
Sample to Insight__
February 2021
QIAseq® Stranded
mRNA Library Kit
Handbook
For enrichment of polyadenylated RNA and
stranded RNAseq libraries for next-generation
sequencing using dual indexing

2
QIAseq Stranded mRNA Library Kit Handbook 02/2021
Contents
Kit Contents ................................................................................................................ 3
Shipping and Storage ................................................................................................. 6
Intended Use .............................................................................................................. 6
Safety Information ....................................................................................................... 7
Quality Control ........................................................................................................... 7
Introduction ................................................................................................................ 8
Principle and procedure .................................................................................... 9
Equipment and Reagents to be Supplied by User .......................................................... 13
Important Notes ........................................................................................................ 14
Protocol: mRNA Enrichment ....................................................................................... 16
Protocol: Fragmentation/FastSelect RNA Removal ........................................................ 19
Protocol: First-strand Synthesis .................................................................................... 21
Protocol: Second-strand Synthesis, End-repair, and A-addition ........................................ 24
Protocol: Strand-specific Ligation ................................................................................. 27
Protocol: CleanStart Library Amplification .................................................................... 31
Recommendations: Library QC, Quantification, and Sequencing .................................... 35
Troubleshooting Guide .............................................................................................. 37
Appendix A: QIAseq Dual-Index Y-Adapters ................................................................ 39
Appendix B: Data Analysis Recommendations .............................................................. 43
Appendix C: mRNA Enrichment in 200 µl Plates .......................................................... 44
Appendix D: General Remarks on Handling RNA ...................................................... 46
Ordering Information ................................................................................................ 48
Document Revision History ......................................................................................... 52

QIAseq Stranded mRNA Library Kit Handbook 02/2021
3
Kit Contents
QIAseq Stranded mRNA Library Kit
Catalog no.
Number of reactions
Component
QIAseq Beads
QIAseq Beads N/A 10 ml 40 ml 40 ml 40 ml 40 ml
QIAseq Stranded mRNA Enrichment Kit
Pure mRNA Beads* Violet 2 x 600 µl 5 x 600 µl 5 x 600 µl 5 x 600 µl 5 x 600 µl
Buffer mRBB (binding buffer)† Clear 2 x 8 ml 4 x 8 ml 4 x 8 ml 4 x 8 ml 4 x 8 ml
Buffer OW2 (wash buffer) Clear 2 x 19 ml 7 x 19 ml 7 x 19 ml 7 x 19 ml 7 x 19 ml
RNase-Free Water Clear 1 x 10 ml 3 x 10 ml 3 x 10 ml 3 x 10 ml 3 x 10 ml
Buffer OEB (elution buffer) Clear 2 x 1.5 ml 5 x 1.5 ml 5 x 1.5 ml 5 x 1.5 ml 5 x 1.5 ml
Quick-Start Protocol N/A 1 1 1 1 1
QIAseq Stranded Total RNA Kit
RT Buffer, 5x Blue 1 x 215 µl 1 x 860 µl 1 x 860 µl 1 x 860 µl 1 x 860 µl
RT Enzyme Violet 1 x 28 µl 1 x 112 µl 1 x 112 µl 1 x 112 µl 1 x 112 µl
RNase Inhibitor Green 1 x 55 µl 1 x 212 µl 1 x 212 µl 1 x 212 µl 1 x 212 µl
Second Strand Buffer, 10x Blue 1 x 275 µl 1 x 1.1 ml 1 x 1.1 ml 1 x 1.1 ml 1 x 1.1 ml
Second Strand Enzyme Mix Violet 1 x 175 µl 1 x 700 µl 1 x 700 µl 1 x 700 µl 1 x 700 µl
DTT, 1 M Clear 1 x 1 ml 1 x 1 ml 1 x 1 ml 1 x 1 ml 1 x 1 ml
Ultralow Input Ligase Orange 2 x 65 µl 1 x 500 µl 1 x 500 µl 1 x 500 µl 1 x 500 µl
Ultralow Input Ligation Buffer, 4x Yellow 1 x 900 µl 3 x 900 µl 3 x 900 µl 3 x 900 µl 3 x 900 µl
CleanStart PCR Mix, 2x Red 1 x 660 µl 2 x 1.4 ml 2 x 1.4 ml 2 x 1.4 ml 2 x 1.4 ml
CleanStart PCR Primer Mix Clear 1 x 36 µl 1 x 150 µl 1 x 150 µl 1 x 150 µl 1 x 150 µl
Ligation Initiator Black 1 x 210 µl 1 x 840 µl 1 x 840 µl 1 x 840 µl 1 x 840 µl
QIAseq UDI Adapter Kit
QIAseq UDI Y-Adapter Plate (24) N/A 1 N/A N/A N/A N/A
QIAseq UDI Y-Adapter Kit A (96) N/A N/A 1 N/A N/A N/A
QIAseq UDI Y-Adapter Kit B (96) N/A N/A N/A 1 N/A N/A
QIAseq UDI Y-Adapter Kit C (96) N/A N/A N/A N/A 1 N/A
QIAseq UDI Y-Adapter Kit D (96) N/A N/A N/A N/A N/A 1
QIAseq Y-Adapter Reference Card N/A 1 1 1 1 1
Quick-Start Protocol N/A 3 3 3 3 3
* Caution: Pure mRNA Beads contain 0.1% sodium azide (NaN3) as a preservative. Dispose of azide-containing
solutions according to your institution’s waste-disposal guidelines. See “Safety Information”, page 7.
Tube cap color Volume
UDI (24)
180440
2424
UDI-A (96)
180441
9696
UDI-B (96)
180442
96
UDI-C (96)
180443
96
UDI-D (96)
180445
96

4
QIAseq Stranded mRNA Library Kit Handbook 02/2021
QIAseq Stranded mRNA Select Kit
Catalog no.
Number of reactions
Component Tube cap color Volume
QIAseq Beads
QIAseq Beads Clear 10 ml 40 ml
QIAseq Stranded mRNA Enrichment Kit
Pure mRNA Beads* Violet 2 x 600 µl 5 x 600 µl
Buffer mRBB (binding buffer)† Clear 2 x 8 ml 4 x 8 ml
Buffer OW2 (wash buffer) Clear 2 x 19 ml 7 x 19 ml
RNase-Free Water Clear 1 x 10 ml 3 x 10 ml
Buffer OEB (elution buffer) Clear 2 x 1.5 ml 5 x 1.5 ml
Quick-Start Protocol N/A 1 1
QIAseq Stranded RNA Lib Enzyme Kit
RT Buffer, 5x Blue 1 x 215 µl 1 x 860 µl
RT Enzyme Violet 1 x 28 µl 1 x 112 µl
RNase Inhibitor Green 1 x 55 µl 1 x 212 µl
Second Strand Buffer, 10x Blue 1 x 275 µl 1 x 1.1 ml
Second Strand Enzyme Mix Violet 1 x 175 µl 1 x 700 µl
DTT, 1 M Clear 1 x 1 ml 1 x 1 ml
Ultralow Input Ligase Orange 2 x 65 µl 1 x 500 µl
Ultralow Input Ligation Buffer, 4x Yellow 1 x 900 µl 3 x 900 µl
CleanStart PCR Mix, 2x Red 1 x 660 µl 2 x 1.4 ml
CleanStart PCR Primer Mix Clear 1 x 36 µl 1 x 150 µl
Ligation Initiator Black 1 x 210 µl 1 x 840 µl
QIAseq CDI Y-Adapter Kit
QIAseq CDI Y-Adapter Plate (24) N/A 1 N/A
QIAseq CD I Y-Adapter Plate (96) N/A N/A 1
QIAseq Y-Adapter Reference Card N/A 1 1
Quick-Start Protocol N/A 3 3
(24)
180773
24
(96)
180775
96
* Caution: Pure mRNA Beads contain 0.1% sodium azide (NaN3) as a preservative. Dispose of azide-containing
solutions according to your institution’s waste-disposal guidelines. See “Safety Information”, page 7.

QIAseq Stranded mRNA Library Kit Handbook 02/2021
5
The QIAseq Stranded Library Kits ship with a QIAseq Y-Adapter plate with either unique dualindex (UDI) adapters or combinatorial dual-index (CDI) adapters. To multiplex more than 96
libraries in a single sequencing run, simply combine the kits with different UDI Y-adapter plates.
For example, to multiplex 384 samples in a single flow cell line, prepare and combine libraries
from the QIAseq Stranded mRNA Library Kit UDI-A (96) with the QIAseq Stranded mRNA
Library Kit UDI-B (96), QIAseq Stranded mRNA Library Kit UDI-C (96), and QIAseq Stranded
mRNA Library Kit UDI-D (96). For more information on QIAseq Y-Adapter plates, please refer
to Appendix A, page 39.

6
QIAseq Stranded mRNA Library Kit Handbook 02/2021
Shipping and Storage
The QIAseq Stranded mRNA Select Kit (cat. nos. 180773, 180775), QIAseq Stranded mRNA
Library Kit UDI (24) (cat. no. 180440), QIAseq Stranded mRNA Library Kit UDI-A (96) (cat.
no. 180441), QIAseq Stranded mRNA Library Kit UDI-B (96) (cat. no. 180442), QIAseq
Stranded mRNA Library Kit UDI-C (96) (cat. no. 180443), and QIAseq Stranded mRNA
Library Kit UDI-D (96) (cat. no. 180445) are shipped in 4 boxes.
In the QIAseq Stranded mRNA Enrichment Kit (cat. nos. 1105688, 1105689), the Buffers
mRBB and Pure mRNA Beads should be stored at 2–8°C (do not freeze). Buffer OW2 (wash
buffer), RNase-Free Water, and Buffer OEB (elution buffer) should be stored at room temperature
(15–25°C).
Store the QIAseq Stranded RNA Lib Enzyme Kit (cat. nos. 1122418, 1122419) at −30 to
−15°C.
Store the QIAseq UDI Y-Adapter Kit (cat. nos. 180312, 180314, 180316, 180318, 180310)
and QIAseq CDI Y-Adapter Kit (cat. nos. 180301, 180303) at −30 to −15°C.
The QIAseq Beads (cat. nos. 1107149, 1107460) should be stored at 2–8°C (do not freeze).
Important: Do not use expired beads as this will significantly reduce library yield
If stored under these conditions, the kit contents are stable until the date indicated on the box
labels.
.
Intended Use
The QIAseq Stranded mRNA Library Kit is intended for molecular biology applications. This
product is not intended for the diagnosis, prevention, or treatment of a disease.
All due care and attention should be exercised in the handling of the products. We recommend
®
all users of QIAGEN
recombinant DNA experiments, or to other applicable guidelines.
products to adhere to the NIH guidelines that have been developed for

QIAseq Stranded mRNA Library Kit Handbook 02/2021
7
Safety Information
When working with chemicals, always wear a suitable lab coat, disposable gloves, and
protective goggles. For more information, please consult the appropriate safety data sheets
(SDSs). These are available online in convenient and compact PDF format at
www.qiagen.com/safety where you can find, view, and print the SDS for each QIAGEN kit
and kit component.
Quality Control
In accordance with QIAGEN’s ISO-certified Quality Management System, each lot of QIAseq
Stranded mRNA Library Kit is tested against predetermined specifications to ensure consistent
product quality.

