QIAGEN DNA, RNA User Manual

Sample to Insight__
August 2020
AllPrep® DNA/RNA
FFPE Handbook
For simultaneous purification of genomic DNA
and total RNA (including small RNAs) from
formalin-fixed, paraffin-embedded tissue
sections

2
AllPrep DNA/RNA FFPE Handbook 08/2020
Contents
Kit Contents................................................................................................................ 4
Storage ..................................................................................................................... 5
Intended Use .............................................................................................................. 5
Quality Control ........................................................................................................... 6
Safety Information ...................................................................................................... 6
Introduction ................................................................................................................ 7
Automated purification of nucleic acids on QIAcube Instruments ............................. 8
Principle and procedure .................................................................................... 9
Solutions for FFPE research .............................................................................. 11
Equipment and Reagents to Be Supplied by User .......................................................... 12
Important Notes........................................................................................................ 13
Starting material ............................................................................................ 13
Deparaffinization ........................................................................................... 14
Copurification of nucleic acids .......................................................................... 15
Eluting pure nucleic acids ................................................................................. 15
Handling of QIAamp and RNeasy MinElute spin columns .................................... 16
Preparation of buffers ..................................................................................... 17
Protocol: Purification of Genomic DNA and Total RNA (Including Small RNAs) from
FFPE Tissue Sections .................................................................................................. 19
Troubleshooting Guide .............................................................................................. 28
Appendix A: General Remarks on Handling RNA ......................................................... 33
Appendix B: Storage, Quantification and Determination of Quality of RNA ...................... 35

AllPrep DNA/RNA FFPE Handbook 08/2020
3
Appendix C: Storage, Quantification and Determination of Quality of Genomic DNA ........ 38
Ordering Information ................................................................................................ 41
Document Revision History ......................................................................................... 44

4
AllPrep DNA/RNA FFPE Handbook 08/2020
Kit Contents
AllPrep DNA/RNA FFPE Kit
Catalog no.
RNeasy® MinElute® Spin Columns (pink) 50
QIAamp® MinElute Spin Columns 50
Collection Tubes (1.5 ml) 100
Collection Tubes (2 ml) 200
Buffer PKD 15 ml
Proteinase K 2 x 1.4 ml
Buffer RLT* 45 ml
Buffer FRN*† (concentrate) 14 ml
Buffer RPE‡ (concentrate) 11 ml
RNase-Free DNase I (lyophilized) 1500 units§
Buffer RDD 2 x 2 ml
RNase-Free Water (for use with RNase-Free DNase I) 1.9 ml
Buffer ATL 14 ml
Buffer AL* 12 ml
Buffer AW1*‡ (concentrate) 19 ml
Buffer AW2ঠ(concentrate) 13 ml
Buffer ATE 20 ml
RNase-Free Water 10 ml
Quick-Start Protocol 1
(50)
80234
* Contains a guanidine salt. Not compatible with disinfectants containing bleach. See page 6 for safety information.
†
Before using for the first time, add 3 volumes of isopropanol (96–100%) as indicated on the bottle and as described
on page 17 to obtain a working solution.
‡
Before using for the first time, add ethanol (96–100%) as indicated on the bottle and as described on page 18 to
obtain a working solution.
§
Kunitz units, defined as the amount of DNase I that causes an increase in
25ºC, pH 5.0, with highly polymerized DNA as the substrate (Kunitz, M. [1950] J. Gen. Physiol. 33, 349 and 363).
¶
Contains sodium azide as a preservative.
A
of 0.001 per minute per milliliter at
260

AllPrep DNA/RNA FFPE Handbook 08/2020
5
Storage
RNase-Free DNase I, Buffer RDD, RNeasy MinElute spin columns, and QIAamp MinElute spin
columns should be immediately stored at 2–8°C upon arrival. The remaining buffers can be
stored at room temperature (15–25°C). Under these conditions, the kit components can be
kept for at least 9 months without any reduction in performance, if not otherwise stated on the
label.
Proteinase K is supplied in a specially formulated storage buffer and is stable for at least 1 year
after delivery when stored at room temperature. If longer storage is required or if ambient
temperatures often exceed 25°C, we recommend storage at 2–8°C.
Intended Use
The AllPrep DNA/RNA FFPE Kit is intended for molecular biology applications. This product
is not intended for the diagnosis, prevention, or treatment of a disease.
QIAcube® Connect is designed to perform fully automated purification of nucleic acids and
proteins in molecular biology applications. The system is intended for use by professional users
trained in molecular biological techniques and the operation of QIAcube Connect.
All due care and attention should be exercised in the handling of the products. We recommend
®
all users of QIAGEN
recombinant DNA experiments, or to other applicable guidelines.
products to adhere to the NIH guidelines that have been developed for

