Page 1

NeuroExplorer Manual
Copyright © 1998-2011 Nex Technologies
Page 2

Revision: 4.095. Date: 9/23/2011.
Page 3

Table of Contents
1. Getting Started 7
1.1. Getting Started with NeuroExplorer 8
1.2. Working with Sentinel Keys 9
1.3. NeuroExplorer Screen Elements 12
1.4. Opening Files and Importing Data 16
1.5. Importing Files Created by Data Acquisition Systems 17
1.6. Importing Data from Text Files 19
1.7. Importing Data from Spreadsheets 20
1.8. Importing Data from Matlab 22
1.9. Reading and Writing NeuroExplorer Data Files 23
1.10. 1D Data Viewer 24
1.11. Analyzing Data 25
1.12. Selecting Variables for Analysis 26
1.13. How to Select Variables for Existing Window 27
1.14. How to Select Variables in Variables Panel 29
1.15. Adjusting Analysis Properties 30
1.16. Analysis Templates 31
1.17. Numerical Results 32
1.18. Post-processing 33
1.19. Saving Results as Power Point Slides 33
1.20. Working with Matlab 35
1.21. Working with Excel 36
1.22. Saving Graphics 37
2. Analysis Reference 38
2.1. Data Types 39
2.1.1. Spike Trains 39
2.1.2. Events 40
2.1.3. Intervals 40
2.1.4. Markers 42
2.1.5. Population Vectors 43
2.1.6. Waveforms 44
2.1.7. Continuously Recorded Data 44
2.2. Data Selection Options 46
2.3. Post-Processing Options 47
2.4. Matlab Options 48
2.5. Excel Options 48
2.6. Confidence Limits for Perievent Histograms 49
2.7. Cumulative Sum Graphs 50
2.8. Rate Histograms 51
2.9. Interspike Interval Histograms 53
2.10. Autocorrelograms 56
2.11. Perievent Histograms 59
2.12. Crosscorrelograms 64
2.13. Shift-Predictor for Crosscorrelograms 69
2.14. Rasters 71
2.15. Perievent Rasters 72
2.16. Joint PSTH 75
2.17. Cumulative Activity Graphs 77
2.18. Instant Frequency 78
2.19. Interspike Intervals vs. Time 79
2.20. Poincare Maps 80
Page 1
Page 4

2.21. Synchrony vs. Time 81
2.22. Trial Bin Counts 83
2.23. Power Spectral Densities 85
2.24. Burst Analysis 88
2.25. Principal Component Analysis 91
2.26. PSTH Versus Time 93
2.27. Correlations with Continuous Variable 95
2.28. Regularity Analysis 97
2.29. Place Cell Analysis 99
2.30. Reverse Correlation 103
2.31. Epoch Counts 106
2.32. Coherence Analysis 108
2.33. Spectrogram Analysis 111
2.34. Perievent Spectrograms 113
2.35. Joint ISI Distribution 115
2.36. Autocorrelograms Versus Time 117
3. Working with Graphics 119
3.1. NeuroExplorer Graphics 119
3.2. Graphics Modes 119
3.3. Positioning the Graphics Objects 121
3.4. Text Labels 122
3.5. Lines 123
3.6. Rectangles 124
4. Working with 3D Graphics 125
4.1. Viewing Multiple Histograms in 3D 126
4.2. 3D Graphics Parameters 127
4.3. Viewing the Neuronal Activity "Movie" 128
4.4. Activity Animation Parameters 130
5. Programming with NexScript 131
5.1. Script Variables 133
5.2. File Variables 134
5.3. Expressions 136
5.4. Flow Control 137
5.5. Functions 139
5.5.1. File Read and Write Functions 140
5.5.1.1. GetFileCount Function 141
5.5.1.2. GetFileName Function 142
5.5.1.3. OpenFile Function 143
5.5.1.4. CloseFile Function 144
5.5.1.5. ReadLine Function 145
5.5.1.6. WriteLine Function 146
5.5.1.7. OpenDocument Function 147
5.5.1.8. NewDocument Function 148
5.5.1.9. CloseDocument Function 149
5.5.1.10. SaveDocument Function 150
5.5.1.11. SaveDocumentAs Function 151
5.5.1.12. SaveNumResults Function 152
5.5.1.13. SaveNumSummary Function 153
5.5.1.14. SaveAsTextFile Function 154
5.5.1.15. MergeFiles Function 155
5.5.1.16. ReadBinary Function 156
5.5.1.17. FileSeek Function 157
5.5.1.18. SelectFile Function 158
5.5.2. Document Properties Functions 159
Page 2
Page 5

5.5.2.1. GetDocPath Function 160
5.5.2.2. GetDocTitle Function 161
5.5.2.3. GetTimestampFrequency Function 162
5.5.2.4. GetDocEndTime Function 163
5.5.2.5. SetDocEndTime Function 164
5.5.2.6. GetDocComment Function 165
5.5.3. Document Variables Functions 166
5.5.3.1. GetVarCount Function 167
5.5.3.2. GetVarName Function 168
5.5.3.3. GetVarSpikeCount Function 169
5.5.3.4. GetVar Function 170
5.5.3.5. DeleteVar Function 171
5.5.3.6. Delete Function 172
5.5.3.7. GetVarByName Function 173
5.5.3.8. NewEvent Function 174
5.5.3.9. NewIntEvent Function 175
5.5.3.10. NewPopVector Function 176
5.5.3.11. GetContNumDataPoints Function 177
5.5.3.12. NewContVar Function 178
5.5.3.13. CopySelectedVarsToAnotherFile Function 179
5.5.4. Variable Selection Functions 180
5.5.4.1. IsSelected Function 181
5.5.4.2. Select Function 182
5.5.4.3. Deselect Function 183
5.5.4.4. SelectVar Function 184
5.5.4.5. DeselectVar Function 185
5.5.4.6. Select Function 186
5.5.4.7. Deselect Function 187
5.5.4.8. SelectAll Function 188
5.5.4.9. DeselectAll Function 189
5.5.4.10. SelectAllNeurons Function 190
5.5.4.11. SelectAllEvents Function 191
5.5.4.12. EnableRecalcOnSelChange Function 192
5.5.4.13. DisableRecalcOnSelChange Function 193
5.5.5. Properties of Variables Functions 194
5.5.5.1. GetName Function 195
5.5.5.2. GetSpikeCount Function 196
5.5.5.3. AddTimestamp Function 197
5.5.5.4. SetNeuronType Function 198
5.5.5.5. AddContValue Function 199
5.5.5.6. AddInterval Function 200
5.5.6. Analysis Functions 201
5.5.6.1. ApplyTemplate Function 202
5.5.6.2. ApplyTemplateToWindow Function 203
5.5.6.3. PrintGraphics Function 204
5.5.6.4. Dialog Function 205
5.5.6.5. ModifyTemplate Function 207
5.5.6.6. RecalculateAnalysisInWindow Function 209
5.5.6.7. EnableRecalcOnSelChange Function 210
5.5.6.8. DisableRecalcOnSelChange Function 211
5.5.6.9. SendGraphicsToPowerPoint Function 212
5.5.6.10. SaveGraphics Function 213
5.5.7. Numerical Results Functions 214
5.5.7.1. GetNumRes Function 215
5.5.7.2. GetNumResNCols Function 216
5.5.7.3. GetNumResNRows Function 217
5.5.7.4. GetNumResColumnName Function 218
5.5.7.5. SendResultsToExcel Function 219
5.5.7.6. GetNumResSummaryNCols Function 220
Page 3
Page 6

5.5.7.7. GetNumResSummaryNRows Function 221
5.5.7.8. GetNumResSummaryColumnName Function 222
5.5.7.9. GetNumResSummaryData Function 223
5.5.7.10. SendResultsSummaryToExcel Function 224
5.5.7.11. SaveNumResults Function 225
5.5.7.12. SaveNumSummary Function 226
5.5.8. Operations on Variables Functions 227
5.5.8.1. Rename Function 229
5.5.8.2. Join Function 230
5.5.8.3. Sync Function 231
5.5.8.4. NotSync Function 232
5.5.8.5. FirstAfter Function 233
5.5.8.6. FirstNAfter Function 234
5.5.8.7. LastBefore Function 235
5.5.8.8. IntervalFilter Function 236
5.5.8.9. SelectTrials Function 237
5.5.8.10. SelectRandom Function 238
5.5.8.11. SelectEven Function 239
5.5.8.12. SelectOdd Function 240
5.5.8.13. ISIFilter Function 241
5.5.8.14. FirstInInterval Function 242
5.5.8.15. LastInInterval Function 243
5.5.8.16. StartOfInterval Function 244
5.5.8.17. EndOfInterval Function 245
5.5.8.18. MakeIntervals Function 246
5.5.8.19. MakeIntFromStart Function 247
5.5.8.20. MakeIntFromEnd Function 248
5.5.8.21. IntOpposite Function 249
5.5.8.22. IntOr Function 250
5.5.8.23. IntAnd Function 251
5.5.8.24. IntSize Function 252
5.5.8.25. IntFind Function 253
5.5.8.26. MarkerExtract Function 254
5.5.8.27. Shift Function 255
5.5.8.28. NthAfter Function 256
5.5.8.29. PositionSpeed Function 257
5.5.8.30. FilterContinuousVariable Function 258
5.5.8.31. LinearCombinationOfContVars Function 259
5.5.8.32. DecimateContVar Function 260
5.5.9. Matlab Functions 261
5.5.9.1. SendSelectedVarsToMatlab Function 262
5.5.9.2. ExecuteMatlabCommand Function 263
5.5.9.3. GetVarFromMatlab Function 264
5.5.9.4. GetContVarFromMatlab Function 265
5.5.9.5. GetContVarWithTimestampsFromMatlab Function 266
5.5.9.6. GetIntervalVarFromMatlab Function 267
5.5.10. Excel Functions 268
5.5.10.1. SetExcelCell Function 269
5.5.10.2. CloseExcelFile Function 270
5.5.11. Power Point Functions 271
5.5.11.1. SendGraphicsToPowerPoint Function 272
5.5.11.2. ClosePowerPointFile Function 273
5.5.12. Running Script Functions 274
5.5.12.1. RunScript Function 275
5.5.12.2. Sleep Function 276
5.5.13. Math Functions 277
5.5.13.1. seed Function 279
5.5.13.2. expo Function 280
5.5.13.3. floor Function 281
Page 4
Page 7

5.5.13.4. ceil Function 282
5.5.13.5. round Function 283
5.5.13.6. abs Function 284
5.5.13.7. sqrt Function 285
5.5.13.8. pow Function 286
5.5.13.9. exp Function 287
5.5.13.10. min Function 288
5.5.13.11. max Function 289
5.5.13.12. log Function 290
5.5.13.13. sin Function 291
5.5.13.14. cos Function 292
5.5.13.15. tan Function 293
5.5.13.16. acos Function 294
5.5.13.17. asin Function 295
5.5.13.18. atan Function 296
5.5.13.19. RoundToTS Function 297
5.5.13.20. GetFirstGE Function 298
5.5.13.21. GetFirstGT Function 299
5.5.13.22. GetBinCount Function 300
5.5.13.23. BitwiseAnd Function 301
5.5.13.24. BitwiseOr Function 302
5.5.13.25. GetBit Function 303
5.5.14. String Functions 304
5.5.14.1. Left Function 305
5.5.14.2. Mid Function 306
5.5.14.3. Right Function 307
5.5.14.4. Find Function 308
5.5.14.5. StrLength Function 309
5.5.14.6. NumToStr Function 310
5.5.14.7. StrToNum Function 311
5.5.14.8. GetNumFields Function 312
5.5.14.9. GetField Function 313
5.5.14.10. CharToNum Function 314
5.5.14.11. NumToChar Function 315
5.5.15. Debug Functions 316
5.5.15.1. Trace Function 317
5.5.15.2. MsgBox Function 318
6. COM/ActiveX Interfaces 319
6.1. Application 320
6.1.1. ActiveDocument Property 321
6.1.2. DocumentCount Property 322
6.1.3. Version Property 323
6.1.4. Visible Property 324
6.1.5. OpenDocument Method 325
6.1.6. Document Medhod 326
6.1.7. Sleep Method 327
6.1.8. RunNexScript Method 328
6.1.9. RunNexScriptCommands Method 329
6.2. Document 330
6.2.1. Path Property 332
6.2.2. FileName Property 333
6.2.3. Comment Property 334
6.2.4. TimestampFrequency Property 335
6.2.5. StartTime Property 336
6.2.6. EndTime Property 337
6.2.7. VariableCount Property 338
6.2.8. NeuronCount Property 339
Page 5
Page 8