8
QIAseq Stranded mRNA Library Kit Handbook 02/2021
Introduction
A typical mammalian cell contains 10–30 pg of total RNA. The majority of RNA within cells
are rRNAs, with mRNA accounting for only 1–5% of the total cellular RNA. Approximately
360,000 mRNA molecules are present in a single mammalian cell, with approximately
12,000 different mRNA transcripts per cell. Some mRNAs comprise as much as 3% of the
mRNA pool, whereas others account for less than 0.01%. These “rare” or “low-abundance”
mRNAs may have a copy number of only 5–15 molecules per cell. However, these rare species
may account for as many as 11,000 different mRNA species, comprising 45% of the mRNA
population. For more information, see Alberts, B., et al. (1994)
3rd ed. New York: Garland Publishing, Inc.
In whole transcriptome next-generation sequencing (NGS) applications, it is of great interest
to maximize the amount of information generated from a single sequencing run. Enriching for
mRNA from the total cellular RNA pool allows scientists to reduce the amount of rRNA and
other non-mRNAs in their RNAseq data and increases the sequencing results from mRNA. The
QIAseq Stranded mRNA Enrichment Kit includes all the necessary reagents and buffers for the
isolation of poly(A)+ mRNA.
Molecular Biology of the Cell
.
NGS library preparation of RNA samples
The QIAseq Stranded mRNA Library Kits enable one-day, accurate stranded NGS library
construction from a broad range of RNA inputs. This kit includes magnetized QIAseq Beads
for fast and efficient reaction cleanups between protocol steps and Y-shaped sample index
adapter plates, which enable sample multiplexing. A total of 384 samples can be sequenced
together per lane on an Illumina NGS instrument by combining different sets of UDI sample
index plates.
Compared to other protocols, many novel advancements are included in the kit. During reverse
transcription, the optimized RT enzyme and buffers do not require the usage of toxic reagents
such as Actinomycin D to enhance strand specificity. In the second-strand synthesis reaction,

QIAseq Stranded mRNA Library Kit Handbook 02/2021
9
a specialized combination of enzymes and optimized buffering not only enables degradation
of the RNA strand, generation of a second cDNA strand, and generation of blunt DNA ends,
but it also guarantees the A-base addition required for the efficient ligation of Illuminacompatible adapters. The novel strand-specific ligation step establishes the strand specificity
of the QIAseq Stranded RNA Kit protocol without additional reagents or laborious and timeconsuming protocol steps. Finally, the CleanStart PCR Mix utilizes a high-fidelity DNA polymerase
to efficiently amplify the RNAseq library irrespective of GC content, while also using a novel method
to degrade previously generated QIAseq Stranded RNAseq libraries to guard against PCR amplicon
contamination.
Globin mRNA removal using QIAseq FastSelect RNA Removal Kits
When working with whole blood samples, globin mRNA represents a substantial library
contaminant. While mRNA enrichment effectively eliminates rRNA, globin mRNA remains a
concern. QIAseq FastSelect RNA Removal Kit (sold separately) is a breakthrough technology
that rapidly and efficiently removes up to 99% of globin mRNA during RNAseq library
preparation. Simply add the QIAseq FastSelect reagent during the NGS library preparation,
and unwanted RNAs are eliminated from the library.
Principle and procedure
The QIAseq Stranded mRNA Library Kit comprises the QIAseq Stranded mRNA Enrichment
Kit, the QIAseq Stranded RNA Lib Enzyme Kit, QIAseq Beads, and QIAseq Y-Adapter Kits.
The QIAseq FastSelect RNA Removal Kit (sold separately) is designed for fast and efficient
removal of globin mRNA during library preparation of whole blood samples. Together, these
kits enable preparation of mRNA-enriched, globin-mRNA–depleted, and strand-specific NGS
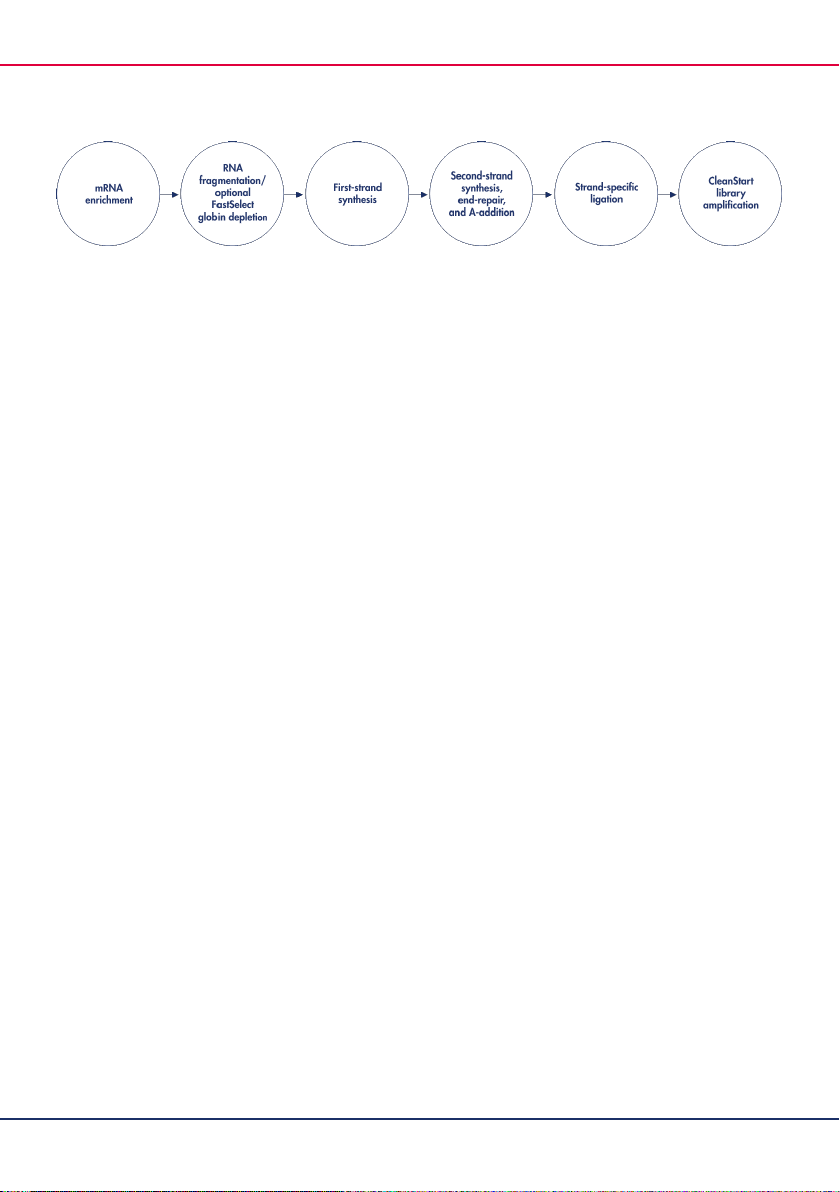
libraries from total RNA in less than 5.5 hours (Figure 1).
10
QIAseq Stranded mRNA Library Kit Handbook 02/2021
Figure 1. QIAseq Stranded mRNA Library Kit workflow. The QIAseq Stranded mRNA Library Kit provides all necessary
reagents for mRNA enrichment and the preparation of strand-specific NGS libraries.
Optional: If you are working with total RNA from whole blood, the QIAseq FastSelect RNA Removal Kit provides all
necessary reagents for the removal of globin mRNA. The CleanStart library amplification step utilizes a high-fidelity
DNA polymerase to amplify the NGS libraries and prevent PCR contamination.
The following reactions are part of the workflow (Figure 2):
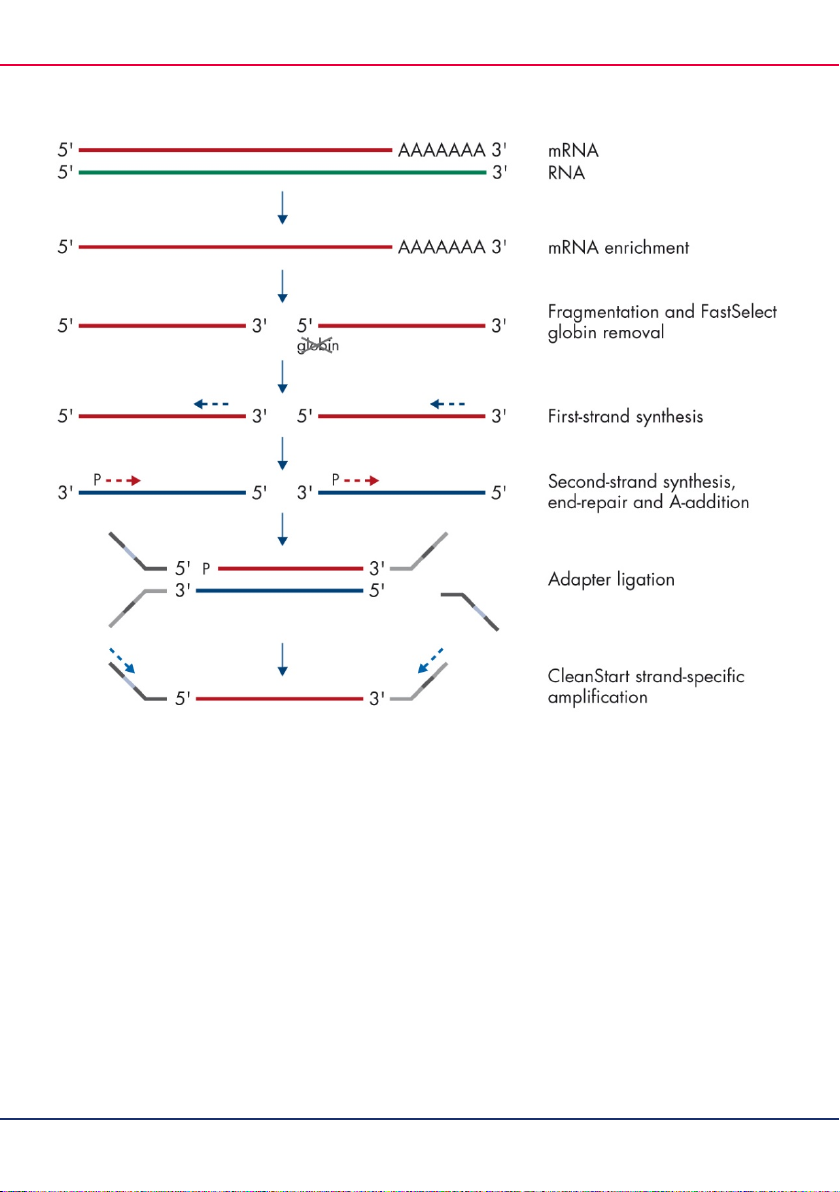
mRNA enrichment
Oligo-dT probes are covalently attached to the surface of the magnetically charged QIAGEN
Pure mRNA Beads. The mRNA binds rapidly and efficiently to the oligo-dT probes in the
presence of Buffer mRBB. The bound mRNA is then washed and eluted, providing a highly
enriched pool of mRNA.
RNA fragmentation/FastSelect RNA removal
Prior to RNA heat fragmentation, the FastSelect reagent is directly combined with the enriched
mRNA and the library prep-specific buffers. Heat fragmentation (if necessary) is then performed
and the reaction temperature is gradually cooled to room temperature (15–25°C).
First-strand synthesis
First-strand synthesis is performed using an RNase H- Reverse Transcriptase (RT) in combination
with random primers.
Second-strand synthesis, end-repair, and A-addition
Second-strand synthesis is performed using 5′ phosphorylated random primers. This enables
subsequent strand-specific ligation, as only one strand of the library is 5′ phosphorylated.
Strand-specific ligation
QIAseq adapters are efficiently asymmetrically ligated to the inserts due to the 5′ phosphate that
results from 5′ phosphorylated second-strand synthesis reaction.
CleanStart library amplification
QIAseq CleanStart PCR reagents use a proprietary PCR reaction, in conjunction with
modification enzymes, to ensure that previously constructed NGS libraries are removed.
QIAseq Stranded mRNA Library Kit Handbook 02/2021
11
Figure 2. QIAseq Stranded mRNA Library Kit with integrated QIAseq FastSelect globin mRNA removal.

