No
interference
No
interference
No
interference
(Continued on next page)

artus
SARS-CoV-2 Prep&Amp UM Kit Instructions for Use (Handbook) 04/2021
35
Table 15 (Continued from previous page)
Pool ID /
Sample ID
Pool 7
Pool 8
Pool 9
Pool 10
Microorganism Source
SARS-CoV-2
Adenovirus T5
Parainfluenza virus 4B
Influenza A H3N2
Switzerland/9715293/13
Streptococcus salivarius
SARS-CoV-2
NATrol Panel RP1 (Influenza A H3N2
(Brisbane/10/07), Influenza A H1N1
(NY/02/2009), Rhinovirus (Type 1A),
Adenovirus T3, Parainfluenza T1,
Parainfluenzavirus T4,
Metapneumovirus (Peru 6-2003)
C. pneumoniae
M. pneumoniae
Coxsackievirus (Type A1)
SARS-CoV-2
NATrol Panel RP2 (Influenza A H1
(New Caledonia/20/99), Influenza B
(Florida/02/06), RSV-A,
Parainfluenza T2, Parainfluenza T3,
Coronavirus HKU recombinant,
Coronaviruses (OC43, NL63, 229E),
Bordetella pertussis
SARS-CoV-2
SARS-CoV-1
(CWL-029),
(M129),
(A639)
Zeptometrix
(NATSARS(COV2)ERC)
Zeptometrix
(0810020CFHI)
Zeptometrix
(0810060BCFHI)
Zeptometrix
(0810511CFHI)
Zeptometrix (BAA1024D-5)
Zeptometrix
(NATSARS(COV2)ERC)
Zeptometrix
(MDZ001)
Zeptometrix
(NATSARS(COV2)ERC)
Zeptometrix
(MDZ001)
Zeptometrix
(NATSARS(COV2)ERC)
Zeptometrix
(NATSARS-ST)
Final
concentration
2.73E+03 cp/ml
6.43E+05 TCID50/ml
7.14E+04 TCID50/ml
2.86E+04 TCID50/ml
1.00E+06 CFU/ml
2.73E+03 cp/ml
Unknown* N/A
2.73E+03 cp/ml
Unknown* N/A
2.73E+03 cp/ml
Unknown* N/A
Unit Result
No
interference
No
interference
No
interference
No
interference
* Concentration not communicated by the supplier.

36
artus
SARS-CoV-2 Prep&Amp UM Kit Instructions for Use (Handbook) 04/2021
Interfering substances
The effect of putative interfering substances (for the substances listed in the Table 16) has been
artus
assessed on the
3 pools of negative nasopharyngeal swabs and in 3 pools of positive nasopharyngeal swabs
spiked at 4 x LoD with inactivated SARS-CoV-2 viral particles (Zeptometrix). The experiments
were performed on the RGQ MDx platform (across 4 instruments) by 1 operator with 1 pilot kit.
Each pool was split into 2 to test either the interfering substance dissolved in a solvent (test
sample) or the solvent alone (control sample). Hit rates in the green and in the red fluorescence
channels were compared between the test and its corresponding control samples. In absence
of interference, the test and its corresponding control samples have the same hit rate.
Table 16 shows that none of the tested substances interfere with the
Prep&Amp UM Kit performance in the green fluorescence channel.
Table 16. List of interfering substances.
SARS-CoV-2 Prep&Amp UM Kit performance. Tests were performed in
artus
SARS-CoV-2
Interfering
substances
Tobramycin
Mupirocin
Fluticasone
Menthol
(Throat lozenges)
Function Tested
Systemic
antibiotic
Nasal antibiotic
ointment
Nasal
corticosteroid
Oral anesthetic
and analgesic
concentration
1 mg/ml
6.6 mg/ml
5% (v/v)
0.5 mg/ml No interference
Results in negative
nasopharyngeal swab
No interference
0/15
No interference
0/15
No interference
0/15
0/15
Results in positive (4x LoD)
nasopharyngeal swab
No interference
0/15
No interference
0/15
No interference
0/15
No interference
0/15
(Continued on next page)