6.2.9. EventCount Property 340
6.2.10. IntervalCount Property 341
6.2.11. MarkerCount Property 342
6.2.12. WaveCount Property 343
6.2.13. ContinuousCount Property 344
6.2.14. Variable Method 345
6.2.15. Neuron Method 346
6.2.16. Event Method 347
6.2.17. Interval Method 348
6.2.18. Marker Method 349
6.2.19. Wave Method 350
6.2.20. Continuous Method 351
6.2.21. DeselectAll Method 352
6.2.22. SelectAllNeurons Method 353
6.2.23. SelectAllContinuous Method 354
6.2.24. ApplyTemplate Method 355
6.2.25. GetNumericalResults Method 356
6.2.26. Close Method 357
6.3. Variable 358
6.3.1. Name Property 359
6.3.2. TimestampCount Property 360
6.3.3. Timestamps Method 361
6.3.4. IntervalStarts Method 362
6.3.5. IntervalEnds Method 363
6.3.6. FragmentTimestamps Method 364
6.3.7. FragmentCounts Method 365
6.3.8. ContinuousValues Method 366
6.3.9. MarkerValues Method 367
6.3.10. WaveformValues Method 368
6.3.11. Select Method 369
6.3.12. Deselect Method 370
Page 6
Page 9

1. Getting Started
Installation
Before you can use NeuroExplorer, you must install NeuroExplorer program files, Sentinel system
drivers and install the Sentinel hardware key.
Running NeuroExplorer Setup
Before you begin installing NeuroExplorer and its components, please exit all currently running
applications. Please follow the following steps to install NeuroExplorer:
•
Insert NeuroExplorer Setup USB flash drive into your computer USB port
•
Navigate to NeuroExplorer4Setup.exe file on the flash drive and double-click on the file.
NeuroExplorer Version 4 setup screen appears
•
Follow the prompts of the setup dialogs and complete the installation
•
At the end of the installation process, Sentinel System Driver Setup will start automatically
•
Follow Sentinel Driver Setup prompts and complete the installation of Sentinel Drivers
•
Reboot your computer
NeuroExplorer Setup will create the following directory structure:
•
The main NeuroExplorer directory: C:\Program Files\Nex Technologies\NeuroExplorer
•
SentinelDrivers subdirectory: C:\Program Files\Nex
Technologies\NeuroExplorer\SentinelDrivers. If you need to reinstall Sentinel Drivers, you can
run SentinelSetup.7.4.0.exe program in this directory.
•
Additional NeuroExplorer files are copied to Windows Application Data directory ( C:\Documents
and Settings\All Users\Application Data\Nex Technologies\NeuroExplorer under Windows XP, or
C:\Application Data\Nex Technologies\NeuroExplorer under Vista or Windows 7). This directory
also contains Scripts and Templates folders where NeuroExplorer scripts and analysis templates
are stored.
Installing Hardware Key
Before you can use NeuroExplorer, you need to install provided Sentinel Hardware Key on your
computer.
To install the USB key:
•
Make sure to run the NeuroExplorer setup and reboot the computer after installing
NeuroExplorer
•
Attach the key to the available USB port
•
If the New Hardware Found wizard is shown, accept the defaults in the wizard
Page 7
Page 10