12
QIAseq Stranded mRNA Library Kit Handbook 02/2021
Next-generation sequencing
Libraries prepared with the QIAseq Stranded mRNA Library Kits can be sequenced with
Illumina
NGS systems (NextSeq® 500/550, HiSeq® 1000, HiSeq 1500, HiSeq 2000, HiSeq
2500, HiSeq 3000/4000, and NovaSeq™ 6000). When using unique dual indexes (cat.
nos. 180440, 180441, 180442, 180443, 180445), 74 bp paired-end reads and dual 10
bp index reads are required. When using combinatorial indexes (cat. no. 180773 or
180775), 76 bp paired-end reads and dual 8 bp index reads are required.
Data analysis
Downstream NGS data can be analyzed with the QIAGEN CLC Genomics Workbench. When
performing read alignment, the QIAseq Stranded Libraries represent the sense strand (or
positive DNA strand) of the RNA sequence due to the strand-specific ligation during the second
strand cDNA synthesis.

QIAseq Stranded mRNA Library Kit Handbook 02/2021
13
Equipment and Reagents to be Supplied by User
When working with chemicals, always wear a suitable lab coat, disposable gloves, and
protective goggles. For more information, consult the appropriate safety data sheets (SDSs),
available from the product supplier.
100% ethanol (ACS grade)
Nuclease-free pipette tips and tubes
PCR tubes (0.2 ml individual tubes or tube strips) (VWR cat. no. 20170–012 or 93001–118) or
plates
1.5 ml LoBind
Ice
Microcentrifuge
Thermal cycler
Vortexer
Magnet for bead cleanups:
Tubes: MagneSphere
Plates: DynaMag™-96 Side Magnet (Thermo Fisher Scientific cat. no. 12331D)
Optional spike-in: ERCC RNA Spike-In Mix (Thermo Fisher Scientific cat. no. 4456740)
Library QC: 2100 Bioanalyzer
(Agilent cat. no. 5067-4626)
Preferred qPCR library quantification method: QIAseq Library Quant Array (cat. no. 333304) or
QIAseq Library Quant Assay Kit (cat. no. 333314)
®
tubes (Eppendorf cat. no. 022431021)
®
Technology Magnetic Separation Stand (Promega cat. no. Z5342)
®
(Agilent cat. no. G2939BA), Agilent High Sensitivity DNA Kit

14
QIAseq Stranded mRNA Library Kit Handbook 02/2021
Important Notes
High-quality RNA is essential for robust library preparation and sequencing. QIAGEN provides
a range of solutions for purification of total RNA (Table 1).
Table 1. Recommended kits for purification of total RNA
Kit Cat. no. Starting material
RNeasy® Micro Kit 74004 Small amounts of cells and tissue
RNeasy Mini Kit 74104, 74106 Animal/human tissues and cells
RNeasy 96 Kit 74181, 74182 Animal/human tissues and cells
RNeasy FFPE Kit 73504 FFPE tissue samples
QIAamp® ccfDNA/RNA Kit 55184 Animal and human plasma and serum
exoRNeasy Midi Kit 77144 Animal and human plasma and serum
exoRNeasy Maxi Kit 77164 Animal and human plasma and serum
Ensure that total RNA samples are of high quality relative to their sample type. For additional
information, see “Appendix D: General Remarks on Handling RNA”.
RNA quantification: The concentration and purity of total RNA isolated from cells and
fresh/frozen tissues should be determined by measuring the absorbance in a
spectrophotometer, such as the QIAxpert
acids are highly dependent on pH, we recommend preparing dilutions and
measuring absorbance in 10 mM Tris·Cl, pH 7.5, instead of RNase-Free Water. Pure
RNA has an
RNA integrity: The integrity and size distribution of total RNA from cells and
A
:
A
ratio of 1.9–2.1 in 10 mM Tris·Cl, pH 7.5.
260
280
fresh/frozen tissue can be confirmed using an automated analysis system (such as the
QIAGEN QIAxcel or Agilent 2100 Bioanalyzer) that assesses RNA integrity by
monitoring the ribosomal RNA bands. Although the RNA integrity number (RIN)
should ideally be ≥8, successful library prep is still possible with samples with RIN
values <8. In situations where the RNA is highly degraded, we recommend to
consider using QIAseq FastSelect in combination with the QIAseq Stranded RNA
Library Kit UDI instead of using an mRNA enrichment strategy, if possible.
®
. Since the spectral properties of nucleic

QIAseq Stranded mRNA Library Kit Handbook 02/2021
15
Indexing recommendations:
The QIAseq Stranded mRNA Library Kits include a QIAseq Y-Adapter plate with either CDI
adapters or UDI adapters. We recommend using the QIAseq Y-Adapter plates delivered with
the kit. Each QIAseq Stranded mRNA Library includes one of the following:
QIAseq Unique Dual-Index (UDI) Y-Adapter Plate (24)
QIAseq Unique Dual-Index (UDI) Y-Adapter Plate A, B, C, or D (96)
QIAseq Combinatorial Dual-Index (CDI) Y-Adapter Plate (24)
QIAseq Combinatorial Dual-Index (CDI) Y-Adapter Plate (96)
Sample multiplexing is one of the most important NGS tools for increasing throughput and
reducing costs. It works by combining multiple samples to be processed together in a single
sequencing run; as a consequence, sequencing reads need to be demultiplexed by reassigning
each single read to its original source library. This is facilitated by the integration of index
sequences into the individual adapter molecules.
CDI adapters use twelve i7 and eight i5 barcode motifs that can be combined to form up to 96
combinatory dual indices. In contrast, QIAseq UDI Adapters use a fixed combination of 2
unique barcode motifs per adapter molecule. Therefore, each single-index motif is only used
once on any UDI adapter plate. To multiplex more than 96 libraries in a single sequencing run,
combine kits with different UDI Y-adapter plates. Importantly, usage of UDI adapters effectively
mitigates the risk of read misassignment due to index hopping. This is enabled by filtering
misassigned reads during the demultiplexing of individual samples, thus generating highly
accurate output data. For this reason, usage of UDI adapters is highly recommended. For more
information on QIAseq Y-Adapter plates, please refer to “Appendix A: QIAseq Dual-Index Y-
Adapters”, page 39.
The protocol can be stopped at several steps and picked up on the following day. The stopping
points are as follows:
End of “Protocol: mRNA Enrichment “
End of “Protocol: First-strand Synthesis”
End of “Protocol: Second-strand Synthesis, End-repair, and A-addition”
End of “Protocol: Strand-specific Ligation”
End of “Protocol: CleanStart Library Amplification”

16
QIAseq Stranded mRNA Library Kit Handbook 02/2021
Protocol: mRNA Enrichment
Important points before starting
This protocol is optimized for enriching RNA originating from all eukaryotic species with a poly-
A tail.
The recommended total RNA input is 100 ng – 1 µg.
See “Appendix C: mRNA Enrichment in 200 µl Plates” for enrichment of mRNA using 200 µl
strip tubes or 96-well plates.
Things to do before starting
All buffers and reagents should be vortexed before use to ensure thorough mixing.
Vortex the Pure mRNA Beads for 3 min before the first use or for 1 min before subsequent uses.
Heat a water bath or heating block to 70°C, and heat Buffer OEB to 70°C.
Unless otherwise indicated, all protocol steps, including centrifugation, should be performed at
room temperature.

QIAseq Stranded mRNA Library Kit Handbook 02/2021
17
Procedure: mRNA enrichment in 1.5 ml tubes
1. Vortex Pure mRNA Beads for 1 min to thoroughly resuspend.
2. Prepare the enrichment reaction according to Table 2. Briefly centrifuge, vortex, and
centrifuge briefly again.
Table 2. Setup of enrichment reaction
Component Volume/reaction
Total RNA (100 ng – 1 µg) Variable
RNase Inhibitor 1 µl
Buffer mRBB 250 µl
Thoroughly resuspended Pure mRNA Beads 25 µl
Nuclease-Free Water Bring total reaction volume to 526 µl
Total volume 526 µl

18
QIAseq Stranded mRNA Library Kit Handbook 02/2021
3. Incubate for 3 min at 70°C, followed by 10 min at room temperature.
4. Briefly centrifuge, and then place the tubes onto a magnetic rack. After the solution has
cleared (~2 min), discard the supernatant.
5. Add 400 µl of Buffer OW2. Vortex, centrifuge briefly, and place the tubes onto a
magnetic rack. After the solution has cleared, discard the supernatant.
6. Repeat step 5.
7. Add 50 µl Buffer OEB. Vortex, centrifuge briefly, and incubate at 70°C for 3 min.
8. Remove the sample from 70°C and place at room temperature for 5 min.
9. Add 50 µl of Buffer mRBB and vortex. Briefly centrifuge, and incubate at room
temperature for 10 min.
10. Briefly centrifuge, and then place the tubes onto a magnetic rack. After the solution has
cleared, carefully discard the supernatant. Leave any residual liquid in the tube to
minimize bead loss.
11. Add 400 µl of Buffer OW2. Vortex, centrifuge briefly, and place tubes onto a magnetic
rack. After the solution has cleared, discard the supernatant.
12. Add 29 µl of Buffer OEB heated to 70°C to the bead pellet, and vortex.
13. Briefly centrifuge, and place the tubes onto a magnetic rack. After the solution has
cleared, transfer 27 µl of the supernatant to a clean tube. The supernatant contains
enriched poly(A)+ RNA.
14. Proceed to “Protocol: Fragmentation/FastSelect RNA Removal”. Alternatively, the
samples can be stored at –90 to –65°C.