6
AllPrep DNA/RNA FFPE Handbook 08/2020
Quality Control
In accordance with QIAGEN’s ISO-certified Quality Management System, each lot of AllPrep
DNA/RNA FFPE Kit is tested against predetermined specifications to ensure consistent product
quality.
Safety Information
When working with chemicals, always wear a suitable lab coat, disposable gloves, and
protective goggles. For more information, please consult the appropriate safety data sheets
(SDSs). These are available online in convenient and compact PDF format at
www.qiagen.com/safety, where you can find, view, and print the SDS for each QIAGEN kit
and kit component.
CAUTION: DO NOT add bleach or acidic solutions directly to the samplepreparation waste.
Buffer RLT and Buffer FRN contain guanidine thiocyanate, and Buffer AL and Buffer AW1
contain guanidine hydrochloride. Guanidine salts can form highly reactive compounds when
combined with bleach. If liquid containing these buffers is spilt, clean with suitable laboratory
detergent and water. If the spilt liquid contains potentially infectious agents, clean the affected
area first with laboratory detergent and water, and then with 1% (v/v) sodium hypochlorite.

AllPrep DNA/RNA FFPE Handbook 08/2020
7
Introduction
The AllPrep DNA/RNA FFPE Kit is specially designed for simultaneous purification of genomic
DNA and total RNA from formalin-fixed, paraffin-embedded (FFPE) tissue sections. DNA and
RNA are released sequentially by differential solubilization of the same precious FFPE sample.
After solubilization, both nucleic acids are treated separately to remove formaldehyde
cross-links and then purified. In contrast to other procedures where either the biological sample
or the purified total nucleic acids is divided into two before being processed separately, with
the AllPrep FFPE procedure, pure DNA and RNA are obtained from the entire sample.
Due to fixation and embedding conditions, nucleic acids in FFPE samples are usually heavily
fragmented and chemically modified by formaldehyde. Therefore, nucleic acids isolated from
FFPE samples are often of a lower molecular weight than those obtained from fresh or frozen
samples. The degree of fragmentation depends on the type and age of the sample, and on
the conditions for fixation, embedding, and storage of the sample. Although formaldehyde
modification cannot be detected in standard quality control assays, such as gel electrophoresis
or lab-on-a-chip analysis, it does strongly interfere with enzymatic analyses.
While the AllPrep DNA/RNA FFPE Kit is optimized to reverse as much formaldehyde
modification as possible without further DNA and RNA degradation, nucleic acids purified
from FFPE samples should not be used in downstream applications that require high-molecular–
weight DNA or full-length RNA. Some applications may require modifications to allow the use
of fragmented nucleic acids (e.g., designing small amplicons for PCR and RT-PCR). For cDNA
synthesis, gene-specific primers should be used instead of oligo-dT primers. If it is not possible
to use gene-specific primers, random primers should be used.

8
AllPrep DNA/RNA FFPE Handbook 08/2020
Automated purification of nucleic acids on QIAcube Instruments
Purification of nucleic acids can be fully automated on QIAcube Connect or the classic
QIAcube. The innovative QIAcube instruments use advanced technology to process QIAGEN
spin columns, enabling seamless integration of automated, low-throughput sample prep into
your laboratory workflow. Sample preparation using QIAcube instruments follows the same
steps as the manual procedure (i.e., lyse, bind, wash, and elute), enabling you to continue
using the AllPrep DNA/RNA FFPE Kit for purification of high-quality nucleic acids.
QIAcube instruments are preinstalled with protocols for purification of plasmid DNA, genomic
DNA, RNA, viral nucleic acids, and proteins, plus DNA and RNA cleanup. The range of
protocols available is continually expanding, and additional QIAGEN protocols can be
downloaded free of charge at www.qiagen.com/qiacubeprotocols.
QIAcube Connect.