1.1. Getting Started with NeuroExplorer
This section, Getting Started with NeuroExplorer, describes the basics of using NeuroExplorer:
•
Working with Sentinel keys
•
NeuroExplorer Screen Elements
•
Opening Files and Importing Data
•
Importing Data from Text Files
•
Importing Data from the Spreadsheets
•
Analyzing Data
•
Selecting Variables for Analysis
•
Adjusting Analysis Properties
•
Analysis Templates
•
Numerical Results
•
Post-processing
•
Working with Matlab
•
Working with Excel
•
Saving Graphics
Additional information is available in the following sections:
•
Working with Graphics
•
NeuroExplorer Analysis Reference
•
Programming with NexScript
NeuroExplorer Technical Support
NeuroExplorer users can get help via e-mail: support@neuroexplorer.com. Please visit
NeuroExplorer Web site http://www.neuroexplorer.com for program updates and latest information
about NeuroExplorer.
NeuroExplorer Updates
To get the latest version of NeuroExplorer, download the file
http://www.neuroexplorer.com/downloads/NeuroExplorer4Setup.exe.
This file is a standard NeuroExplorer setup executable that is included in the NeuroExplorer CD. This
setup file will execute the complete install – it will create folders, menu items in Start\Programs and
create a desktop icon for NeuroExplorer.
Page 8
Page 11
1.2. Working with Sentinel Keys
General
NeuroExplorer requires a Sentinel key to operate. Sentinel drivers need to be installed so that
NeuroExplorer can communicate with the Sentinel keys. These drivers are installed when you run
NeuroExplorer setup (NeuroExplorer4Setup.exe).
Running NeuroExplorer via Remote Desktop
If you would like to run NeuroExplorer via remote desktop, you will need to use Sentinel Protection
Server on the server (the computer that runs NeuroExplorer). - Install NeuroExplorer on the server Open Safenet downloads page: http://www.safenetinc.com/support_and_downloads/download_drivers/ - Download Sentinel Protection Installer - Run
Sentinel Protection Installer on the server and choose Complete option - Reboot the server
Error Messages and Troubleshooting
Error Message Description and Probable
Unable to initialize Sentinel
library
Cause
Sentinel drivers are not
installed or driver version is
not supported.
Troubleshooting
Make sure that Sentinel drivers version
7.4.0 or later are installed:
- Open Control Panel | Add or Remove
Programs and verify that Sentinel
System Driver Installer 7.4.0 is listed in
Currently Installed Programs.
- If an older version (prior to 7.4.0) of
Sentinel Driver installer is listed,
remove this version and reboot the
computer.
- To install the 7.4.0 drivers, run the
driver installer:
C:\Program Files\Nex
Technologies\NeuroExplorer\SentinelD
rivers\SentinelSetup.7.4.0.exe
- Reboot the computer after installing
the drivers.
Unable to find Sentinel key NeuroExplorer Sentinel key
is not attached to the
computer or there is a
problem communicating with
the key.
Page 9
1) Verify that you are using a
NeuroExplorer key. The code
SRB10491 should be printed on the
key. The newer keys also have
Neuroexplorer printed on them.
2) Make sure that Sentinel drivers
version 7.4.0 or later are installed.
- Open Control Panel | Add or Remove
Programs and verify that Sentinel
System Driver Installer 7.4.0 is listed in
Currently Installed Programs.
- If an older version (prior to 7.4.0) of
Sentinel Driver installer is listed,
Page 12
remove this version and reboot the
computer.
- To install the 7.4.0 drivers, run the
driver installer:
C:\Program Files\Nex
Technologies\NeuroExplorer\SentinelD
rivers\SentinelSetup.7.4.0.exe
- Reboot the computer after installing
the drivers.
3) If the problem persists, you may
need to use the Cleanup utility.
- Uninstall NeuroExplorer and all the
Sentinel software items listed in
Control Panel | Add or Remove
Programs
- Download Sentinel cleanup utility
(SSD Cleanup) from this page:
http://www.safenetinc.com/Support_and_Downloads/Dow
nload_Drivers/Sentinel_Drivers.aspx
- Unzip and run the utility. Make sure
you let the utility to finish running. It
may take several minutes.
- Reboot the computer
- Run NeuroExplorer4Setup.exe.
Make sure not to cancel Sentinel Driver
Setup. Accept defaults in Sentinel
Driver Setup.
- Reboot the computer
Please contact NeuroExplorer
technical support (
mailto:support@neuroexplorer.com ) if
you still have problems with the
Sentinel key.
Unable to read Sentinel key
data
Sentinel key does not have
version 4 license
Malfunctioning or damaged
Sentinel key.
You are using NeuroExplorer
version 3 Sentinel key.
Page 10
Please contact NeuroExplorer
technical support (
mailto:support@neuroexplorer.com )
If you purchased NeuroExplorer
version 4, please contact
NeuroExplorer technical support (
mailto:support@neuroexplorer.com )
If you would like to upgrade to
NeuroExplorer version 4, please
contact NeuroExplorer support
(support@neuroexplorer.com) or one
Page 13
of the NeuroExplorer resellers listed in
the Purchase page of the
NeuroExplorer web site:
http://www.neuroexplorer.com/purchas
e.htm
Page 11
Page 14
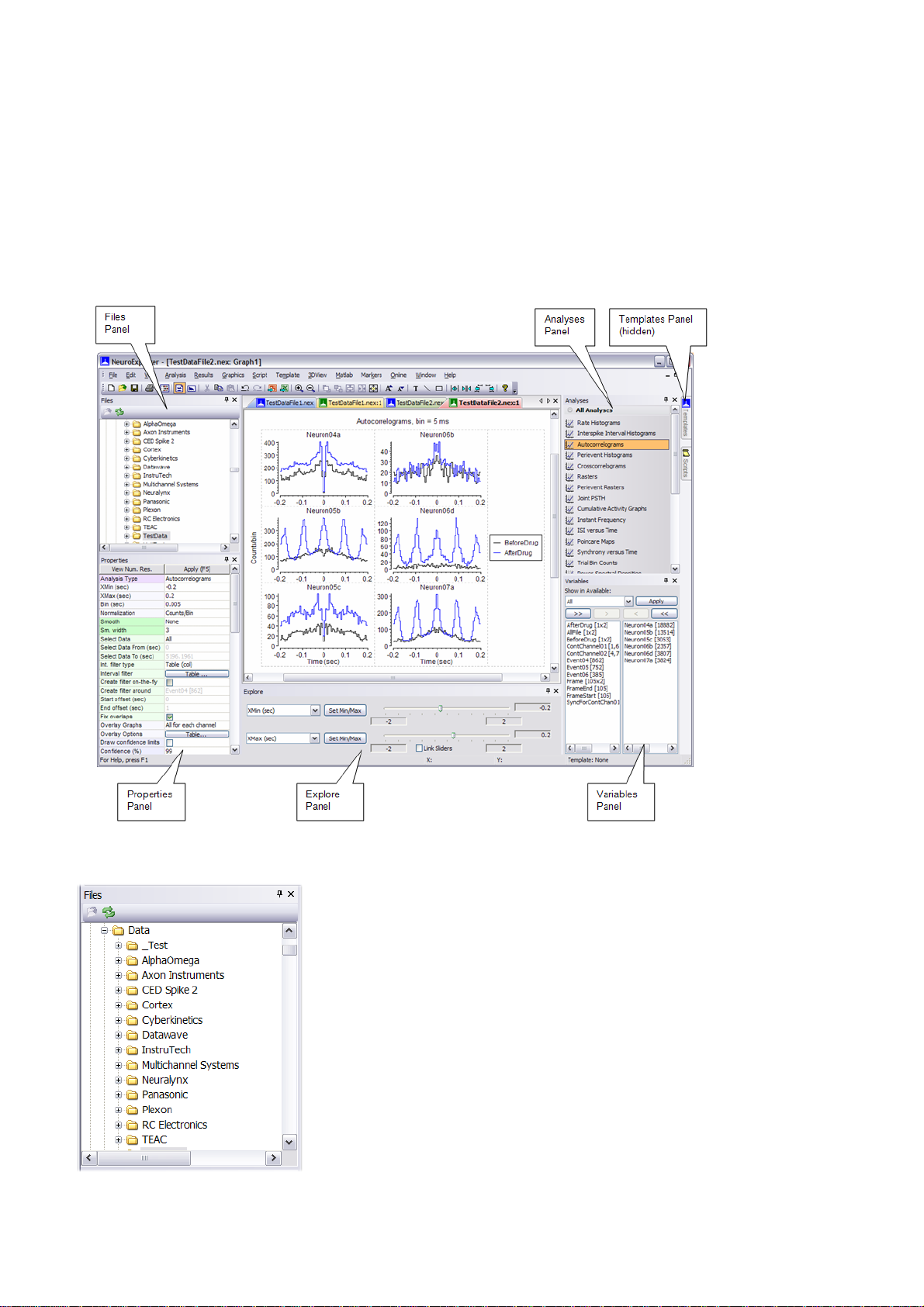
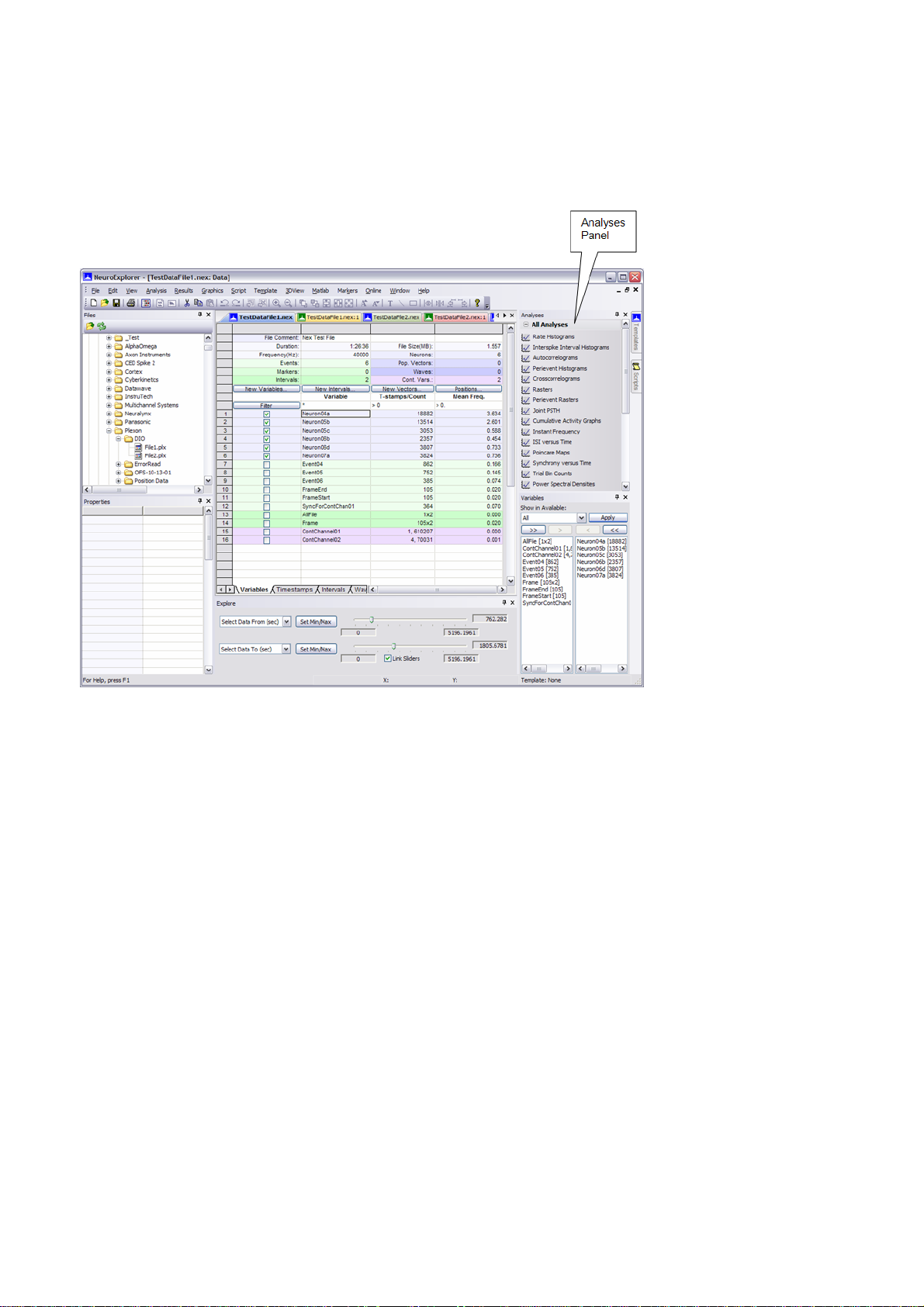
1.3. NeuroExplorer Screen Elements
NeuroExplorer user interface consists of the main window surrounded by several panels. These
panels allow you to select files, select analyses and specify analysis properties. The figure below
shows the default layout of NeuroExplorer window. Files, Properties, Analyses, Variables and Explore
panels are visible and Templates and Scripts panels are hidden. To view a hidden panel, click at the
panel tab or use View menu command.
You can reposition panels by dragging them with your mouse and you can save and restore window
layouts using Window menu commands.
Files Panel
This view allows you to quickly browse through your data files. When you select (single-click) one of
the data files, NeuroExplorer displays the file header information in the Properties panel. To open the
Page 12
Page 15
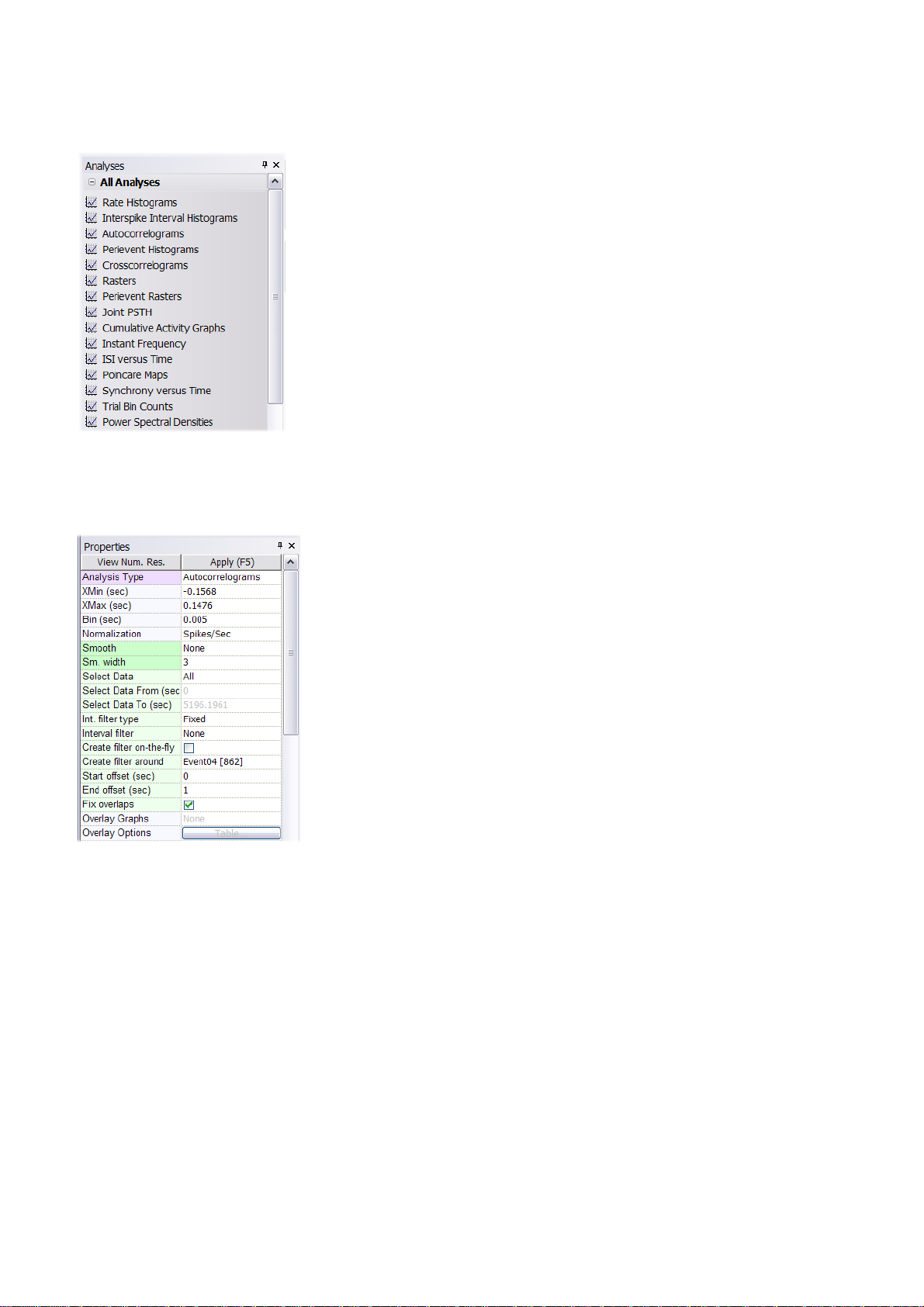
data file, simply double-click the file name.
Analyses Panel
This view allows you to quickly select one of the analyses available in NeuroExplorer. To apply
analysis to the active data file, click at the analysis name.
Properties Panel
The left column of the Properties Panel lists the names of adjustable parameters for the selected
object. The right column contains various controls that can be used to change the parameter values.
To apply the changes, press the Apply button or hit the F5 key.
Templates Panel
The Templates View can be used to quickly execute an Analysis Template. Double-click the template
name and the corresponding template will be immediately executed.
You can create subfolders in your template directory and then NeuroExplorer will allow you to
navigate through the templates tree within the Templates View.
Page 13
Page 16
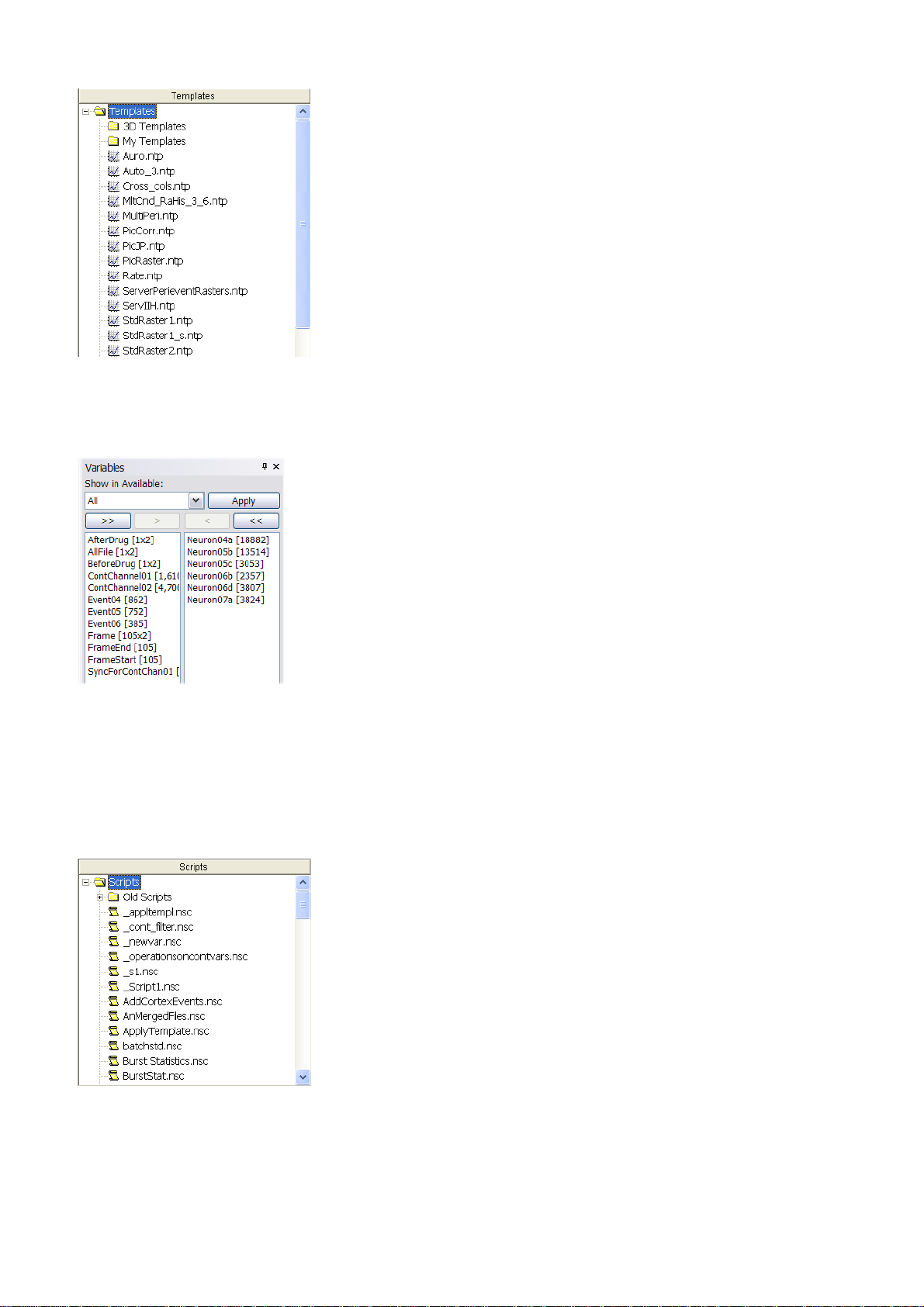
Variables Panel
You can analyze all the variables in your data file, a subset of the variables, or may be just one
variable. Variables Panel allows you to quickly select and deselect the variables used for analysis:
The left column shows the variables that are not currently selected; the right column shows the
selected variables. To move variables from column to column, first select them, then press ">"
(Select) or "<" (Deselect) buttons.
Scripts Panel
This panel can be used to select a script to be executed. Double-click the script name to run the
selected script.
You can create subfolders in your script directory and then NeuroExplorer will allow you to navigate
through the scripts tree within the Scripts View.
Explore Panel
Page 14
Page 17