QIAseq Stranded mRNA Library Kit Handbook 02/2021
19
Protocol: Fragmentation/FastSelect RNA Removal
Important points before starting
The entire 27 µl product from “Protocol: mRNA Enrichment” is the starting material for
fragmentation/FastSelect RNA removal.
FastSelect removal of globin mRNA (sold separately, see “Ordering Information”) is
recommended when mRNA has been enriched from whole blood samples.
To generate optimal insert sizes, fragmentation time needs to be determined for each
experiment, depending on the quality and origin of the RNA. Follow the recommendations in
Table 4.
Procedure
1. Thaw previously enriched mRNA on ice. Gently mix, then briefly centrifuge to collect
residual liquid from the sides of the tubes, and return to ice.
2. Prepare the reagents required for the RNA fragmentation and QIAseq FastSelect globin
mRNA removal.
2a. Thaw RT Buffer, 5x, Nuclease-Free Water, and the tube(s) from the appropriate
QIAseq FastSelect RNA Removal Kits at room temperature.
2b. Mix by vortexing and then briefly centrifuge to collect residual liquid from the sides
of the tubes.
3. On ice, prepare the fragmentation/RNA removal reaction according to Table 3. Briefly
centrifuge, mix by pipetting up and down 10 times, and centrifuge briefly again.
Note: If setting up more than one reaction, prepare a volume of master mix 10% greater
than what is required for the total number of reactions.

20
QIAseq Stranded mRNA Library Kit Handbook 02/2021
Table 3. Setup of fragmentation/RNA removal reactions
Component Volume/reaction
mRNA enrichment reaction (already in tube) 27 µl
RT Buffer, 5x 8 µl
QIAseq FastSelect –Globin* 1 µl
ERCC Control† Optional
Nuclease-Free Water 1 µl
Total volume 37 µl
* From QIAseq FastSelect RNA Removal Kit. If not performing, add 1 µl of Nuclease-Free Water instead.
†
ERCC Control RNA (see “Equipment and Reagents to be Supplied by User”) can be added according to the
concentrations specified by the manufacturer. If added, replace the Nuclease-Free Water (1 µl) with ERCC.
4. Incubate as described in Table 4, according to your input RNA quality and approximate
insert size.
Table 4. Fragmentation/RNA removal protocol
Input RNA quality Step Insert size ~150–250 bp Insert size ~350 bp
High quality (RIN >9) 1* 15 min at 95°C 3 min at 95°C
Moderate quality (RIN 5–6) 1* 10 min at 95°C 3 min at 95°C
FFPE or degraded sample (RIN <3) 1* No fragmentation† No fragmentation†
Important: Only include steps 2–9 if
performing FastSelect Globin mRNA
Removal
Steps 2–9 are performed when using
FastSelect –Globin, regardless of Input
RNA quality. They need to be performed
whether the RNA is high quality,
moderate quality, FFPE, or degraded.
* Choose one option for the Step 1 time, according to the input RNA quality and desired insert size.
†
Also suitable for exosomal RNA or RNA of other origin with a size between 80–500 bp.
2 2 min at 75°C 2 min at 75°C
3 2 min at 70°C 2 min at 70°C
4 2 min at 65°C 2 min at 65°C
5 2 min at 60°C 2 min at 60°C
6 2 min at 55°C 2 min at 55°C
7 2 min at 37°C 2 min at 37°C
8 2 min at 25°C 2 min at 25°C
9 Hold at 4°C Hold at 4°C
5. Proceed immediately to “Protocol: First-strand Synthesis”.

QIAseq Stranded mRNA Library Kit Handbook 02/2021
21
Protocol: First-strand Synthesis
Important points before starting
The entire 37 µl product from “Protocol: Fragmentation/FastSelect RNA Removal” is the starting
material for first-strand synthesis.
Set up first-strand synthesis on ice.
Do not vortex any first-strand synthesis reagents or reactions.
Use a thermal cycler with a heated lid.
Ensure that QIAseq Beads are brought to room temperature before using.
Ensure that the QIAseq Beads are thoroughly mixed at all times. This necessitates working
quickly and resuspending the beads immediately before use. If a delay in the protocol occurs,
simply vortex the beads.
Procedure
1. Prepare the reagents required for first-strand synthesis.
1a. Thaw 1 M DTT at room temperature.
1b. Mix by flicking the tube.
1c. Centrifuge to collect residual liquid from the sides of the tube.
Note: RT Enzyme and RNase Inhibitor should be removed from the freezer just before use
and placed on ice. After use, immediately return the enzymes to the freezer.
2. Dilute 1 M DTT to 0.4 M for use in Table 5.
3. On ice, prepare the first-strand reaction according to Table 5. Briefly centrifuge, mix by
pipetting up and down 10 times, and centrifuge briefly again.
Note: If setting up more than one reaction, prepare a volume of master mix 10% greater
than what is required for the total number of reactions.

22
QIAseq Stranded mRNA Library Kit Handbook 02/2021
Table 5. Setup of first-strand reaction
Component Volume/reaction
Fragmentation/RNA removal reaction (already in tube) 37 µl
Diluted DTT (0.4 M) 1 µl
RT Enzyme 1 µl
RNase Inhibitor 1 µl
Total volume 40 µl
4. Incubate as described in Table 6.
Table 6. First-strand protocol
Step Temperature Incubation time
1 25 °C 10 min
2 42°C 15 min
3 70°C 15 min
4 4°C Hold
5. Add 56 µl of resuspended QIAseq Beads. Vortex for 3 s and briefly centrifuge.
6. Incubate for 5 min at room temperature.
7. Place the tubes/plate onto a magnetic rack. After the solution has cleared (~10 min or
longer), carefully remove and discard the supernatant.
Important: Do not discard the beads, because they contain the DNA of interest.
8. With the tubes still on the magnetic stand, add 200 µl of 80% ethanol. Rotate the
tubes/plate 3 times to wash the beads. Carefully remove and discard the wash.
9. Repeat the ethanol wash.
Important: Completely remove all traces of the ethanol after this second wash. Briefly
centrifuge and return the tubes to the magnetic stand. Remove the ethanol with a 200 µl
pipette first, and then use a 10 µl pipette to remove any residual ethanol.

QIAseq Stranded mRNA Library Kit Handbook 02/2021
23
10. With the tubes/plate (caps opened) still on the magnetic stand, air-dry at room
temperature for 5–10 min.
Note: Visually inspect that the pellet is completely dry and that all residual ethanol has
evaporated.
11. Remove the tubes/plate from the magnetic stand. Eluate the DNA from the beads by
adding 40 µl Nuclease-Free Water. Mix well by pipetting.
12. Return the tubes/plate to the magnetic rack until the solution has cleared.
13. Transfer 38.5 µl supernatant to clean tubes/plate.
14. Proceed to “Protocol: Second-strand Synthesis, End-repair, and A-addition”. Alternatively,
the samples can be stored at –30 to –15°C in a constant-temperature freezer.

24
QIAseq Stranded mRNA Library Kit Handbook 02/2021
Protocol: Second-strand Synthesis, End-repair,
and A-addition
Important points before starting
The entire 38.5 µl product from “Protocol: First-strand Synthesis” is the starting material for the
second-strand synthesis, end-repair, and A-addition procedure.
Set up reaction on ice.
Do not vortex any second-strand synthesis, end-repair, and A-addition reagents or reactions.
Use a thermal cycler with a heated lid.
Ensure the QIAseq Beads are brought to room temperature before using.
Ensure the QIAseq Beads are thoroughly mixed at all times. This necessitates working quickly
and resuspending the beads immediately before use. If a delay in the protocol occurs, simply
vortex the beads.
Procedure
1. Prepare required reagents.
1a. Thaw Second Strand Buffer, 10x, at room temperature.
1b. Mix by vortexing.
1c. Centrifuge to collect residual liquid from the sides of the tubes.
Note: Second Strand Enzyme Mix should be removed from the freezer just before use
and placed on ice. After use, immediately return the enzyme to the freezer.
2. On ice, prepare the first-strand reaction according to Table 7. Briefly centrifuge, mix by
pipetting up and down 10 times, and centrifuge briefly again.
Note: If setting up more than one reaction, prepare a volume of master mix 10% greater
than what is required for the total number of reactions.

QIAseq Stranded mRNA Library Kit Handbook 02/2021
25
Table 7. Setup of second-strand synthesis, end repair, and A-addition reaction
Component Volume/reaction
Product from “Protocol: First-strand Synthesis” 38.5 µl
Second Strand Buffer, 10x 5 µl
Second Strand Enzyme Mix 6.5 µl
Total volume 50 µl
3. Incubate as described in Table 8.
Table 8. Second-strand synthesis, end-repair, and A-addition protocol
Step Temperature Incubation time
1 25 °C 30 min
2 65°C 15 min
3 4°C Hold
4. Add 70 µl of resuspended QIAseq Beads. Vortex for 3 s and briefly centrifuge.
5. Incubate for 5 min at room temperature.
6. Place the tubes/plate onto a magnetic rack. After the solution has cleared (~10 min or
longer), carefully remove and discard the supernatant.
Important: Do not discard the beads, because they contain the DNA of interest.
7. With the tubes still on the magnetic stand, add 200 µl of 80% ethanol. Rotate the
tubes/plate 3 times to wash the beads. Carefully remove and discard the wash.
8. Repeat the ethanol wash.
Important: Completely remove all traces of the ethanol after this second wash. Briefly
centrifuge and return the tubes to the magnetic stand. Remove the ethanol with a 200 µl
pipette first, and then use a 10 µl pipette to remove any residual ethanol.