AllPrep DNA/RNA FFPE Handbook 08/2020
9
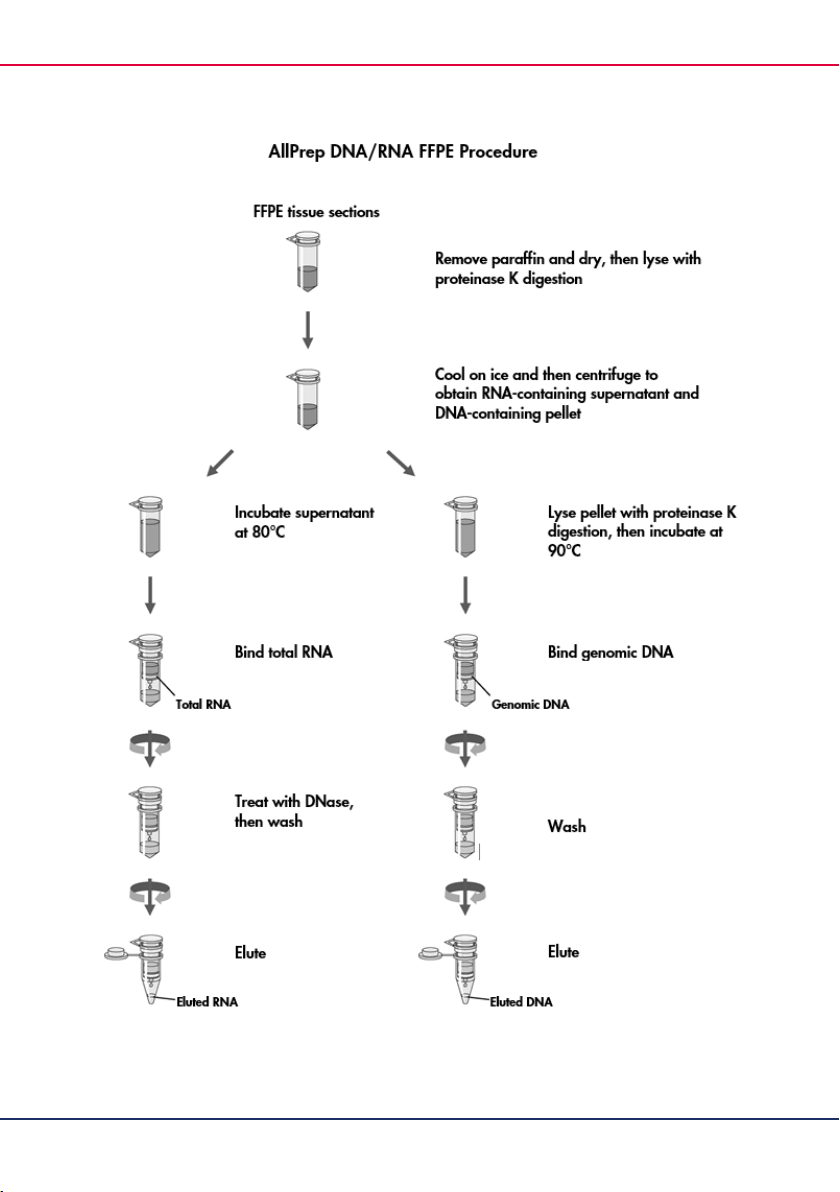
Principle and procedure
The AllPrep DNA/RNA FFPE procedure integrates well-established QIAamp and RNeasy technologies
for DNA and RNA purification with QIAGEN’s novel technology for selective removal of DNA and
RNA from a single FFPE sample. Specially optimized lysis conditions allow the differential release of
DNA and RNA from the same FFPE sample and avoid the need for overnight proteinase K incubation.
Freshly cut FFPE tissue sections are incubated in an optimized lysis buffer that contains proteinase K.
Under these conditions, RNA is released into solution, whereas genomic DNA and other insoluble
material are precipitated. The sample is then centrifuged to give an RNA-containing supernatant
and a DNA-containing pellet, which then undergo separate purification procedures.
The RNA-containing supernatant is incubated at 80°C to partially reverse formalin
cross-linking. This incubation step helps to improve RNA yield and quality, as well as RNA
performance in downstream enzymatic assays. The supernatant is then mixed with Buffer RLT
and ethanol to provide appropriate binding conditions for RNA. The sample is applied to an
RNeasy MinElute spin column, where RNA binds specifically to the silica membrane. The
bound RNA is treated with DNase to digest contaminating genomic DNA, washed with Buffer
FRN and Buffer RPE to remove contaminants and then eluted in 14–30 µl of RNase-free water.
Small RNAs, such as microRNA, are either present or absent in the purified RNA, depending
on the volume of ethanol used earlier in the procedure to adjust RNA binding conditions.
The DNA-containing pellet is lysed further in the presence of proteinase K and then incubated
at 90°C. Incubation at this high temperature partially reverses formalin cross-linking, helping
to improve DNA yield and quality and DNA performance in downstream applications. The
sample is mixed with Buffer AL and ethanol to provide optimal DNA binding conditions and
then applied to a QIAamp MinElute spin column. Genomic DNA binds specifically to the silica
membrane and is washed with Buffer AW1, Buffer AW2, and ethanol to remove contaminants.
Pure DNA is then eluted in 30–100 µl of Buffer ATE.
The purified RNA and DNA samples are ready to use in downstream applications.
Alternatively, they can be stored at –30 to –15°C.
10
AllPrep DNA/RNA FFPE Handbook 08/2020

AllPrep DNA/RNA FFPE Handbook 08/2020
11
Solutions for FFPE research
QIAGEN’s dedicated products for FFPE samples enable easy deparaffinization and efficient
recovery of DNA, RNA, miRNA, and protein (see ordering information, page 41). Solutions
for reliable downstream analysis include dedicated chemistry for PCR amplification of small
fragments.
QIAGEN’s comprehensive FFPE portfolio provides:
Maximum data output with minimum sample consumption
Prep technologies that reverse cross-links for higher yields
DNA, RNA, and protein purification without further compromising analyte integrity
Optimized chemistries for analysis of lower quality FFPE analytes
FFPE research data you can trust