Explore panel allows you to quickly explore analysis parameter ranges. Select a parameter you want
to explore from a list and drag a slider. NeuroExplorer will keep recalculating analysis with the new
parameter values while you drag the slider, thus creating an “analysis animation”.
One of the best uses for this panel is to explore analysis properties over a sliding window within your
file:
•
Specify Use Time Range using Analysis | Edit menu command, then Data Selection tab
•
Select ‘Select Data From’ and ‘Select Data To’ parameters in the list boxes of the Explore panel
•
Click on Link Sliders check box
Now, when you drag one of the sliders, the other one will follow allowing you to visualize analysis
results over a sliding window in time.
Page 15
Page 18
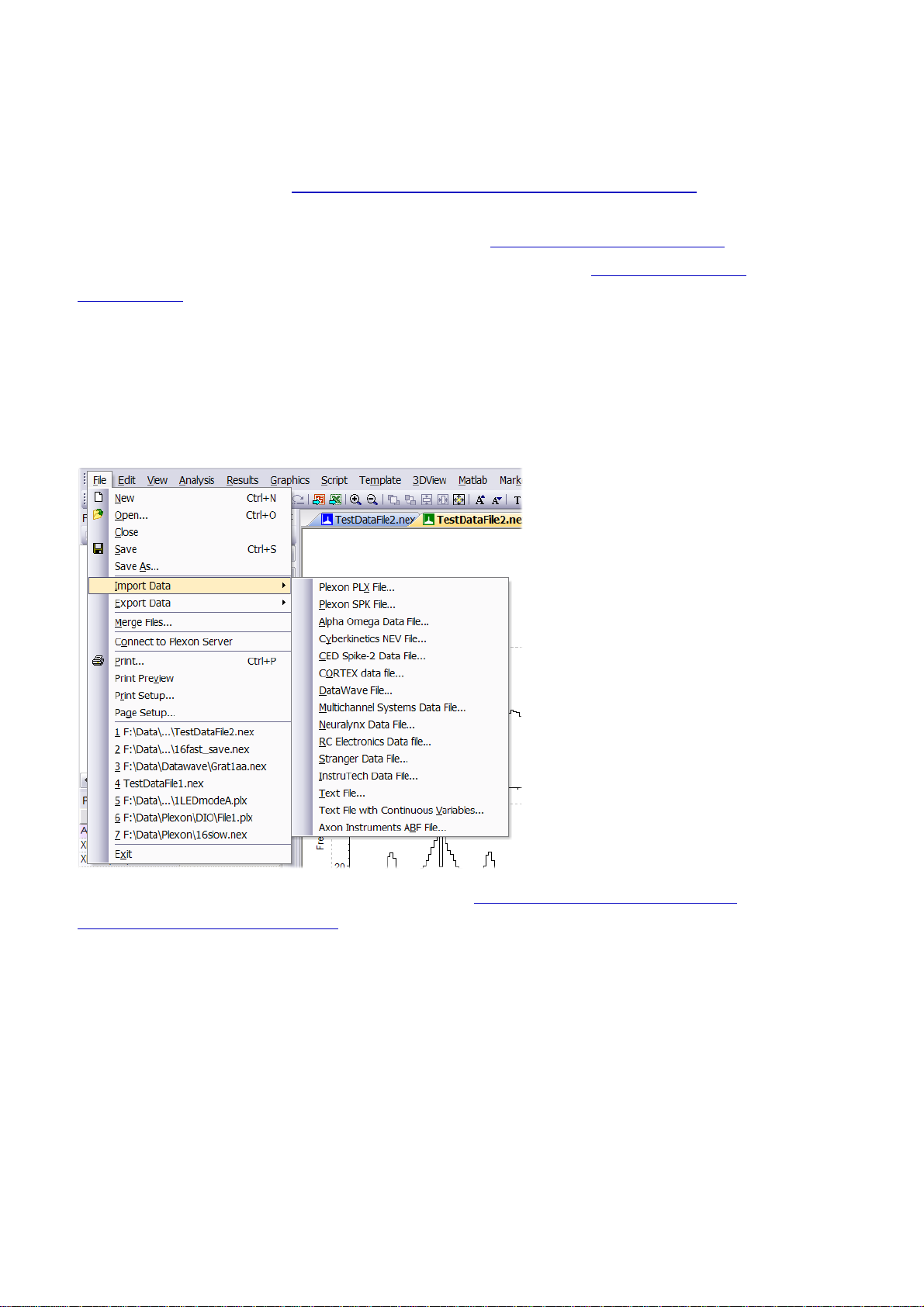
1.4. Opening Files and Importing Data
NeuroExplorer can read native data files created by popular data acquisition systems (Alpha Omega,
CED Spike-2, Cortex, Blackrock Microsystems, DataWave, Multi Channel Systems, Neuralynx,
Plexon, RC Electronics. See Importing Files Created By Data Acquisition Systems for more
information).
NeuroExplorer can also import data from text files (see Importing Data from Text Files ).
You can import the data from spreadsheets using the clipboard (see Importing Data from
Spreadsheets ).
NeuroExplorer has its own data format and by default saves the data in the binary file with the
extension.nex.
To open a NeuroExplorer data file,
•
press File Open toolbar button , or
•
select File | Open... menu command.
To import data in any of the supported file formats, select the corresponding File | Import command:
You can also paste data directly into Data View. See Importing Data from the Text Files and
Importing Data from Spreadsheets for more information.
Page 16
Page 19

1.5. Importing Files Created by Data Acquisition Systems
NeuroExplorer can read native data files created by popular data acquisition systems (Alpha Omega,
CED Spike-2, Cortex, DataWave, Blackrock Microsystems, Multi Channel Systems, Neuralynx,
Plexon, RC Electronics).
Since NeuroExplorer does not provide spike sorting capabilities yet, it is assumed that you have
already sorted (clustered) the waveforms.
Alpha Omega Files
NeuroExplorer can import Alpha Omega MAP, ISI, NDA, MAT and LSM files. All the spike trains, DIO
events and continuous variables are imported. The waveforms can be imported as an option (see File
Import Options below).
Blackrock Microsystems Files
NeuroExplorer can import Blackrock Microsystems NEV files. All the spike trains and DIO events
Analog channels can be imported as an option. The waveforms are imported when using Neuroshare
DLL to import NEV files.
DataWave Files
NeuroExplorer can import both Discovery and Workbench files. In general, all the sorted spike trains,
DIO events and trial descriptors are imported. The waveforms are not imported. Analog channels can
be imported as an option (see Options below).
The following UFF types are imported from the Discovery files:
S, E and U UFF types are imported as spike trains
B UFF type records are imported as events
T UFF type records are imported as markers
P (position) UFF type records are imported as 4 continuous variables.
The following record types are imported from the Workbench files:
Analysis records (with appropriate subtypes) are imported as spike trains
Event records are imported as events
Trial records are imported as markers
CED Spike-2 Files
NeuroExplorer can import Spike-2 *.smr files. All the sorted spike trains, events and markers are
imported. The waveforms are not imported. Analog (Adc) channels can be imported as an option (see
File Import Options below).
Plexon Files
NeuroExplorer can read *.plx and *.ddt Plexon files. All the sorted spike trains and external events are
imported. The waveforms are not imported. Analog (slow) channels can be imported as an option
(see File Import Options below).
RC Electronics Files
NeuroExplorer can import both 12-bit RC Electronics files and 16-bit DATAMAX files. Since the
current version of NeuroExplorer stores all the data in RAM, NeuroExplorer can import only relatively
small (about the size of RAM on your PC) RC Electronics files.
Page 17
Page 20