26
QIAseq Stranded mRNA Library Kit Handbook 02/2021
9. With the tubes/plate (caps opened) still on the magnetic stand, air-dry at room
temperature for 5–10 min.
Note: Visually inspect that the pellet is completely dry and that all ethanol has
evaporated.
10. Remove the tubes/plate from the magnetic stand, and elute the DNA from the beads by
adding 52 µl Nuclease-Free Water. Mix well by pipetting.
11. Return the tubes/plate to the magnetic rack until the solution has cleared.
12. Transfer 50 µl supernatant to clean tubes/plate.
13. Proceed to “Protocol: Strand-specific Ligation”. Alternatively, the samples can be stored
at –30 to –15°C in a constant-temperature freezer.

QIAseq Stranded mRNA Library Kit Handbook 02/2021
27
Protocol: Strand-specific Ligation
Important points before starting
The entire 50 µl product from “Protocol: Second-strand Synthesis, End-repair, and A-addition” is
the starting material for the strand-specific ligation.
Set up reaction on ice.
Do not vortex any strand-specific ligation enzymes or reactions.
Use a thermal cycler without a heated lid.
For UDI adapter plates, the layout of the 24-plex and 96-plex plates is described in “Appendix
A: QIAseq Dual-Index Y-Adapters”. The index motifs used in the QIAseq Unique Dual-Index Kits
are listed at www.qiagen.com.
For CDI adapter plates, the layout and barcode sequences are described in Appendix A.
Unused, undiluted adapters can be stored at –30 to –15°C. If desired, residual diluted adapter
can be removed and discarded before plate storage.
Important: Do not reuse diluted adapter due to the risk of barcode cross-contamination
and lower-than-expected adapter concentration after storage of diluted material.
Ensure the QIAseq Beads are brought to room temperature before using.
Ensure the QIAseq Beads are thoroughly mixed at all times. This necessitates working quickly
and resuspending the beads immediately before use. If a delay in the protocol occurs, simply
vortex the beads.
Procedure
1. Prepare the required reagents.
1a. Thaw Ultralow Input Ligation Buffer, 4x, and Ligation Initiator at room temperature.
1b. Mix by vortexing.
1c. Centrifuge to collect residual liquid from the sides of the tubes.
Note: Ultralow Input Ligase should be removed from the freezer just before use and
placed on ice. After use, immediately return the enzyme to the freezer.

28
QIAseq Stranded mRNA Library Kit Handbook 02/2021
2. Prepare the adapter plate as follows. The layouts of the 24-plex and 96-plex single-use
adapter plates are displayed in Appendix A.
Note: If multiplexing 1–6 samples, consult the
Protocols
document from Illumina to select the ideal adapter combinations.
Low-Plex Pooling Guidelines for Enrichment
2a. Thaw the adapter plate at room temperature. Vortex and centrifuge briefly before
use.
2b. Remove the clear protective adapter plate lid and carefully pierce only the foil seal
for each adapter well to be used, using a fresh tip to pierce each well.
2c. Remove an aliquot and dilute the adapters as suggested in Table 9.
Table 9. Dilution of QIAseq adapters
Total RNA input amount Adapter dilution
100 ng 1:100
500 ng 1:25
1 µg 1:12.5
5 µg 1:5
2d. Replace the plate lid and freeze unused, undiluted adapter at –30 to –15°C.
Remove and discard residual diluted adapter before plate storage.
3. On ice, prepare the strand-specific ligation reaction according to Table 10. Briefly
centrifuge. Mix by pipetting up and down 15–20 times and centrifuge briefly again.
Note: If setting up more than one reaction, prepare a volume of master mix 10% greater
than what is required for the total number of reactions.
Important: Pipet slowly to mix. The Ligation Initiator and the reaction mix are very
viscous.

QIAseq Stranded mRNA Library Kit Handbook 02/2021
29
Table 10. Setup of Strand-specific ligation reaction
Component Volume/reaction
Product from “Protocol: Second-strand Synthesis, End-repair, and A-addition” 50 µl
Diluted adapter* 2 µl
Ultralow Input Ligation Buffer, 4x 25 µl
Ultralow Input Ligase 5 µl
Ligation Initiator 6.5 µl
Nuclease-Free Water 11.5 µl
Total volume 100 µl
* Choose a unique adapter for each sample.
4. Incubate at 25°C for 10 min.
Important: Do not use a heated lid.
5. Add 80 µl of resuspended QIAseq Beads. Vortex for 3 s and briefly centrifuge.
6. Incubate for 5 min at room temperature.
7. Place the tubes/plate onto a magnetic rack. After the solution has cleared (~10 min or
longer), carefully remove and discard the supernatant.
Important: Do not discard the beads, because they contain the DNA of interest.
8. With the tubes still on the magnetic stand, add 200 µl of 80% ethanol. Rotate the
tubes/plate 3 times to wash the beads. Carefully remove and discard the wash.
9. Repeat the ethanol wash.
Important: Completely remove all traces of the ethanol after this second wash. Briefly
centrifuge and return the tubes to the magnetic stand. Remove the ethanol with a 200 µl
pipette first, and then use a 10 µl pipette to remove any residual ethanol.
10. With the tubes/plate (caps opened) still on the magnetic stand, air-dry at room
temperature for 5–10 min.
Note: Visually inspect that the pellet is completely dry and that all ethanol has
evaporated.

30
QIAseq Stranded mRNA Library Kit Handbook 02/2021
11. Remove the tubes from the magnetic stand, and elute the DNA from the beads by adding
92 µl Nuclease-Free Water. Mix well by pipetting.
12. Return the tubes/plate to the magnetic rack until the solution has cleared.
13. Transfer 90 µl supernatant to clean tubes/plate.
14. Add 108 µl of resuspended QIAseq Beads. Vortex for 3 s and briefly centrifuge.
15. Incubate for 5 min at room temperature.
16. Place the tubes/plate onto a magnetic rack. After the solution has cleared (~10 min or
longer), carefully remove and discard the supernatant.
Important: Do not discard the beads, because they contain the DNA of interest.
17. With the tubes still on the magnetic stand, add 200 µl of 80% ethanol. Rotate the
tubes/plate 3 times to wash the beads. Carefully remove and discard the wash.
18. Repeat the ethanol wash.
Important: Completely remove all traces of the ethanol after this second wash. Briefly
centrifuge and return the tubes to the magnetic stand. Remove the ethanol with a 200 µl
pipette first, and then use a 10 µl pipette to remove any residual ethanol.
19. With the tubes/plate (caps opened) still on the magnetic stand, air-dry at room
temperature for 5–10 min.
Note: Visually inspect that the pellet is completely dry and that all ethanol has
evaporated.
20. Remove the tubes/plate from the magnetic stand, and elute the DNA from the beads by
adding 25 µl Nuclease-Free Water. Mix well by pipetting.
21. Return the tubes/plate to the magnetic rack until the solution has cleared.
22. Transfer 23.5 µl of supernatant to clean tubes/plate.
23. Proceed to “Protocol: CleanStart Library Amplification
”. Alternatively, the samples can be
stored at −30 to –15°C in a constant-temperature freezer.

QIAseq Stranded mRNA Library Kit Handbook 02/2021
31
Protocol: CleanStart Library Amplification
Important points before starting
The entire 23.5 µl product from “Protocol: Strand-specific Ligation” is the starting material for the
CleanStart Library Amplification.
QIAseq CleanStart PCR reagents use a proprietary PCR reaction, in conjunction with
modification enzymes, to ensure that previously constructed NGS libraries are removed.
Important: If a previously amplified CleanStart Library needs to be reamplified – for
instance, when an additional library is needed to replace a failed NGS run – omit the
decontamination step of the PCR protocol (incubation for 15 min at 37°C) to disable
selective degradation.
Set up reaction on ice.
Do not vortex any CleanStart library amplification reagents or reactions.
Use a thermal cycler with a heated lid.
Ensure the QIAseq Beads are brought to room temperature before using.
Ensure the QIAseq Beads are thoroughly mixed at all times. This necessitates working quickly
and resuspending the beads immediately before use. If a delay in the protocol occurs, simply
vortex the beads.

32
QIAseq Stranded mRNA Library Kit Handbook 02/2021
Procedure
1. Prepare the reagents required for CleanStart library amplification.
1a. Thaw CleanStart PCR Primer Mix at room temperature and thaw CleanStart PCR
Mix, 2x, on ice.
1b. Mix by flicking the tube.
1c. Centrifuge to collect residual liquid from the sides of the tubes.
2. On ice, prepare the library amplification reaction according to Table 11. Briefly
centrifuge, mix by pipetting up and down 10 times, and centrifuge briefly again.
Note: If setting up more than one reaction, prepare a volume of master mix 10% greater
than that required for the total number of reactions.
Table 11. Setup of library amplification
Component Volume/reaction
Product from “Protocol: Strand-specific Ligation” 23.5 µl
CleanStart PCR Mix, 2x 25 µl
CleanStart PCR Primer Mix 1.5 µl
Total volume 50 µl
3. Select the number of PCR cycles, based on total RNA input, according to Table 12.
Important: When removing globin mRNA with QIAseq FastSelect –Globin Kit, 2
additional cycles of library amplification need to be performed.
Table 12. Recommended number of PCR cycles, based on total RNA input
Total RNA input Number of amplification cycles*
100 ng 14–16†
500 ng 11–13†
1 µg 9–11†
5 µg 7–9†
* Use selected number of cycles for amplification in Table 13.
†
Important: When removing globin mRNA with QIAseq FastSelect -Globin RNA Removal, 2 additional cycles of
library amplification need to be performed.

QIAseq Stranded mRNA Library Kit Handbook 02/2021
33
4. Incubate as described in Table 13.
Table 13. CleanStart library amplification cycling conditions
Step Time Temperature Number of cycles
CleanStart decontamination* 15 min 37°C 1
Initial denaturation 2 min 98°C 1
PCR 20 s
30 s
30 s
Final extension 1 min 72°C 1
Hold ∞ 4°C Hold
* For the reamplification of libraries, omit the CleanStart decontamination step and start with incubation at 98°C for
2 min.
98°C
60°C
72°C
See Table 12
5. After amplification, add 60 µl of QIAseq Beads. Vortex for 3 s and briefly centrifuge.
6. Incubate for 5 min at room temperature.
7. Place the tubes/plate onto a magnetic rack. After the solution has cleared (~10 min or
longer), carefully remove and discard the supernatant.
Important: Do not discard the beads, because they contain the DNA of interest.
8. With the tubes/plate still on the magnetic stand, add 200 µl of 80% ethanol. Rotate the
tube 3 times to wash the beads. Carefully remove and discard the wash.
9. Repeat the ethanol wash.
Important: Completely remove all traces of the ethanol after this second wash. Briefly
centrifuge, and then return the tubes to the magnetic stand. Remove the ethanol with a
200 µl pipette first, and then use a 10 µl pipette to remove any residual ethanol.
10. With the tubes/plate (caps opened) still on the magnetic stand, air-dry at room
temperature for 5–10 min.
Note: Visually inspect that the pellet is completely dry and that all ethanol has
evaporated.