artus
SARS-CoV-2 Prep&Amp UM Kit Instructions for Use (Handbook) 04/2021
37
Table 16. (Continued from previous page)
Interfering
substances
Oxymetazoline
Oseltamivir
Mucin
(Bovine submaxillary
gland type I-S)
Whole Blood
* An amplification corresponding to an artefact has been detected.
Function Tested
Nasal spray 10% (v/v)
Anti-viral drug 3.3 mg/ml
concentration
2.5 mg/ml
4% (v/v)
Results in negative
nasopharyngeal swab
No interference
0/15
No interference
0/15
No interference
0/15
No interference
1/15*
Results in positive (4x LoD)
nasopharyngeal swab
No interference
0/15
No interference
0/15
No interference
0/15
No interference
0/15
Precision
The Precision study assessed the reproducibility (the same sample is repeated in different runs and
conditions: 5 days, 3 kit lots, 3 operators, and 2 instruments) and the repeatability (the same sample
is repeated in the same run and condition). Tests were performed on negative nasopharyngeal
samples and negative nasopharyngeal samples spiked at 5 x LoD on the RGQ MDx.
For each dilution level, 204 data points were collected. Repeatability and reproducibility data
were used to determine the standard deviation (SD) and the coefficient of variation (%CV) for the
SARS-CoV-2 targets in the green, yellow, and red channels. Table 17 shows that the
SARS-CoV-2 Prep&Amp UM Kit has an overall precision of 0.63 SD (2.03% CV) in the green
channel, 0.54 SD (2.22 %CV) in the yellow channel, and 1.28 SD (4.10 %CV) in the red
channel.
artus

38
artus
SARS-CoV-2 Prep&Amp UM Kit Instructions for Use (Handbook) 04/2021
Table 17. Standard deviation and coefficient of variation of the
Samples and
detection channel
Negative NPS
Yellow channel
Negative NPS
Red channel
Spiked NPS
Green channel
Spiked NPS
Yellow channel
Spiked NPS
Red channel
Total
0.54
(2.22)
1.15
(3.68)
0.63
(2.03)
0.47
(1.93)
1.28
(4.10)
Day-to-
day
0.09
(0.37)
0.0
(0.00)0
0.18
(0.59)
0.13
(0.53)
0.12
(0.37)
Batch-to-
batch
0.10
(0.42)
0.55
(1.76)
0.31
(1.00)
0.24
(0.98)
0.58
(1.84)
artus
SARS-CoV-2 Prep&Amp UM Kit
Operator-to-
operator
Standard deviation (SD)
(Coefficient of variation (%CV))
0.06
(0.27)
0.00
(0.00)
0.00
(0.00)
0.05
(0.20)
0.11
(0.34)
Instrument-to-
instrument
0.11
(0.47)
0.12
(0.40)
0.08
(0.25)
0.18
(0.73)
0.00
(0.00)
Run-to-
run
0.09
(0.36)
0.39
(1.26)
0.00
(0.00)
0.00
(0.00)
0.49
(1.57)
Clinical performance
The clinical performance of the
retrospective nasopharyngeal swab specimens in transport medium, consisting of:
98 SARS-CoV-2 RNA negative specimens
artus
SARS-CoV-2 UM Prep&Amp Kit was evaluated using
Within
run
0.50
(2.05)
0.92
(2.96)
0.51
(1.64)
0.33
(1.38)
1.02
(3.27)
52 SARS-CoV-2 RNA positive nasopharyngeal specimens
All specimens were collected from patients with signs and symptoms of infection who were
suspected of COVID-19 and were stored frozen until use.
The clinical validation was performed on the ABI 7500 Fast Dx. Table 18 reports the
artus
performance of the
SARS-CoV-2 Prep&Amp UM Kit against a n RT-PCR reference method.

artus
SARS-CoV-2 Prep&Amp UM Kit Instructions for Use (Handbook) 04/2021
39
Table 18. Clinical performance of the
Reference
method result
Positive 52 98.1
Negative 98
Of note, samples with discordant results were evaluated by a third method. The overall clinical performance results did
not change following the discordant testing.
artus
SARS-CoV-2 Prep&Amp UM Kit against a reference method
N % Positive 95% CI % Negative 95% CI
–
88.7 – 97.8
(51/52)
1.9
(1/52)
89.9 – 99.7 5.1
–
(5/98)
94.9
(93/98)
Listed below are the positive percent agreement (sensitivity) and negative percent agreement
(specificity):
Positive Percent Agreement (PPA%): 51/52 = 98.1% (95% CI: 89.9% - 99.7%)
Negative Percent Agreement (NPA%): 93/98 = 94.9% (95% CI: 88.6% - 97.8%)

40
artus
SARS-CoV-2 Prep&Amp UM Kit Instructions for Use (Handbook) 04/2021
References
1. CUI J
2. Gagneur
3. HU
et al.
(2019) Origin and evolution of pathogenic coronaviruses. Nat Rev Microbiol
17, 181-192.
et al.
(2002) Infections nosocomiales à coronavirus humains chez le nouveau-
né. Arch Pédiatr 9, 61-69.
et al.
(2020) Characteristics of SARS-CoV-2 and COVID-19. Nat Rev Microbiol 6:1-14.