Multi Channel Systems Files
NeuroExplorer can import standard MCS data files. All the sorted spike trains and external events are
imported. The waveforms are not imported. Analog channels can be imported as an option (see File
Import Options below).
Neuralynx Files
NeuroExplorer can import Neuralynx data files (tt*.dat, se*.dat, etc.) as well as the files created by
MClust spike sorter (*.t files). All the sorted spike trains are imported. External events are imported
as markers. Analog channels and waveforms can be imported as an option (see File Import Options
below).
File Import Options
The file import options can be set in the Data Import dialog (use View | Data Import Options menu
command to invoke the dialog).
Page 18
Page 21
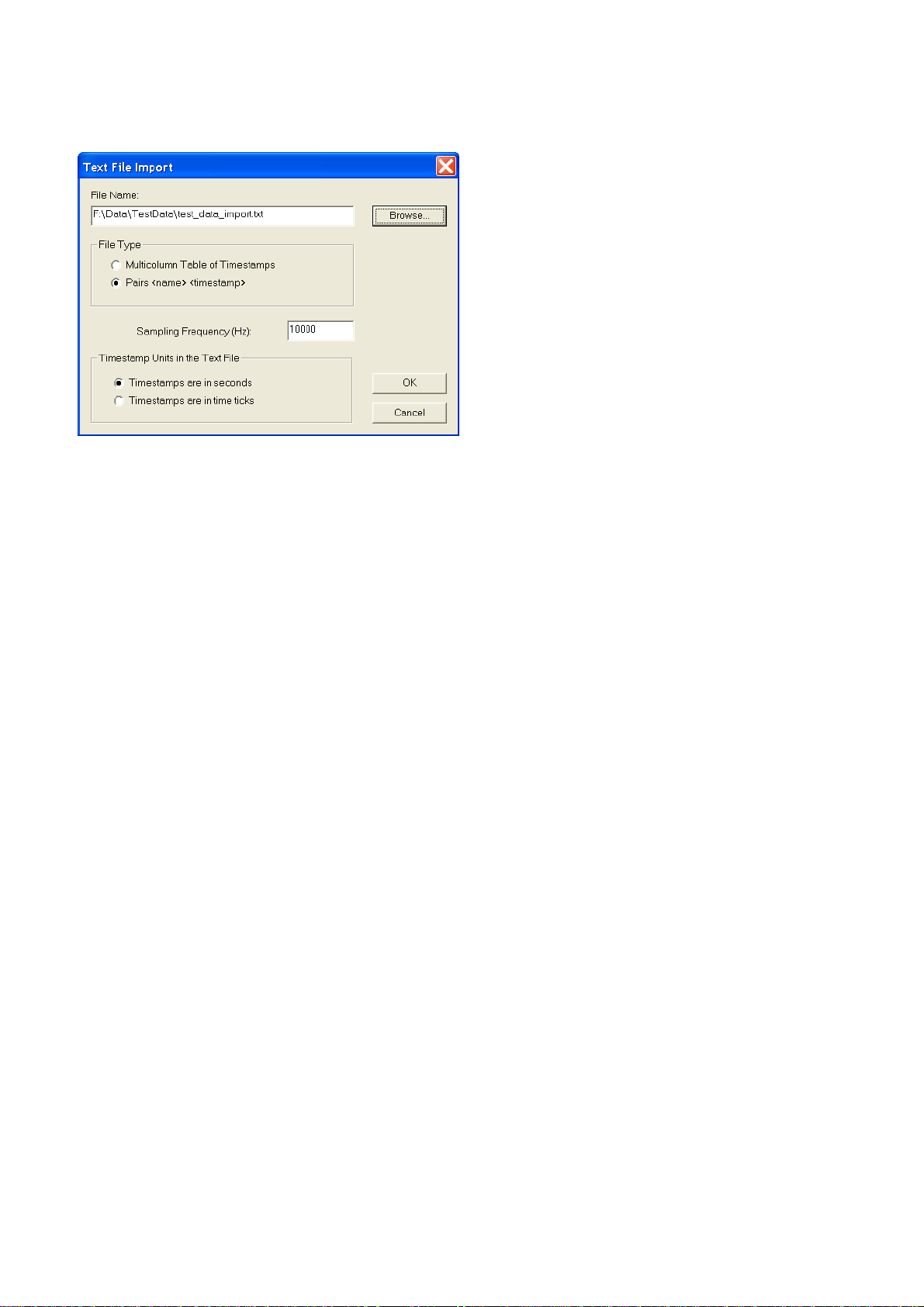
1.6. Importing Data from Text Files
You can specify text import options in the Text Import dialog:
Sampling Frequency - this parameter defines the internal representation of the timestamps that
NeuroExplorer will use for this file. Internally, the timestamps are stored as integers representing the
number of time ticks from the start of the experiment. The time tick is equal to
1./ Sampling_Frequency
Timestamp Units - this parameter defines how the numbers representing the timestamps are treated
by NeuroExplorer. If the timestamps are in time ticks, they are stored internally exactly as they are in
the text file. If the timestamps are in seconds, NeuroExplorer converts them to time ticks:
Internal_Timestamp = Timestamp_In_Seconds * Sampling_Frequency
NeuroExplorer can import timestamped data stored in the following text formats:
1. Multicolumn table of timestamps
In this format, each column in the text file contains the timestamps of a neuron. The first element in
each column is a neuron name, that is the first line of the file contains names of all the variables.
Each name should be less than 64 characters long and should contain only letters, digits or the
underscore sign. The first character of the name should be a letter. The timestamps are numbers
representing the neuron firing time (or event time) in seconds or in time ticks. NeuroExplorer assumes
that the columns are separated by tabs.
Here is an example of a text file with the timestamps represented in seconds:
Neuron01 Neuron02
0.01 0.001
0.3 0.05
0.5 0.1
0.4
0.6
NeuroExplorer can also export data in this format (use File | Save Data | As a Text File menu
command).
2. Pairs <name> <timestamp>
The text file in this format should contain the pairs of the type
<name><timestamp>
where
<name> is a character string that is less that 64 characters long and contains only letters, digits and
the underscore sign. The first character of the name should be a letter. If the number is used for the
Page 19
Page 22
name, NeuroExplorer will add "event" at the beginning of the name.
<timestamp> is a number representing the neuron firing time (or event time) in seconds or in time
ticks.
Here is an example of a text file with the timestamps represented in seconds:
Neuron01 0.01
Neuron01 0.3
Neuron02 0.001
Neuron02 0.05
Neuron01 0.5
Neuron02 0.1
Neuron02 0.4
3. Multicolumn text files with continuous data
NeuroExplorer can also import continuous data from a multicolumn text file where each column
corresponds to a continuous variable:
ContChannel1 ContChannel2
114.74609 -63.47656
56.15234 -358.88672
-187.98828 -63.47656
-48.82813 388.18359
-26.85547 285.64453
The columns may be separated by any number of spaces, tabs or commas.
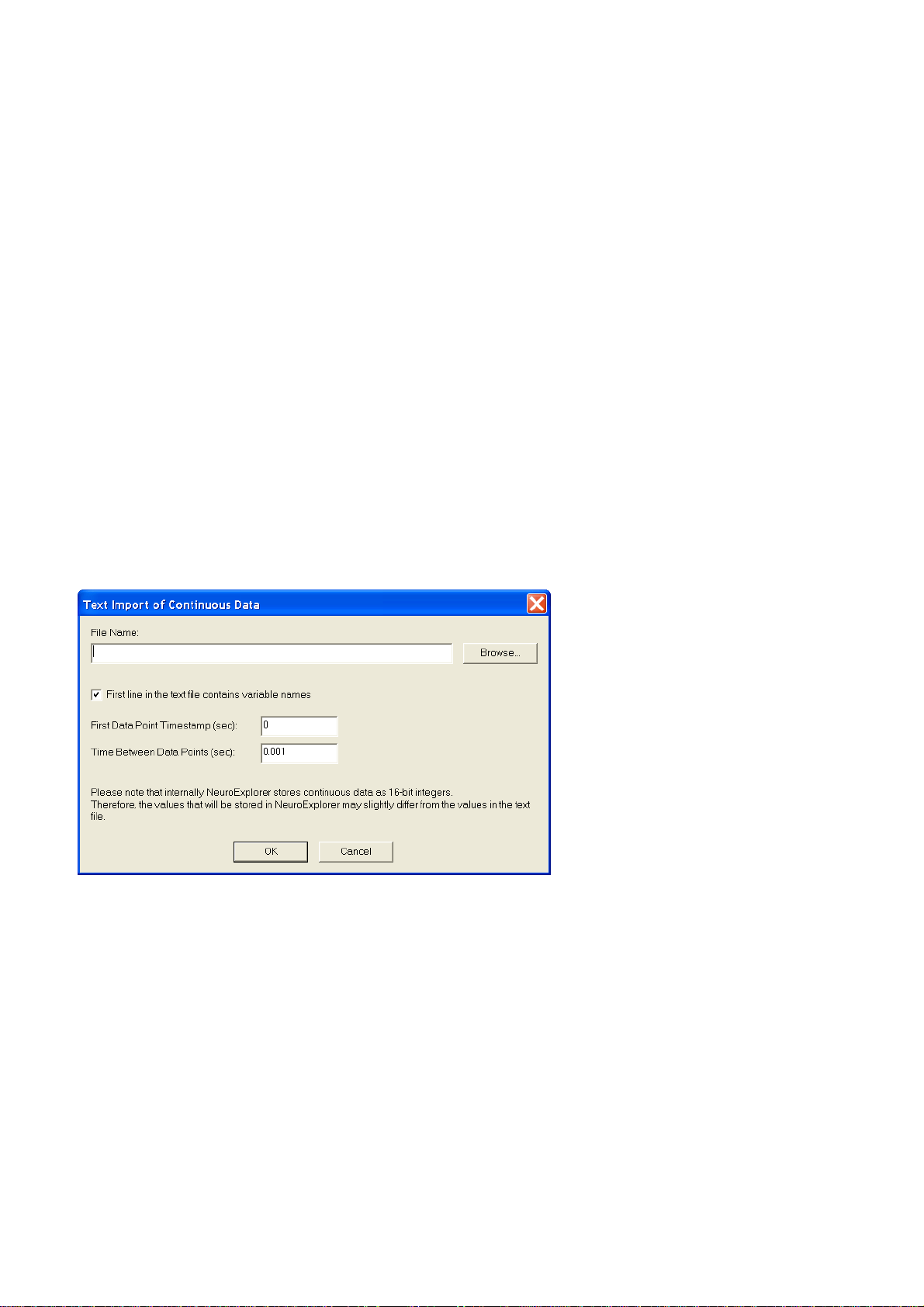
When you import continuous data from text files, the following dialog is shown:
First line contains variable names – if this option is selected, the fields of the first line of the text file
are used as variable names. The second line in the file is the first row of data.
First Data Point Timestamp specifies the timestamp of the first data point in each continuous
variable.
Time Between Data Points specifies the time step used in calculation of the timestamp for each row
of data. For data row N (N = 1, 2, …) in the text file, the timestamp is calculated using the following
formula:
DataRowTimestamp = FirstDataPoint + (N-1)*TimeBetweenDataPoints
1.7. Importing Data from Spreadsheets
Page 20
Page 23
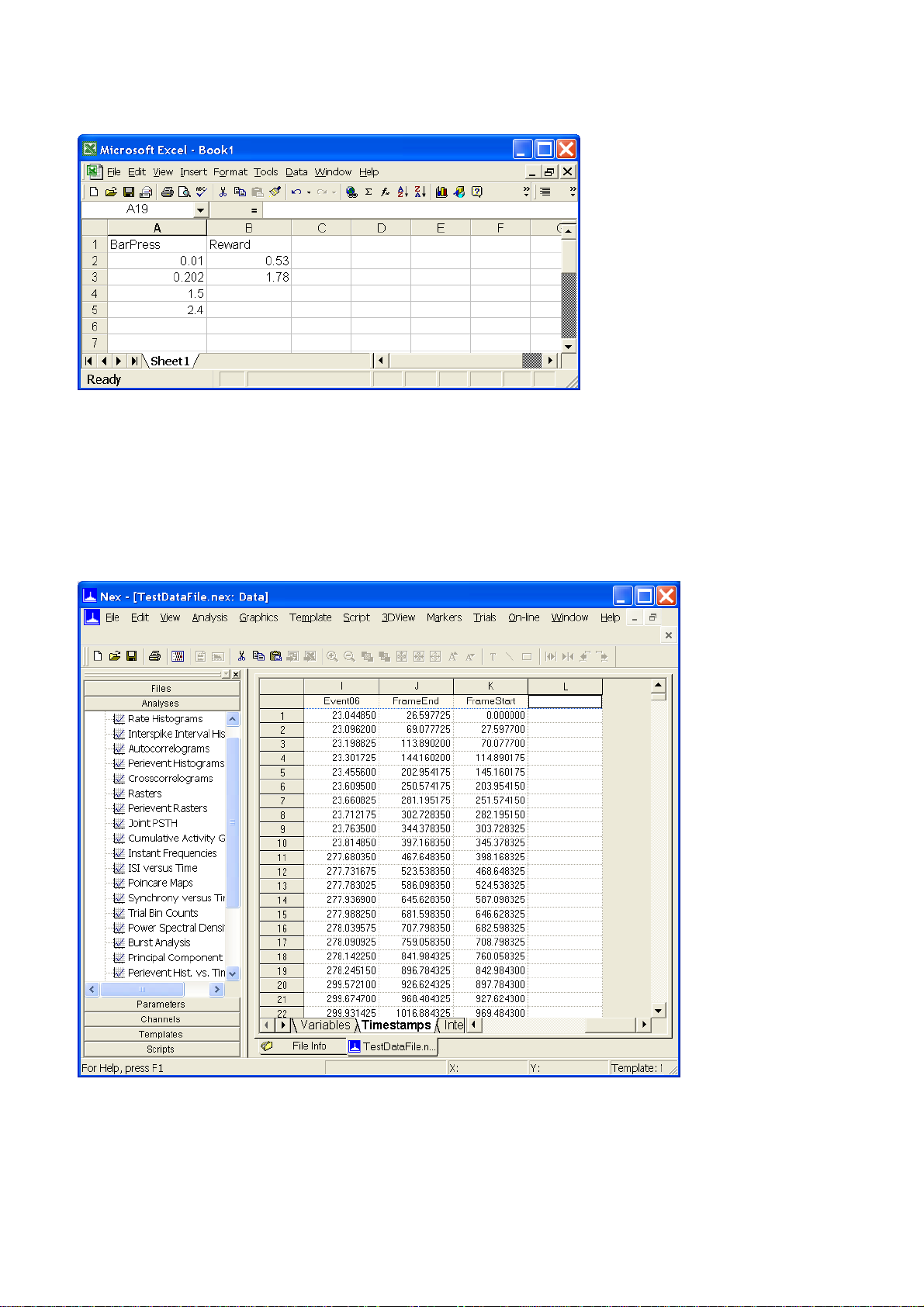
Timestamped data can be pasted directly into the NeuroExplorer data table. To paste the following
two timestamped variables (BarPress and Reward) from a spreadsheet to NeuroExplorer:
In Excel:
•
Using the mouse, select the cell range with both variables (A1 to B5)
•
Select Edit | Copy menu command
In NeuroExplorer:
•
Select Data view of the file in NeuroExplorer
•
Select the Timestamps tab of the Data view
•
Scroll the timestamps window to the right and select the topmost cell of the empty column:
Select Edit | Paste menu command. NeuroExplorer will create two new Event variables and add
them to the file. You need to save the file (use File | Save or File | SaveAs menu command) to make
this change permanent.
Page 21
Page 24