34
QIAseq Stranded mRNA Library Kit Handbook 02/2021
11. Remove the tubes/plate from the magnetic stand, and elute the DNA from the beads by
adding 22 µl Nuclease-Free Water. Mix well by pipetting.
12. Return the tubes/plate to the magnetic rack until the solution has cleared.
13. Transfer 20 µl to clean tubes/plate. This is the QIAseq Stranded Sequencing Library.
14. Proceed to “Recommendations: Library QC, Quantification, and Sequencing”.
Alternatively, the samples can be stored at –30 to –15°C in a constant-temperature
freezer.

QIAseq Stranded mRNA Library Kit Handbook 02/2021
35
Recommendations: Library QC, Quantification,
and Sequencing
NGS library QC
QC can be performed with the Agilent Bioanalyzer or TapeStation. Check for the correct size
distribution of library fragments (~300–500 bp median size) and for the absence of adapters
or adapter-dimers (~130 bp). Figure
libraries prepared from UHRR input material. Both results show no traces of adapter-dimers.
The 1 ng input RNAseq library shows optimal size distribution, while the 50 ng input RNAseq
library shows broader library size distribution (but does not affect the NGS sequencing
quality).
3 shows the library size distributions for 1 ng and 50 ng
Figure 3. QIAseq Stranded RNAseq library size distributions measured with the Agilent Bioanalyzer High Sensitivity
DNA Chip using standard protocol conditions with different input amounts. Left: 1 ng, right: 50 ng UHRR input material.
Important: If excessive adapter-dimers (~130 bp) are prominent after library QC (greater than
1–2% of total library yields), perform a second purification with QIAseq Beads. This can be
accomplished by bringing the sample to a final volume of 55 µl and repeating steps 5–13 of
the CleanStart library amplification.

36
QIAseq Stranded mRNA Library Kit Handbook 02/2021
Preferred library quantification method
The library yield measurements from the Bioanalyzer or TapeStation rely on fluorescence dyes
that intercalate into DNA or RNA. These dyes cannot discriminate between cDNA with or
without adapter sequences, so only complete QIAseq Stranded mRNA Select libraries with full
adapter sequences will be sequenced. As a result, QIAGEN’s QIAseq Library Quant Array Kit
or Assay Kit, which contains laboratory-verified forward and reverse primers, together with a
DNA standard, is highly recommended for accurate quantification of the prepared QIAseq
Stranded mRNA Select library.
Sample dilution, pooling, sequencing, and data analysis
When the QIAseq Stranded libraries have been quantified with the QIAseq Library Quant
Array or Assay Kit, typical QIAseq Stranded library yields are approximately 8–10 nM in
20 µl volume, depending on the quality of the input starting RNA used. This yield is sufficient
for an NGS sequencing run. Dilute the individual QIAseq Stranded libraries to a concentration
of 4 nM. Then, combine libraries with different sample indexes in equimolar amounts. The
recommended starting concentration of the pooled QIAseq Stranded libraries to load onto a
®
MiSeq
is 9 pM, while it is 1.6 pM on a NextSeq.
The recommended starting point for mRNA-enriched samples is 25 M reads/sample. When
using UDIs, 74 bp paired-end reads and dual 10 bp index reads are required. When using
CDIs, 76 bp paired-end reads and dual 8 bp index reads are required. Longer paired-end
reads (UDI libraries: 149 bp paired-end reads and dual 10 bp index reads, and CDI libraries:
151 bp paired-end reads and dual 8 bp index reads) are recommended for rare/novel
transcripts, splice site isoforms, and fusion gene detection.
Data analysis recommendations are outlined in “Appendix B: Data Analysis
Recommendations”.

QIAseq Stranded mRNA Library Kit Handbook 02/2021
37
Comments and suggestions
Low library yields
a) Suboptimal PCR cycle
An increased number of PCR cycles in the CleanStart PCR enrichment step
b) Insufficient RNA input amount
Higher RNA input amounts can lead to higher library yields; however, more RNA
c) Not enough adapter
Decrease adapter dilution. An increased number of adapter molecules during
d) Low mRNA enrichment
Use higher-quality RNA or increase the amount of total RNA input for the mRNA
High Bioanalyzer peak at 120–140 bp (adapter-dimers)
a) Increase adapter dilution
Higher adapter dilutions decrease the adapter-dimer formation during the ligation
b) Decrease QIAseq Beads
Lower QIAseq Bead volumes in cleanup step after ligation (e.g., 0.6x/1.1x instead
c) Increase RNA input amount
Higher RNA input amounts can lead to higher library yields.
Bioanalyzer peaks at higher molecular weight (>1000 bp; PCR overamplification)
Reduce PCR cycle number in the
Single-stranded library products can self-anneal after too many PCR cycles
Troubleshooting Guide
This troubleshooting guide may be helpful in solving any problems that may arise. For more
information, see also the Frequently Asked Questions page in our Technical Support Center:
www.qiagen.com/FAQ/FAQList.aspx. The scientists in QIAGEN Technical Services are
always happy to answer any questions you may have about either the information or protocols
in this handbook (for contact information, visit support.qiagen.com).
number
molecules in ligation (only if
no adapter-dimers are visible)
performance
during ligation
volume in cleanup after
ligation
CleanStart PCR enrichment step
can increase library yields.
input could lead to a higher adapter dilution (see “Appendix C: mRNA
Enrichment in 200 µl Plates”).
ligation can increase ligation efficiency and library yields but can also increase
adapter-dimer formation.
enrichment procedure, to increase resulting mRNA material for the RNAseq library
preparation.
step.
of the standard volumes with 0.8x/1.2x) can increase adapter-dimer depletion but
can lead to lower library yields.
when free PCR primers are no longer available. Reduced PCR cycle numbers
are only necessary when the molarity of the high molecular peak is
significantly elevated (>50% compared to library yields <700 bp).

38
QIAseq Stranded mRNA Library Kit Handbook 02/2021
Comments and suggestions
Strong bias in transcript coverage plots after NGS data analysis
a) Keep input RNA on ice as
Degraded RNA can lead to stronger transcript coverage bias after NGS data
b) Add additional RNase
In cases of strong RNAseq contaminants, add additional RNase Inhibitor into the
Very broad library size distributions
Increase fragmentation time
If the library size distribution is very broad with library sizes >700 bp, increase
much as possible
Inhibitor into reactions
analysis. Degradation can be caused by RNase contamination or prolonged
storage of RNA at elevated temperatures (>4°C).
reactions to inhibit enzymatic activity of QIAseq Stranded RNA library enzymes.
RNA fragmentation time to 20 min, depending on the used RNA input material.

QIAseq Stranded mRNA Library Kit Handbook 02/2021
39
Appendix A: QIAseq Dual-Index Y-Adapters
Generation of sample sheets for Illumina instruments
Index sequences for QIAseq UDI and CDI Y-Adapters are available for download at
www.qiagen.com. Sequencing on the NextSeq, HiSeq X™, or HiSeq 3000/4000 system
follows a different dual-indexing workflow than other Illumina systems. If you are manually
creating sample sheets for these instruments, enter the reverse complement of the i5 index
adapter sequence. If you are using Illumina Experiment Manager, BaseSpace, or Local Run
Manager for run planning, the software will automatically reverse complement index
sequences when necessary.
Ready-to-use sample sheets containing all QIAseq CDI and UDI Y-Adapter barcode sequences
®
are available for MiSeq, NextSeq, MiniSeq
conveniently downloaded from the product pages on www.qiagen.com. These can be
imported and edited using the Illumina Experiment Manager Software, Illumina Local Run
Manager, or any text editor. Make sure to download the appropriate sample sheet for
NextSeq, HiSeq X, or HiSeq 3000/4000 systems depending on whether you are using Local
Run Manager or manually configuring the sequencing run.
, and HiSeq instruments. These can be
Unique Dual-Index Y-Adapters
The layout of the 24-plex and 96-plex (A/B/C/D) single-use UDI adapter plate is shown in
Figure 4 and Figure 5. The index motifs used in the QIAseq Unique Dual-Index Kits are listed
at www.qiagen.com. To make sequencing preparation more convenient, you can download
Illumina-compatible sample sheets for different sequencing instruments at www.qiagen.com.

40
QIAseq Stranded mRNA Library Kit Handbook 02/2021
Figure 4. QIAseq UDI Y-Adapter Plate (24) layout (UDI 1–24).
Figure 5. QIAseq UDI Y-Adapter Plates: Plate A (96) layout (UDI 1–96), Plate B (96) layout (UDI 97–192), Plate C (96)
layout (UDI 193–288), and Plate D (96) layout (UDI 289–384).