artus
SARS-CoV-2 Prep&Amp UM Kit Instructions for Use (Handbook) 04/2021
41
Comments and suggestions
Weak or No Green signal (FAM) in Positive Control (PC)
a) The selected fluorescence channel for
For data analysis, select the fluorescence channel FAM (green) for the
b) Incorrect programming of the
Compare the RT-PCR program with the protocol.
c) Incorrect configuration of the PCR
Verify your work steps through the pipetting scheme and repeat the
d) The storage conditions for one or
Follow the storage conditions and verify the reagents' expiration date
e) Incorrect configuration of the qPCR
Apply the recommended configurations related to your qPCR platform
f) The PCR was inhibited.
Follow the good practices in molecular biology laboratory to avoid the
Green signal (FAM) in the No Template Control or in the No Extraction Control
Contamination with SARS-CoV-2
Repeat the RT-PCR with new reagents.
Troubleshooting Guide
This troubleshooting guide may help solve any problems that may arise. For more information,
see also the Frequently Asked Questions page at our Technical Support Center:
www.qiagen.com/FAQ/FAQList.aspx.
RT-PCR data analysis does not
comply with the protocol.
temperature profile.
reaction.
more kit components did not comply
with the instructions, or the
SARS-CoV-2 RT-PCR Kit has expired.
platform during the data
configuration.
sequences occurred during the RT-PCR
plate preparation.
artus
analytical SARS-CoV-2 RT-PCR targets, the fluorescence channel
HEX/VIC/JOE (yellow) for the sampling control, and the Cy5/Atto (red)
for the internal control.
PCR, if necessary.
and use a new kit, if necessary.
that are described in this manual.
introduction of contaminants.
Make sure that the workspace and instruments are decontaminated at
regular intervals.
Follow the protocol mentioned in this manual. Check the expiration date
of the reagent and use a new kit, if necessary. Repeat the assay with
another sample.
Follow the good practices in molecular biology laboratory to avoid the
introduction of contaminants. Follow the protocol mentioned in this
handbook.
Make sure that the workspace and instruments are decontaminated at
regular intervals.

42
artus
SARS-CoV-2 Prep&Amp UM Kit Instructions for Use (Handbook) 04/2021
Comments and suggestions
a) An interferent has been introduced in
Follow the good practices in molecular biology laboratory to avoid the
b) The internal control is degraded.
Follow the good practices in molecular biology laboratory to avoid the
c) Incorrect configuration of the qPCR
Apply the recommended configurations related to your qPCR platform
Weak or no yellow signal (VIC/HEX) of the sampling control
a) The clinical sample is degraded.
Follow the recommendations provided by the collection device supplier
b) The specimen was not properly
Follow the recommendations provided by the collection device supplier
c) Incorrect configuration of the qPCR
Apply the configurations related to your qPCR platform that are
Weak or no red signal (Cy5/Atto) from the Internal control
the RT-PCR reaction. The PCR is
inhibited.
platform during the data
configuration.
collected. Not enough human cells
were collected on the swab or
transferred in the transport media.
introduction of contaminants.
Make sure that the workspace and instruments are decontaminated at
regular intervals.
Follow the protocol mentioned in this manual.
Repeat the experiment with a sample newly collected.
introduction of RNAses. Follow the recommendations mentioned in this
manual.
Make sure that the workspace and instruments are decontaminated at
regular intervals.
Follow the storage conditions and check the reagents' expiration date
and use a new kit, if necessary.
that are described in this manual.
for their storage, handling, and transport.
Follow the protocol mentioned in this manual, including the sample
preparation steps with the SARS-CoV-2 UM Prep buffer.
Follow the storage conditions and check the reagents' expiration date,
such as the SARS-CoV-2 UM Prep buffer, and use a new kit, if
necessary.
for the specimen collection and the specimen handling.
platform during the data
configuration.
described in this manual.

artus
SARS-CoV-2 Prep&Amp UM Kit Instructions for Use (Handbook) 04/2021
43
Symbol
Symbol definition
n
R is for revision of the Instructions for Use, and n is the
revision number
℞
Prescription Use Only
Symbols
The following symbols may appear in the instructions for use or on the packaging and labeling:
Contains reagents sufficient for 768 or 3072 reactions
Use by
In vitro diagnostic medical device
Catalog number
Lot number
Components
Contains
Number
Global Trade Item Number
Rn
Temperature limitation
Manufacturer
Consult instructions for use
Keep away from sunlight
Warning/caution