1.8. Importing Data from Matlab
You can create spike trains (1xN or Nx1 matrices with timestamps in seconds) or continuous
variables in Matlab and transfer them to NeuroExplorer on the fly. Here is how to do this:
•
Select File | New menu command.
•
Select Matlab | Get Data From Matlab... | Open Matlab As Engine menu command.
NeuroExplorer will open Matlab as engine.
•
In opened Matlab, run your Matlab scripts and create (or load from file) spike train variables or
continuous variables.
•
To import timestamp variables, in NeuroExplorer, select Matlab | Get Data From Matlab... | Get
Timestamp Variables... menu command. NeuroExplorer will open a dialog with the list of
available Matlab variables.
•
In the dialog, select the variables you want to transfer to NeuroExplorer and press OK.
•
To import continuous variables, in NeuroExplorer, select Matlab | Get Data From Matlab... |
Get Continuous Variables... menu command. NeuroExplorer will open a dialog with the list of
available Matlab variables.
•
In the dialog, select one of the variables you want to transfer to NeuroExplorer and press OK.
NeuroExplorer will open a dialog where you will specify the variable options.
Page 22
Page 25
1.9. Reading and Writing NeuroExplorer Data Files
NeuroExplorer Data file has the following structure:
file header
variable header 1
variable header 2
...
data for variable i
data for variable j
...
Each variable header contains the size of the array that stores the variable data as well as the
location of this array in the file.
The pseudo-code for reading NeuroExplorer data file looks like this:
open file in binary mode
read file header
for each variable
read variable header
end for
for each variable header
seek to the file offset specified in the variable header
read variable data
end for
The C++ source code of the program that reads and writes NeuroExplorer data files is available at
NeuroExplorer web site:
http://www.neuroexplorer.com/updates/HowToReadAndWriteNexFiles.zip
There is also a Matlab script that reads NeuroExplorer data files:
http://www.neuroexplorer.com/updates/HowToReadNexFilesInMatlab.zip
Page 23
Page 26
1.10. 1D Data Viewer
NeuroExplorer provides a window that displays graphically all the selected variables. To open this
window, use View | 1D Data Viewer Window menu command.
You can also use 1D view to manually add events to the data file. Simply press the left mouse button
when the pointer is in the 1D view and NeuroExplorer will add a new timestamp to the variable
ManualEvent. You can also specify to what Event variable NeuroExplorer will add timestamps when
you click in 1D view (use On mouse click parameter in the Properties Panel).
Mouse wheel can be used to quickly navigate in 1D View:
•
Click in 1D View so set 1D View as an active window in NeuroExplorer
•
Press Shift and rotate mouse wheel to shift 1D View horizontally
•
Press Ctrl and rotate mouse wheel to increase time range (zoom out) and decrease time range
(zoom in)
Page 24
Page 27
1.11. Analyzing Data
NeuroExplorer provides a variety of spike train analysis methods. Each method has a number of
parameters and options. You can apply any available analysis by clicking at the corresponding
analysis in the Analyses Panel (you can also use Analysis | Select Analysis menu commands):
After you click at one of the analysis items, NeuroExplorer will open a dialog that will allow you to edit
the analysis parameters. When you click OK in this dialog, NeuroExplorer will apply the specified
analysis to all the selected variables.
Any combination of analysis parameters and options (together with all the graphics options) can be
saved as a template. To save the current configuration as a template, right-click in the Graph Window
and select Save As New Template menu command. The template names are shown in the
Templates view of the control panel.
When you open a new data file, you can simply double-click on the template name to apply the
specific analysis with the selected parameter values.
Page 25
Page 28
1.12. Selecting Variables for Analysis
NeuroExplorer can analyze at once any group of variables in the file.
To select the variables to be analyzed, use one of the following methods:
•
Select the variables using the Variable Panel (see How to select variables using Variables Panel
)
•
Select the variables using Analysis | Select Variables menu command. This command will
invoke the Variable Selection dialog similar to Variables Panel.
•
Select the variables directly in the Variables Window by using the check boxes located next to
the variable names. Please note that variable selection in Variables Window is applied only
when a new Graph Window or 1D View is created. To change the list of selected variables
in existing Graph Window, use Variables Panel or Analysis | Select Variables menu
command. See How to Select Variables for Existing Analysis Window for details.
Page 26
Page 29
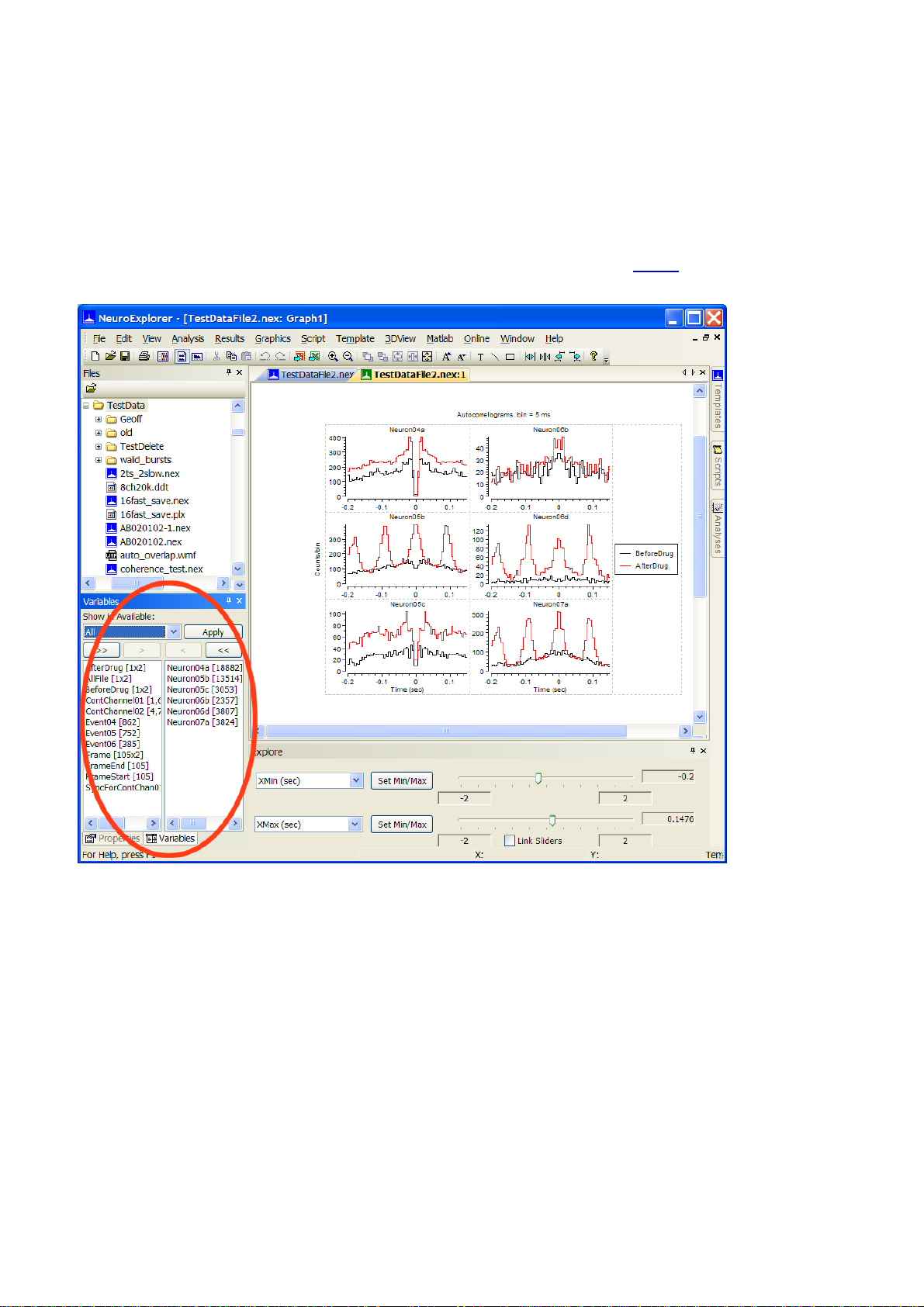
1.13. How to Select Variables for Existing Window
Variables selected in the Variables spreadsheet of data window are used to populate variables lists of
new analysis windows.
If you would like to change the list of variables for existing analysis window, please follow these
steps:
•
Click anywhere in analysis window to activate it
•
If Variables panel is visible, make changes in selected variables list ( how? ) and press Apply
button:
•
If Variables panel is hidden, click at panel tab to open it:
Page 27
Page 30
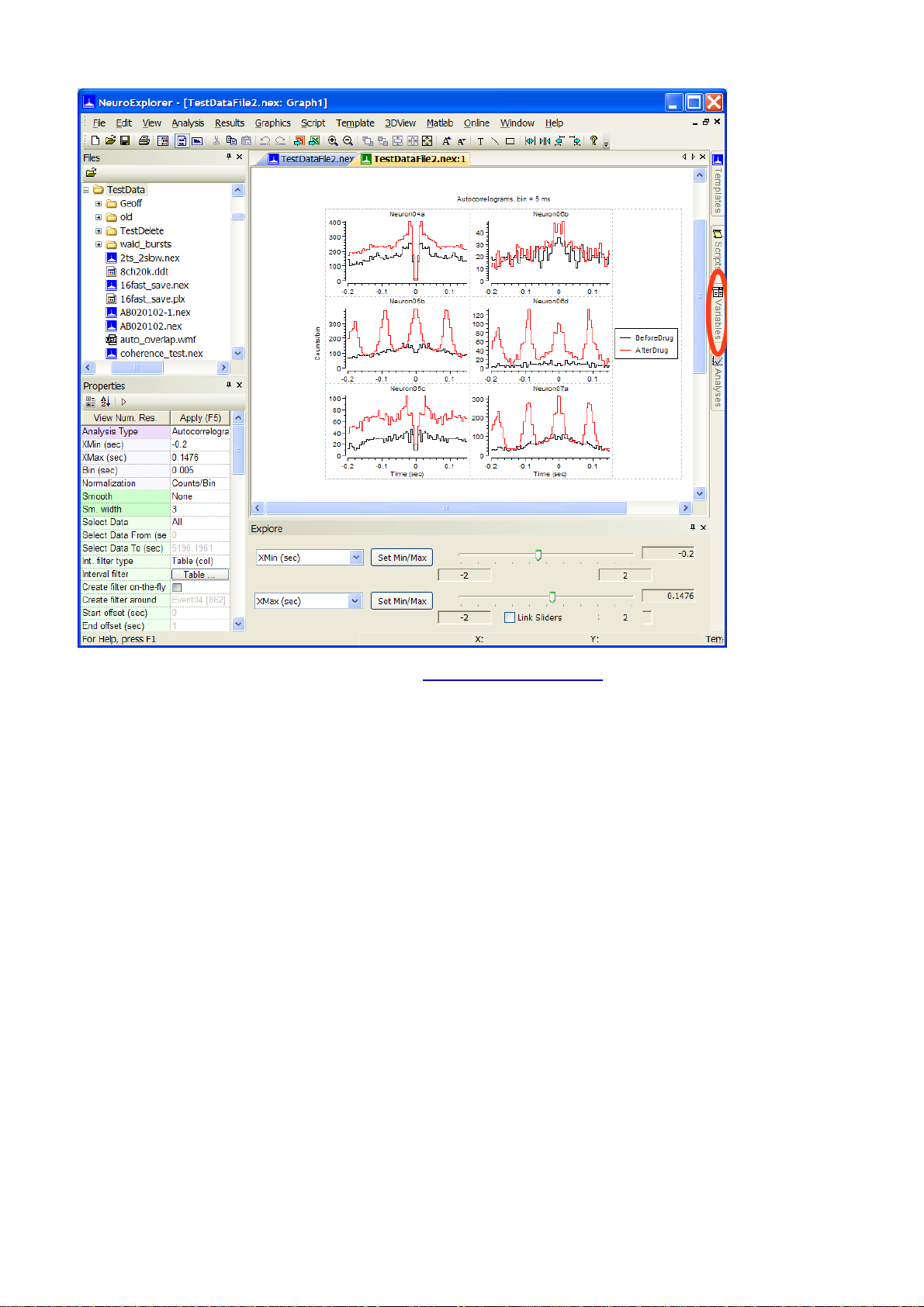
•
Make changes in selected variables list ( how to make changes? ) and press Apply button
•
If Variables panel or its tab is not visible, use View | Variables Panel menu command
Page 28
Page 31