12
AllPrep DNA/RNA FFPE Handbook 08/2020
Equipment and Reagents to Be Supplied by User
When working with chemicals, always wear a suitable lab coat, disposable gloves and
protective goggles. For more information, consult the appropriate safety data sheets (SDSs),
available from the product supplier.
Sterile, RNase-free pipet tips (to avoid cross-contamination, we recommend pipet tips
with aerosol barriers)
1.5 ml Safe-Lock microcentrifuge tubes (available from Brinkmann, cat. no. 022363204,
or Eppendorf, cat. no. 0030 120.086) or 1.5 ml SafeSeal microcentrifuge tubes
(Sarstedt, cat. no. 72.706)*
If purifying RNA containing miRNAs: 2 ml Safe-Lock microcentrifuge tubes (available
from Brinkmann, cat. no. 022363352, or Eppendorf, cat. no. 0030 120.094) or 2 ml
SafeSeal microcentrifuge tubes (Sarstedt, cat. no. 72.695)*
Microcentrifuge (with rotor for 2 ml tubes)
Vortexer
96–100% ethanol
†
96–100% isopropanol
For deparaffinization of FFPE tissue sections:
Deparaffinization Solution (cat. no. 19093), or
99–100% heptane, methanol and 96–100% ethanol, or
99–100% xylene and 96–100% ethanol
Thermal mixer, heated orbital incubator, heating block, or water bath capable of
incubation at 56°C, 80°C, and 90°C
Optional: RNase A (100 mg/ml, cat. no. 19101)
* This is not a complete list of suppliers and does not include many important vendors of biological supplies.

AllPrep DNA/RNA FFPE Handbook 08/2020
13
Important Notes
Starting material
Standard formalin-fixation and paraffin-embedding procedures always result in significant
fragmentation and cross-linking of nucleic acids. To limit the extent of nucleic acid
fragmentation and cross-linking, be sure to:
Fixate tissue samples in 4–10% formalin as quickly as possible after surgical removal
Use a maximum fixation time of 24 hours (longer fixation times lead to over-fixation and
more severe nucleic acid fragmentation, resulting in poor performance in downstream
assays)
Thoroughly dehydrate samples prior to embedding, as residual formalin can inhibit
proteinase K digestion
Use low-melting paraffin for embedding, as high temperatures during embedding can
cause nucleic acid fragmentation
Store FFPE samples at low temperatures (2–8°C); storage at room temperature
(15−25°C) can lead to nucleic acid degradation
The starting material for nucleic acid purification should be freshly cut sections of FFPE tissue,
each with a thickness of 10–20 µm. Thicker sections may result in lower nucleic acid yields,
even after prolonged incubation with proteinase K. Thinner sections can be used but are more
difficult to pellet. Up to 4 sections, each with a thickness of 10 µm and a surface area of up
2
to 150 mm
150 mm
, or 2 sections each with a thickness of 20 µm and a surface area of up to
2
, can be combined in one preparation.
Avoid using too much starting material, as this affects lysis efficiency and purification and can
lead to reduced yields and nucleic acid fragmentation.

14
AllPrep DNA/RNA FFPE Handbook 08/2020
If there is no information about the nature of your starting material or if the surface area of the
sample is high, we recommend starting with one 10–20 µm thick section per preparation.
Do not overload the QIAamp and RNeasy MinElute spin columns, as this will significantly
reduce DNA/RNA yield and quality.
Deparaffinization
Prior to nucleic acid purification, the paraffin in an FFPE sample needs to be removed to allow
exposure of the sample to proteinase K. One of the following deparaffinization procedures
should be used.
Deparaffinization using Deparaffinization Solution
Deparaffinization Solution (cat. no. 19093) dissolves paraffin efficiently and allows
deparaffinization without further washing steps. Deparaffinization Solution is colored to
enable easy visibility for removal of supernatant during the procedure. The volume of
Deparaffinization Solution needed for deparaffinization depends on the amount of sample
material. One set of Deparaffinization Solution (2 x 8 ml, cat. no. 19093) is sufficient for
deparaffinization of 50 samples consisting of up to two 10 µm sections or one 20 µm section.
For larger samples, the volume of Deparaffinization Solution required is doubled.
Deparaffinization using heptane and methanol
Deparaffinization with heptane and methanol is very efficient. After precipitation of the sample
and removal of the supernatant, residual heptane is removed by washing with ethanol. This
procedure usually gives more compact pellets than other procedures and allows good results
in DNA and RNA applications.
 Loading...
Loading...