44
artus
SARS-CoV-2 Prep&Amp UM Kit Instructions for Use (Handbook) 04/2021
Contact Information
For technical assistance and more information, please contact the QIAGEN Technical Services
at support.qiagen.com.

artus
SARS-CoV-2 Prep&Amp UM Kit Instructions for Use (Handbook) 04/2021
45
Product
time PCR cycler with 5 channels,
, software,
Ordering Information
Contents Cat. no.
artus
SARS-CoV-2
Prep&Amp UM Kit (768)
artus
SARS-CoV-2
Prep&Amp UM Kit (3072)
For 768 reactions: Preparation Buffer, ROX
dye, Master Mix, Primers and Probes,
Internal Control, Water (NTC), and Positive
Control
For 3072 reactions: Preparation Buffer,
ROX dye, Master Mix, Primers and Probes,
Internal Control, Water (NTC), and Positive
Control
4511440
4511449
Instrument and accessories
PCR tubes, 0.1 ml for
Rotor-Gene Q MDx
Rotor-Gene Q software Rotor-Gene Q software v2.3.1 (or higher)
Rotor-Gene Q MDx
Loading Block 72 x 0.1 ml tubes 9018901
For up-to-date licensing information and product-specific disclaimers, see the respective
QIAGEN kit handbook or user manual. QIAGEN kit handbooks and user manuals are
available at www.qiagen.com or can be requested from QIAGEN Technical Services or your
local distributor.
For use with 72-well rotor, Strip tubes, and
caps
Realhigh-resolution melt analyzer
laptop computer, and accessories; 1-year
warranty on parts and labor, installation
981103
9002035
9002036
or

46
artus
SARS-CoV-2 Prep&Amp UM Kit Instructions for Use (Handbook) 04/2021
Document Revision History
Revision Description
R1, April 2021 Initial release.

artus
SARS-CoV-2 Prep&Amp UM Kit Instructions for Use (Handbook) 04/2021
47
Limited License Agreement for
Use of this product signifies the agreement of any purchaser or user of the product to the following terms:
1. The product may be used solely in accordance with the protocols provided with the product and this handbook and for use with components contained in the
panel only. QIAGEN grants no license under any of its intellectual property to use or incorporate the enclosed components of this panel with any components not
included within this panel except as described in the protocols provided with the product, this handbook, and additional protocols available at www.qiagen.com.
Some of these additional protocols have been provided by QIAGEN users for QIAGEN users. These protocols have not been thoroughly tested or optimized by
QIAGEN. QIAGEN neither guarantees them nor warrants that they do not infringe the rights of third-parties.
2. Other than expressly stated licenses, QIAGEN makes no warranty that this panel and/or its use(s) do not infringe the rights of third-parties.
3. This panel and its components are licensed for one-time use and may not be reused, refurbished, or resold.
4. QIAGEN specifically disclaims any other licenses, expressed or implied other than those expressly stated.
5. The purchaser and user of the panel agree not to take or permit anyone else to take any steps that could lead to or facilitate any acts prohibited above.
QIAGEN may enforce the prohibitions of this Limited License Agreement in any Court, and shall recover all its investigative and Court costs, including attorney fees, in
any action to enforce this Limited License Agreement or any of its intellectual property rights relating to the panel and/or its components.
For updated license terms, see www.qiagen.com.
Trademarks: QIAGEN
®
CLSI
(Clinical and Laboratory Standards Institute, Inc ); Zeptometrix®, NATtrol™ (Cole-Parmer); Excel® (Microsoft Corporation); ABI®, MicroAmp™, Thermo Fisher Scientific®
(Thermo Fisher Scientific or its Subsidiaries). Registered names, trademarks, etc. used in this document, even when not specifically marked as such, are not to be considered
unprotected by law.
04/2021 HB-2851-003 R1 © 2021 QIAGEN, all rights reserved.
artus
SARS-CoV-2 Prep&Amp UM Kit
®
, Sample to Insight®,
®
artus
, Rotor-Gene® (QIAGEN Group); ATCC® (American Type Culture Collection); Clinical and Laboratory Standards Institute®,

48
artus
SARS-CoV-2 Prep&Amp UM Kit Instructions for Use (Handbook) 04/2021
Ordering
04/2021
www.qiagen.com/shop | Technical Support support.qiagen.com | Website www.qiagen.com
 Loading...
Loading...