1.14. How to Select Variables in Variables Panel
Select variables for analysis by moving them from the left (available) to the right (selected) list box. In
the figure below, 6 variables (Neuron04a, Neuron05b, ..., Neuron07a) are selected for analysis:
To move variables from one listbox to another:
Select variables using the mouse (in the figure above two variables in Available listbox are
•
selected -- ContChannel01 and ContChannel02)
Press one of four buttons just above the listboxes
•
The drop-down box above the buttons can be used to specify what kind of variables are shown in the
available window:
Page 29
Page 32

1.15. Adjusting Analysis Properties
There are several methods to adjust the analysis parameters:
•
Use Analysis | Edit Parameters menu command to invoke the Analysis Parameters dialog
•
Right-click in the Graph Window and choose Analysis Parameters item in the floating menu
The floating menu is the fastest way to adjust any analysis or graphics parameters. Double-click (or
right-click) anywhere in the Graph Window to invoke this menu:
This menu allows you to go directly to analysis properties, graph properties, axes properties, etc. You
can also use it to save the current template and save the current analysis configuration as a new
template.
•
Single-click in the graph area of the Graph Window and adjust parameters in the Properties
Panel:
Page 30
Page 33

1.16. Analysis Templates
In any analysis in NeuroExplorer, you can adjust a large number of parameters:
•
Analysis type (Rate Histograms, IIH, etc)
•
Analysis parameters (Bin, XMin, XMax, etc.)
•
Graph parameters (graph type, graph color, grid lines, etc)
•
Parameters of X and Y axes (labels, numerics, lines, colors, etc.)
•
Graph and page labels and other elements
NeuroExplorer allows you to save all these parameters so that when you open another data file, you
can easily reproduce exactly the same analysis with the same axes, labels and so on.
The set of all the analysis parameters is called the Template.
To save the current analysis as a template:
•
Select the menu command Template | Save As New Template, or
•
Right-click in the graph and choose the Save As New Template command in the floating menu.
After you saved the template, the name of the template appears in the Templates Window (see
NeuroExplorer Screen Elements ). You can apply the template to the currently opened file by double-
clicking the template name in the Templates Window.
NeuroExplorer stores template files in Windows Application Data directory:
C:\Documents and Settings\All Users\Application Data\Nex Technologies\NeuroExplorer\Templates
under Windows XP, or
C:\Application Data\Nex Technologies\NeuroExplorer\Templates under Vista or Windows 7.
Page 31
Page 34

1.17. Numerical Results
To view the analysis numerical results, select View | Numerical Results menu command.
You can also press the "View. Num. Res." button in the upper-left corner of the Properties Panel:
NeuroExplorer will open Numerical Results Window:
Note that the window has two Excel-style sheets -- the Results sheet and the Summary sheet. The
Results sheet usually contains the bin counts or other histogram values. The Summary sheet
contains summary statistics of the analyses such as the mean firing rate, the number of spikes used
in the analysis, etc.
Page 32
Page 35

1.18. Post-processing
For the histogram-style analyses, NeuroExplorer has an optional Post-Processing analysis step.
You can select post-processing options using Post-processing tab in the Analysis Parameters dialog
(double-click in Graph window and select Analysis Parameters menu item):
You can smooth the resulting histogram with Gaussian or Boxcar filters. You can also add bin
information to the matrix of results. See Post-processing Options for details.
See Saving Results as Power Point Slides, Working with Matlab and Working with Excel for more
information on NeuroExplorer post-processing options.
1.19. Saving Results as Power Point Slides
NeuroExplorer version 3 provides a powerful new option that allows you to save graphics, analysis
parameters and your annotations as slides in a Power Point Presentation.
To save your results to Power Point, press a toolbar button , or select Graphics | Export Graphics |
Export to Power Point menu command. NeuroExplorer will start Power Point if Power Point is not
running, and will add a new slide to the specified presentation file. The slide will contain:
Copy of the graphics
•
Analysis parameters
•
Your comment
•
Data file name
•
Current date and time
•
Page 33
Page 36

Power point presentation then becomes your lab notebook where you can save your NeuroExplorer
analysis results:
Page 34
Page 37

1.20. Working with Matlab
NeuroExplorer can interact with Matlab via COM interface using Matlab as a powerful postprocessing engine. Matlab can also control NeuroExplorer and exchange data with NeuroExplorer via
NeuroExplorer COM interfaces. See COM/ActiveX Interfaces for details.
Immediately after calculating the histograms, NeuroExplorer can send the resulting matrix of
histograms to Matlab and then ask Matlab to execute any series of Matlab commands.
Use the Matlab tab in the Analysis parameters dialog to specify the matrix name and the Matlab
command string.
NeuroExplorer can also send its data variables to Matlab. To transfer the variables to Matlab, use
Matlab | Send Selected Variables to Matlab menu command.
In general, a continuous variable may contain several fragments of data. Each fragment may be of a
different length. NeuroExplorer does not store the timestamps for all the A/D values since they would
use too much space. Instead, for each fragment, it stores the timestamp of the first A/D value in the
fragment and the index of the first data point in the fragment. Therefore, for a continuous variable
(named ContVar1), the following 3 vectors will be created in Matlab:
•
ContVar1 - vector (or matrix with 2 columns) of all A/D values in milliVolts. If "Optimize transfer
for small amounts of data" is selected in View | Options | Matlab, Contvar1 is a matrix with 2
columns: the first column contains A/D values in millivolts, the second column contains
timestamps in seconds.
•
ContVar1_ts - vector of fragment timestamps in seconds. Each timestamp is for the first A/D
value of the fragment.
•
ContVar1_ind - array of indexes. Each index is the position of the first data point of the
fragment in the A/D array. If ContVar1_ind (2) = 201, it means that the second fragment is
ContVar1 [201], ContVar1 [202], etc.
•
ContVar1_ts_step - digitizing step in seconds.
You can generate all the timestamps for continuous variable using this Matlab script:
function [ ts ] = MakeTs( fragmentInd, fragmentTs, numValues, step )
% MakeTs: makes timestamps for continuous variable based on the fragment
% information
% INPUT: fragmentInd - vector of fragment indexes
% fragmentTs - vector of fragment timestamps
% numValues - total number of values in all fragments
% step - digitizing step of continuous variable
%
% Example: NeuroExplorer sent continuous variable FP01 via file transfer
% The following values were sent:
% FP01 - vector of continuous values
% FP01_ind - fragment indexes
% FP01_ts - fragment timestamps
% FP01_ts_step - digitizing step
%
% to generate all timestamps for FP01, use:
%
% ts = MakeTs(FP01_ind, FP01_ts, size(FP01,1), FP01_ts_step);
%
ts = [];
numFr = size(fragmentTs);
% add fake index for the last fragment
fragmetInd = [fragmentInd; numValues+1];
for i=1:numFr
valuesInFragment = fragmetInd(i+1)-fragmentInd(i);
Page 35
Page 38

ts = [ts; (fragmentTs(i) + (0:(valuesInFragment-1))*step)'];
end
end
You can also import timestamped and continuous variables directly from Matlab. See Importing Data
From Matlab for details.
See Also
Importing Data From Matlab
COM/ActiveX Interfaces
1.21. Working with Excel
The easiest way to transfer data and numerical results from NeuroExplorer to Excel is by using the
clipboard:
•
select a range of cells in NeuroExplorer and choose Edit| Copy
•
switch to Excel and use the Paste command (select a cell and choose Edit | Paste ).
NeuroExplorer can also send numerical results directly to Excel. There two ways to send the results
to Excel:
•
Use Send to Excel button on the toolbar or menu command File | Save Numerical Results |
Send Results to Excel
•
Use Excel tab in the Analysis Parameters dialog to specify what to transfer to Excel and the
location of the top-left cell for the data from NeuroExplorer.
Page 36
Page 39