QIAseq Stranded mRNA Library Kit Handbook 02/2021
41
Combinatorial Dual-Index Y-Adapters
The layout for the 96-plex and 24-plex single-use CDI adapter plates is shown in Figure 6 and
Figure 7. The barcode sequences used in the QIAseq Combinatorial Dual-Index Kits are listed
in Table 14. Indices 501-508 and 701-712 correspond to the respective Illumina adapter
®
barcodes, Illumina TruSeq
more convenient, you can download Illumina-compatible sample sheets for different
sequencing instruments from the product pages on www.qiagen.com
Follow
Low-Plex Pooling Guidelines for Enrichment Protocols
combinations of D50x/D70x adapters for the corresponding instrument if loading between
1–6 samples onto one flow-cell lane.
Table 14. CDI adapter barcodes sequences used in the QIAseq CDI Y-Adapter Kits (24- and 96-plex Adapter Plates)
D50X barcode name i5 barcode sequence* D70X barcode name i7 barcode sequence
D501 TATAGCCT D701 ATTACTCG
D502 ATAGAGGC D702 TCCGGAGA
D503 CCTATCCT D703 CGCTCATT
D504 GGCTCTGA D704 GAGATTCC
D505 AGGCGAAG D705 ATTCAGAA
D506 TAATCTTA D706 GAATTCGT
D507 CAGGACGT D707 CTGAAGCT
D508 GTACTGAC D708 TAATGCGC
D709 CGGCTATG
D710 TCCGCGAA
D711 TCTCGCGC
D712 AGCGATAG
CD indexes (formerly TruSeq HT). To make sequencing preparation
.
from Illumina to choose the correct
* Sequencing on the MiniSeq, NextSeq, and HiSeq 3000/4000 systems follow a different dual-indexing workflow
than other Illumina systems, which requires the reverse complement of the i5 index adapter sequence.
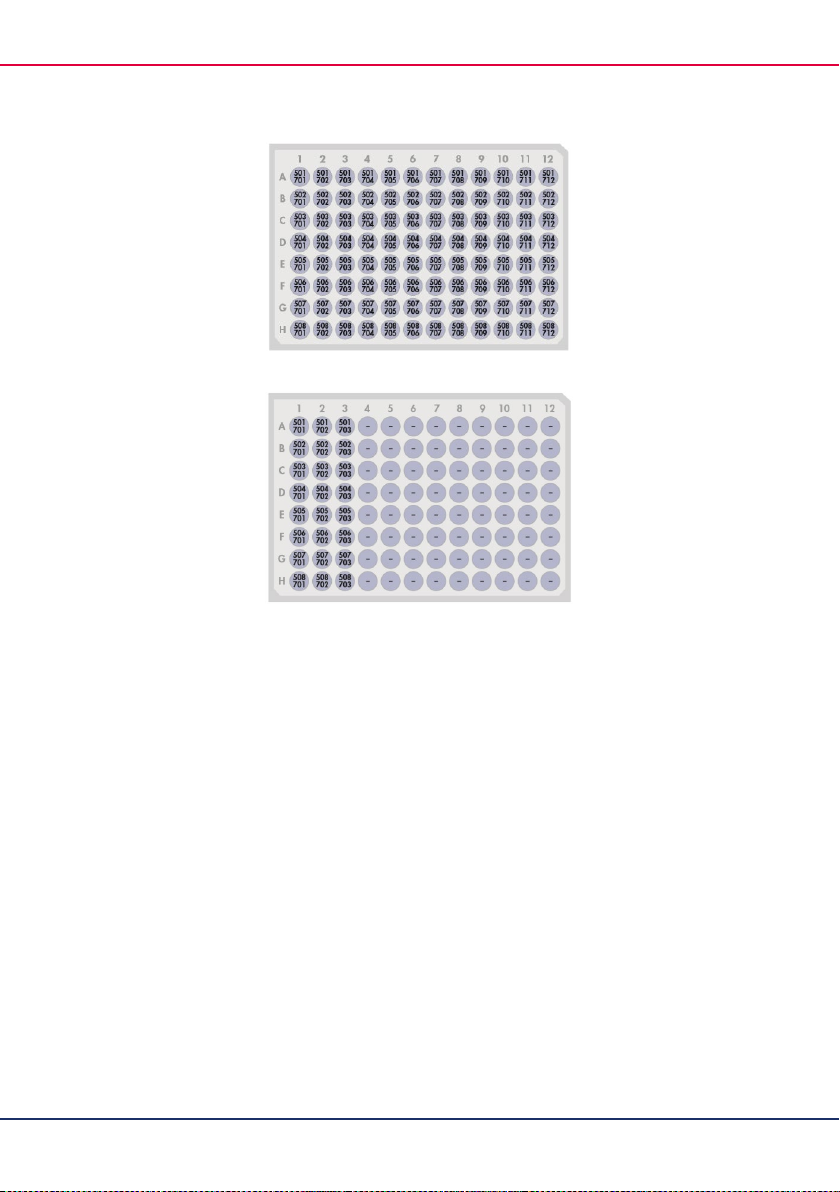
42
QIAseq Stranded mRNA Library Kit Handbook 02/2021
Figure 6. QIAseq CDI Y-Adapter Plate (96) layout (CDI 1–96).
Figure 7. QIAseq CDI Y-Adapter Plate (24) layout (CDI 1–24).

QIAseq Stranded mRNA Library Kit Handbook 02/2021
43
Appendix B: Data Analysis Recommendations
RNAseq alignment
Downstream NGS data can be analyzed with CLC Genomics Workbench (see
digitalinsights.qiagen.com). When doing alignment, the QIAseq Stranded libraries represent
the sense strand of the RNA sequence.
CLC Genomics Workbench is a comprehensive analysis package for the analysis and
visualization of data from all major NGS platforms. The workbench supports and seamlessly
integrates into a typical NGS workflow. CLC Genomics Workbench is available for
®
Windows
algorithms, CLC Genomics Workbench supports key NGS features within genomics,
transcriptomics, and epigenomics research fields. Additionally, it includes all the classical
analysis tools of CLC Main Workbench.
Gene expression interpretation
Ingenuity® Pathway Analysis (IPA®) is an all-in-one, web-based software application that
enables analysis, integration, and understanding of data from gene expression, miRNA, and
SNP microarrays, as well as metabolomics, proteomics, and RNAseq experiments. This
application is a great tool for interpreting the data you generate from the new QIAseq
Stranded RNAseq kit. IPA is the market leader in gene expression analysis, having been cited
in over 18,000 scientific publications to date.
, Mac OS® X, and Linux® platforms. Incorporating cutting-edge technology and
You will find that IPA data analysis and search capabilities help you understand the
significance of data, specific targets, or candidate biomarkers in the context of larger
biological or chemical systems. The software is backed by the Ingenuity Knowledge Base of
highly structured, detail-rich biological and chemical findings. For information on IPA, visit
digitalinsights.qiagen.com.

44
QIAseq Stranded mRNA Library Kit Handbook 02/2021
Appendix C: mRNA Enrichment in 200 µl Plates
This protocol is used to enrich poly(A)+ RNA from total RNA using 200 µl strip tubes or 96-well
plates.
Important points before starting
The recommended total RNA input is 100 ng to 1 µg.
Things to do before starting
All buffers and reagents should be vortexed before use to ensure thorough mixing.
Vortex the Pure mRNA Beads for 3 min before the first use or for 1 min before subsequent uses.
Heat a water bath or heating block to 70°C, and heat Buffer OEB to 7°C.
Unless otherwise indicated, all protocol steps – including centrifugation – should be performed at
room temperature.
Procedure
1. Vortex Pure mRNA Beads for 1 min to thoroughly resuspend.
2. Prepare the enrichment reaction according to Table 15. Briefly centrifuge, vortex, and
centrifuge briefly again.
Table 15. Setup of enrichment reaction for 200 µl plates
Component Volume/reaction
Total RNA (100 ng – 1 µg) Variable
RNase Inhibitor 1 µl
Buffer mRBB 71 µl
Thoroughly resuspend Pure mRNA Beads 25 µl
Nuclease-Free Water Bring total reaction volume to 150 µl
Total volume 150 µl

QIAseq Stranded mRNA Library Kit Handbook 02/2021
45
3. Incubate for 3 min at 70°C, followed by 10 min at room temperature (15–25°).
4. Briefly centrifuge, and then place tubes onto a magnetic rack. After the solution has
cleared (~2 min), discard the supernatant.
5. Add 150 µl Buffer OW2. Vortex, centrifuge briefly, and place the tubes onto a magnetic
rack. After the solution has cleared, discard the supernatant.
6. Repeat step 5.
7. Add 50 µl Buffer OEB. Vortex, centrifuge briefly, and incubate at 70°C for 3 min.
8. Remove the sample from 70°C and place at room temperature for 5 min.
9. Add 50 µl of Buffer mRBB and vortex. Briefly centrifuge, and incubate at room
temperature for 10 min.
10. Briefly centrifuge, and then place the tubes/plates onto a magnetic rack. After the
solution has cleared, carefully discard the supernatant.
11. Add 150 µl of Buffer OW2. Vortex, centrifuge briefly, and place the tubes/plates onto a
magnetic rack. After the solution has cleared, discard the supernatant.
12. Add 29 µl of Buffer OEB heated to 70°C to the bead pellet and vortex.
13. Briefly centrifuge, and place the tubes/plates onto a magnetic rack. After the solution has
cleared, transfer 27 µl of the supernatant to a clean tube. The supernatant contains
enriched, poly(A)+ RNA.
14. Proceed to Protocol: Fragmentation/FastSelect RNA Removal or store the samples at
−90 to –65°C.

46
QIAseq Stranded mRNA Library Kit Handbook 02/2021
Appendix D: General Remarks on Handling
RNA
Handling RNA
Ribonucleases (RNases) are very stable and active enzymes that generally do not require
cofactors to function. Since RNases are difficult to inactivate and even minute amounts are
sufficient to degrade RNA, do not use any plasticware or glassware without first eliminating
possible RNase contamination. Care should be taken to avoid inadvertently introducing
RNases into the RNA sample during or after the purification procedure. To create and maintain
an RNase-free environment, the following precautions must be taken during the pretreatment
and use of disposable and nondisposable vessels and solutions while working with RNA.
General handling
Proper microbiological, aseptic technique should always be used when working with RNA.
Hands and dust particles may carry bacteria and molds and are the most common sources of
RNase contamination. Always wear latex or vinyl gloves while handling reagents and RNA
samples, to prevent RNase contamination from the surface of the skin or from dusty laboratory
equipment. Change gloves frequently, and keep tubes closed whenever possible. Keep
purified RNA on ice when aliquots are pipetted for downstream applications.
For removal of RNase contamination from bench surfaces, nondisposable plasticware, and
laboratory equipment (e.g., pipettes and electrophoresis tanks), general laboratory reagents
can be used. To decontaminate plasticware, rinse with 0.1 M NaOH, 1 mM EDTA* followed
by RNase-Free Water, or rinse with chloroform* if the plasticware is chloroform-resistant. To
decontaminate electrophoresis tanks, clean with detergent (e.g., 0.5% SDS),* rinse with
RNase-Free Water, rinse with ethanol (if the tanks are ethanol-resistant), and allow to dry.
* When working with chemicals, always wear a suitable lab coat, disposable gloves, and protective goggles. For more
information, consult the appropriate material data sheets (SDSs), available from the product supplier.

QIAseq Stranded mRNA Library Kit Handbook 02/2021
47
Disposable plasticware
The use of sterile, disposable polypropylene tubes is recommended throughout the procedure.
These tubes are generally RNase-free and do not require pretreatment to inactivate RNases.
Glassware
Glassware should be treated before use to ensure that it is RNase-free. Glassware used for
RNA work should be cleaned with a detergent,* thoroughly rinsed, and oven baked at 240°C
for 4 hours or more (overnight, if more convenient) before use. Autoclaving alone will not fully
inactivate many RNases. Alternatively, glassware can be treated with DEPC* (diethyl
pyrocarbonate), as described in “Solutions” below.
Solutions
Solutions (water and other solutions)* should be treated with 0.1% DEPC. DEPC is a strong,
but not absolute, inhibitor of RNases. It is commonly used at a concentration of 0.1% to
inactivate RNases on glass or plasticware or to create RNase-free solutions and water. DEPC
inactivates RNases by covalent modification. Add 0.1 ml DEPC to 100 ml of the solution to be
treated and shake vigorously to bring the DEPC into solution. Let the solution incubate for
12 hours at 37ºC. Autoclave for 15 minutes to remove any trace of DEPC. DEPC will react
with primary amines and cannot be used directly to treat Tris* buffers. DEPC is highly unstable
in the presence of Tris buffers and decomposes rapidly into ethanol and CO
Tris buffers, treat water with DEPC first, and then dissolve Tris to make the appropriate buffer.
Trace amounts of DEPC will modify purine residues in RNA by carbethoxylation.
Carbethoxylated RNA is translated with very low efficiency in cell-free systems. However, its
ability to form DNA:RNA or RNA:RNA hybrids is not seriously affected unless a large fraction
of the purine residues have been modified. Residual DEPC must always be eliminated from
solutions or vessels by autoclaving or heating to 100ºC for 15 minutes.
. When preparing
2
* When working with chemicals, always wear a suitable lab coat, disposable gloves, and protective goggles. For more
information, consult the appropriate material data sheets (SDSs), available from the product supplier.

48
QIAseq Stranded mRNA Library Kit Handbook 02/2021
Product
Contents
Cat. no.
Ordering Information
QIAseq Stranded mRNA Library UDI Kits
QIAseq Stranded mRNA Library
Kit UDI-A (96)
QIAseq Stranded mRNA Library
Kit UDI-B (96)
For 96 stranded RNAseq sequencing
library prep reactions: mRNA
enrichment of total RNA,
fragmentation, reverse transcription,
second-strand synthesis + end-repair +
A-addition, adapter ligation,
CleanStart PCR enrichment and
QIAseq Beads for library cleanups; for
library cleanups for use with Illumina
instruments; includes a plate containing
96 adapters with different barcodes
(pierceable foil seal allowing usage of
defined parts of the plate)
For 96 stranded RNAseq sequencing
library prep reactions: mRNA
enrichment of total RNA,
fragmentation, reverse transcription,
second-strand synthesis + end-repair +
A-addition, adapter ligation,
CleanStart PCR enrichment and
QIAseq Beads for library cleanups; for
library cleanups for use with Illumina
instruments; includes a plate containing
96 adapters with different barcodes
(pierceable foil seal allowing usage of
defined parts of the plate)
180441
180442

QIAseq Stranded mRNA Library Kit Handbook 02/2021
49
Product
A-addition, adapter ligation,
Contents Cat. no.
QIAseq Stranded mRNA Library
Kit UDI-C (96)
QIAseq Stranded mRNA Library
Kit UDI-D (96)
For 96 stranded RNAseq sequencing
library prep reactions: mRNA
enrichment of total RNA,
fragmentation, reverse transcription,
second-strand synthesis + end-repair +
A-addition, adapter ligation,
CleanStart PCR enrichment and
QIAseq Beads for library cleanups; for
library cleanups for use with Illumina
instruments; includes a plate containing
96 adapters with different barcodes
(pierceable foil seal allowing usage of
defined parts of the plate)
For 96 stranded RNAseq sequencing
library prep reactions: mRNA
enrichment of total RNA,
fragmentation, reverse transcription,
second-strand synthesis + end-repair +
A-addition, adapter ligation,
CleanStart PCR enrichment and
QIAseq Beads for library cleanups; for
library cleanups for use with Illumina
instruments; includes a plate containing
96 adapters with different barcodes
(pierceable foil seal allowing usage of
defined parts of the plate)
180443
180445
QIAseq Stranded mRNA Library
Kit UDI (24)
For 24 stranded RNAseq sequencing
library prep reactions: mRNA
enrichment of total RNA,
fragmentation, reverse transcription,
second-strand synthesis + end-repair +
180440

50
QIAseq Stranded mRNA Library Kit Handbook 02/2021
Product
QIAseq Y
Contents Cat. no.
CleanStart PCR enrichment and
QIAseq Beads for library cleanups; for
library cleanups for use with Illumina
instruments; includes a plate containing
96 adapters with different barcodes
(pierceable foil seal allowing usage of
defined parts of the plate)
QIAseq FastSelect Kits
QIAseq FastSelect –Globin Kit Globin mRNA removal reagent:
supports human, mouse, and rat;
available in 24, 96, or 384 reactions
QIAseq
FastSelect –rRNA/Globin Kit
Cytoplasmic and mitochondrial rRNA
removal reagent and globin mRNA
removal reagent: supports human,
mouse, and rat; available in 24, 96, or
384 reactions
-Adapter Kits for Illumina
QIAseq UDI Y-Adapter Kit A (96) Unique Dual-Index Adapters for
Illumina (1–96)
QIAseq UDI Y-Adapter Kit B (96) Unique Dual-Index Adapters for
Illumina (97–192)
QIAseq UDI Y-Adapter Kit C (96) Unique Dual-Index Adapters for
Illumina (193–288)
QIAseq UDI Y-Adapter Kit D (96) Unique Dual-Index Adapters for
Illumina (289–384)
334376
334377
334378
335376
335377
335378
180312
180314
180316
180318
QIAseq UDI Y-Adapter Kit (24) Unique Dual-Index Adapters for
Illumina (1–24)
180310

QIAseq Stranded mRNA Library Kit Handbook 02/2021
51
Product
Contents Cat. no.
QIAseq Library Quantification Kits for use with Illumina instruments
QIAseq Library Quant Array Kit
Plate containing dried assay reagents
333304
for quantification of libraries prepared
®
for Illumina; SYBR
Green Master Mix
(1.35 ml x 2)
QIAseq Library Quant Assay Kit Laboratory-verified forward and
333314
reverse primers for 500 x 25 µl
reactions (500 µl); DNA Standard
(100 µl); Dilution Buffer (30 ml); SYBR
Green Master Mix (1.35 ml x 5)
For up-to-date licensing information and product-specific disclaimers, see the respective
QIAGEN kit handbook or user manual. QIAGEN kit handbooks and user manuals are
available at www.qiagen.com or can be requested from QIAGEN Technical Services or your
local distributor.

52
QIAseq Stranded mRNA Library Kit Handbook 02/2021
Document Revision History
Date Changes
10/2018 Handbook extended to include supplementary protocol (Appendix C).
06/2019 Revised second header in cover page; added 5x to RT Buffer in “Kit Contents” and revised
05/2020 Corrected the volume of QIAseq Beads for step 5 in “Protocol: CleanStart Library
02/2021 Changed the main title to “QIAseq Stranded mRNA Library Kit Handbook”. Incorporated
table notes to delete cautions about sodium azide and Buffer mRBB; updated “Safety
Information” section to delete sodium azide and Buffer mRBB information; added “Globin
mRNA removal” topic; revised “Principle and Workflow” section; revised RNA integrity item
in “Important Notes”; additional items in “Equipments and Reagents Supplied by User”;
renamed and revised “Protocol: Enrichment of Poly(A)+ RNA” to “Protocol: mRNA
Enrichment”; renamed and revised “Protocol: NGS Library Preparation” to “Protocol:
Fragmentation/FastSelect RNA Removal”; revised “Protocol: First-Strand Synthesis” section
to update procedure; revised “Protocol: Second-Strand Synthesis, End Repair, and Aaddition” section to update procedure; revised “Protocol: Strand-specific Ligation section” to
update procedure; revised “Protocol: CleanStart Library Amplification” section to update
procedure; replaced high quality Poly(A)+ RNA isolated with libraries prepared in
Recommendations; updated sample dilution, pooling, sequencing, and data analysis topic;
updated appendices A and B; added new “Appendix C: mRNA Enrichment in 200 µl”;
deleted “Appendix E: Supplementary Protocol”; updated “Ordering Information” section.
Amplification”.
the UDIs in this handbook. Added QIAseq Stranded mRNA Library Kit in the “Kit Contents”
section. Revised the “Shipping and Storage”, “Introduction”, “Important Notes” sections.
Added 100% ethanol (ACS grade) in the “Equipment and Reagents to Be Supplied by User”
section. Updated tables and added new tables. Deleted Figure 3 and added new figures.
Updated “Procedure” in “Protocol: First-strand Synthesis” and “Protocol: Second-strand
Synthesis, End-repair, and A-addition”. Revised “Important points before starting” in
“Protocol: Strand-specific Ligation” and “Protocol: CleanStart Library Amplification”.
Updated Appendix A and changed the title to “QIAseq Dual-Index Y-Adapters”. Updated
the “Ordering Information” section.

QIAseq Stranded mRNA Library Kit Handbook 02/2021
53
Notes

54
QIAseq Stranded mRNA Library Kit Handbook 02/2021
Limited License Agreement for the QIAseq Stranded mRNA Library Kit
Use of this product signifies the agreement of any purchaser or user of the product to the following terms:
1. The product may be used solely in accordance with the protocols provided with the product and this handbook and for use with components contained in the kit
only. QIAGEN grants no license under any of its intellectual property to use or incorporate the enclosed components of this kit with any components not included
within this kit except as described in the protocols provided with the product, this handbook, and additional protocols available at www.qiagen.com. Some of
these additional protocols have been provided by QIAGEN users for QIAGE N users. These protocols have not been thoroughly tested or optimized by QIAGEN.
QIAGEN neither guarantees them nor warrants that they do not infringe the rights of third-parties.
2. Other than expressly stated licenses, QIAGEN makes no warranty that this kit and/or its use(s) do not infringe the rights of third-parties.
3. This kit and its components are licensed for one-time use and may not be reused, refurbished, or resold.
4. QIAGEN specifically disclaims any other licenses, expressed or implied other than those expressly stated.
5. The purchaser and user of the kit agree not to take or permit anyone else to take any steps that could lead to or facilitate any acts prohibited above. QIAGEN
may enforce the prohibitions of this Limited License Agreement in any Court, and shall recover all its investigative and Court costs, including attorney fees, in any
action to enforce this Limited License Agreement or any of its intellectual property rights relating to the kit and/or its components.
For updated license terms, see www.qiagen.com.
Trademarks: QIAGEN
TapeStation
(Illumina, Inc.); Linux
trademarks, etc. used in this document, even when not specifically marked as such, are not to be considered unprotected by law.
02/2021 HB-2464-005 © 2021 QIAGEN, all rights reserved.
®
, Sample to Insight®, QIAamp®, QIAseq®, QIAxcel®, QIAxpert®, Ingenuity®, IPA®, RNeasy®, (QIAGEN Group); Agilent®, Bioanalyzer®,
®
(Agilent Corp.); Mac OS® (Apple, Inc.); LoBind® (Eppendorf AG); HiSeq®, HiSeq X™, Illumina
®
(Linus Torvalds); Windows® (Microsoft Corporation); DynaMag™, SYBR® (Thermo Fisher Scientific or its subsidiaries). Registered names,
®
MiniSeq®, MiSeq®, NextSeq®, NovaSeq™, TruSeq®
,

QIAseq Stranded mRNA Library Kit Handbook 02/2021
55
Ordering
HB-2464-005 02/2021
www.qiagen.com/shop | Technical Support support.qiagen.com | Website www.qiagen.com
 Loading...
Loading...