1.22. Saving Graphics
NeuroExplorer can save the contents of the Graph window in a file using the Windows Metafile
format. The file can then be opened in any program that can use the.wmf format files (a word
processor, graphics editor, etc.).
To save the graphics in a metafile, use File | Save Graphics menu command.
You can also copy the graphics to the clipboard and then paste in another application. To copy the
graphics, single-click in the graph area and then select Edit | Copy menu command.
See also Saving Results as Power Point Slides.
Page 37
Page 40

2. Analysis Reference
This section, NeuroExplorer Analysis Reference, describes general pre-processing and postprocessing analysis options, analysis parameters and computational algorithms used in analyses.
•
Data Types
•
Data Selection Options
•
Post-Processing Options
•
Matlab Options
•
Excel Options
•
Confidence Limits for Perievent Histograms
•
Rate Histograms
•
Interspike Interval Histograms
•
Autocorrelograms
•
Perievent Histograms
•
Crosscorrelograms
•
Rasters
•
Perievent Rasters
•
Joint PSTH
•
Cumulative Activity Graphs
•
Instant Frequency Graphs
•
Interspike Intervals vs Time
•
Poincare Maps
•
Spike Distance vs Time
•
Trial Bin Counts
•
Power Spectral Densities
•
Burst Analysis
•
Principal Component Analysis
•
Perievent Histograms vs. Time
•
Correlations with Continuous Variables
•
Regularity Analysis
•
Place Cell Analysis
•
Reverse Correlation
•
Epoch Counts
•
Coherence Analysis
Page 38
Page 41

2.1. Data Types
NeuroExplorer supports several data types -- spike trains, behavioral events, time intervals,
continuously recorded data and other data types. The topics in this section describe the data types
used in NeuroExplorer, list the properties of data types, and show how to view and modify various
data types in NeuroExplorer.
2.1.1. Spike Trains
In NeuroExplorer, the most commonly used data type is a spike train. A spike train in NeuroExplorer
does not contain the spike waveforms, it represents only the spike timestamps (the times when the
spikes occurred). A special Waveform data type is used to store the spike waveform values.
Events are very similar to spike trains. The only difference between spike trains and events is that
spike trains may contain additional information about recording sites, electrode numbers, etc. (for
example, spike trains contain positions of neurons used in the 3D activity "movie" ).
Internally, the timestamps are stored as 32-bit signed integers. These integers are usually the
timestamps recorded by the data acquisition system and they represent time in the so-called time
ticks. For example, the typical time tick for the Plexon system is 25 microseconds, so an event
recorded at 1 sec will have the timestamp equal to 40000.
Limitations
The maximum number of timestamps (in each spike train) in NeuroExplorer is 2,147,483,647. In
reality, the limiting factor is the amount of virtual memory on your machine, since the timestamps of
all the variables of a data file should fit into memory. NeuroExplorer requires that in each spike train
all the timestamps are in ascending order, that is
timestamp[i+1] > timestamp[i] for all i.
NeuroExplorer also requires that
timestamp[i] >= 0 and timestamp[i] < 2,147,483,647 for all i.
The upper timestamp limit defines the maximum experiment length that can be safely analyzed in
NeuroExplorer. Thus, for the standard Plexon setup, the maximum duration of the experiment is
2147483647/40000 seconds, or 14 hours and 54 minutes.
Timestamped Variables in NexScript
You can get access to any timestamp in the current file. For example, to assign the value 0.5 (sec) to
the third timestamp of the variable DSP01a, you simply write:
doc = GetActiveDocument()
doc.DSP01a[3] = 0.5
Viewers
You can view the timestamps of the selected variables in graphical display ( View | 1D Data Viewer
menu command):
Page 39
Page 42

Numerical values of the timestamps (in seconds) are shown in the Timestamps sheet of the Data
view:
2.1.2. Events
Event data type in NeuroExplorer is used to represent the series of timestamps recorded from
external devices (for example, stimulation times recorded as pulses produced by the stimulus
generator). Event data type stores only the times when the external events occurred. A special
Marker data type is used to store the timestamps together with other stimulus or trial information.
Events are very similar to spike trains. The only difference between spike trains and events is that
spike trains may contain additional information about recording sites, electrode numbers, etc.
Internally, event timestamps are represented exactly as Spike Train timestamps. See Spike Trains for
more information about the timestamps in NeuroExplorer.
Creating Event Variables
You can add events to the data file using Copy | Paste command in the Timestamps view (see
Importing Data from Spreadsheets for details).
You can also create additional events based on the events in the data file. Use Edit | Operations on
Data Variables menu command and then select one of the operations in the Operations dialog.
2.1.3. Intervals
Intervals are usually used in NeuroExplorer to select the data from a specified time periods (see Data
Selection Options for details).
Each interval variable can contain multiple time intervals. For example, the Frame variable in the
following figure has two intervals:
Page 40
Page 43

Creating Interval Variables
You can create intervals in NeuroExplorer using Edit | Add Interval Variable menu command.
You can also create intervals based on existing Event or Interval variables. Use Edit | Operations on
Data Variables menu command and then select one of the following operations:
MakeIntervals
MakeIntFromStart
MakeIntFromEnd
IntOpposite
IntAnd
IntOr
IntSize
IntFind
Burst analysis creates interval variables (one for each neuron) that contain all the detected bursts as
time intervals.
Interval Variables in NexScript
IntVar[i,1] gives you the access to the start of the i-th interval,
IntVar[i,2] gives you the access to the end of the i-th interval.
For example, the following script creates a new interval variable that has two intervals: from 0 to 100
seconds and from 200 to 300 seconds:
doc = GetActiveDocument()
doc.MyInterval = NewIntEvent(doc, 2)
doc.MyInterval[1,1] = 0.
doc.MyInterval[1,2] = 100.
doc.MyInterval[2,1] = 200.
doc.MyInterval[2,2] = 300.
Limitations
Internally, NeuroExplorer stores the beginning and the end of each interval as a timestamp (see
Spike Trains for more information about timestamps in NeuroExplorer).
Viewers
You can view the intervals of the selected variables in graphical display ( View | 1D Data Viewer
menu command, see figure above).
The intervals (in seconds) are shown in the Intervals sheet of the Data view:
Page 41
Page 44

2.1.4. Markers
Marker data type in NeuroExplorer is used to represent the series of timestamps recorded from
external devices (for example, stimulation times recorded as pulses produced by the stimulus
generator) together with other stimulus or trial information. Event data type stores only the times
when the external events occurred.
NeuroExplorer creates Marker variables when it imports strobed events from Plexon data files, when
it imports Trial Descriptor Records from the Datawave files and when DIO events with flags are
loaded from Alpha Omega files.
Viewers
You can view both the marker timestamps and additional marker information in the Markers tab of the
Data window:
Extracting Events
Usually you will need to extract events with specific trial information from the marker variables. To do
this, use Marker | Split... and Marker | Extract... menu commands.
For example, you may want to extract from the marker variable shown in the figure above only the
markers with DIOValue 131 or 132. Marker | Extract... menu command will open the Extract dialog
box in which you can specify multiple criteria for extracting events from the marker variable:
Page 42
Page 45

2.1.5. Population Vectors
Population vectors in NeuroExplorer can be used to display the linear combinations of histograms in
some of the analyses. For example, you can calculate perievent histograms for each individual
neuron recorded in a data file. If you want to calculate the response of the whole population of
neurons (that is, create an average PST histogram) you need to use a population vector.
Population vector assigns a weight to each spike train or event variable in the file. You can then use
population vectors in the following analyses:
Rate Histograms
•
Perievent Histograms
•
Trial Counts
•
If you select the population vector for analysis and then run one of the three analyses listed above,
the histogram corresponding to the population vector will be calculated as:
histogram_of_neuron_1 * weight_1 + histogram_of_neuron_2 * weight_2...
where the weights are defined in the population vector. For example, to calculate an average
histogram for 6 neurons, the following population vector should be used:
Creating Population Vectors
You can create new population vectors in NeuroExplorer using Edit | Add Population Vector menu
command.
Page 43
Page 46

Principal Component Analysis creates a set of population vectors based on correlations between
activities of individual neurons.
2.1.6. Waveforms
The waveform data type is used in NeuroExplorer to store the spike waveform values together with
the spike timestamps. The following figure shows the waveform variable sig006a_wf together with the
corresponding spike train sig006a:
The waveform data may not be imported by default from the data files created by the data acquisition
systems. Use View | Data Import Options menu command to specify which data types
NeuroExplorer will import from the data files.
The waveform variables can be used in the following analyses:
•
Rate Histograms
•
Rasters
•
Perievent Histograms (instead of PST, NeuroExplorer calculates spike-triggered average for a
waveform variable)
Viewers
You can view the waveforms of the selected variables in the graphical display ( View | 1D Data
Viewer menu command, see figure above).
Numerical values of the waveforms and their timestamps are shown in the Waveforms sheet of the
Data view.
To select a different waveform variable, click in the cell located in the row 1 of the Variable column
(the cell shows sig001i_wf in the figure above).
Waveform values are shown in columns labeled 1, 2, etc.
Page 44
Page 47

2.1.7. Continuously Recorded Data
The continuous data type is used in NeuroExplorer to store the data that has been continuously
recorded (digitized).
The following figure shows continuous variable AD01 together with the spike train sig001a:
Continuous data may not be imported by default from the data files created by the data acquisition
systems. Use View | Data Import Options menu command to specify which data types
NeuroExplorer will import from the data files.
Continuous variables can be used in the following analyses:
•
Rate Histograms
•
Rasters
•
Perievent Histograms (instead of PST, NeuroExplorer calculates the spike-triggered average for
a continuous variable)
•
Perievent Rasters (NeuroExplorer shows the values of continuous variable using color)
•
Spectral Densities
•
Correlations with Continuous Variable
•
Coherence Analysis
Limitations
Internally, each data point of the continuous variable is stored as a 2-byte integer. The maximum
number of values (in each continuous variable) in NeuroExplorer is 2,147,483,647. In reality, the
limiting factor is the amount of virtual memory on your machine, since all the values should fit into
virtual memory.
Viewers
You can view continuous variables in the graphical display ( View | 1D Data Viewer menu command,
see figure above).
Numerical values of the continuous variables are shown in the Continuous sheet of the Data view.
Page 45
Page 48

2.2. Data Selection Options
Before performing the analysis, NeuroExplorer can select the timestamped events that are inside the
specified time intervals:
For example, you may want to analyze only the first 10 minutes of the recording session. To do this,
choose the Data Selection tab in the Analysis Parameters dialog, click Use only the data from the
following time range and specify From and To parameters in seconds:
You can also choose an interval variable (at the bottom of the Data Selection page) as an additional
data filter. NeuroExplorer then will select data from one or more time intervals as specified by the
interval variable.
Some analyses (Perievent Histograms, Perievent Rasters and Crosscorrelograms) allow you to
specify several interval filters, so that a different filter will be used for a column or for a row of graphs.
Page 46
Page 49

2.3. Post-Processing Options
For the histogram-style analyses, NeuroExplorer has an optional Post-processing analysis step.
You can select post-processing options using Post-processing tab in the Analysis Parameters dialog.
(double-click in the Graph window and select Analysis Parameters menu item):
You can smooth the resulting histogram with Gaussian or Boxcar filters.
The boxcar filter is a filter with equal coefficients. If f[i] is i-th filter coefficient and w is the filter width,
then
f[i] = 1/w for all i
Thus for the filter of width 3, the boxcar filter is
f[-1] = 0.33333, f[0] = 0.33333, f[1] = 0.33333.
and the smoothed histogram sh[i] is calculated as
sh[i] = f[-1]*h[i-1] + f[0]*h[i] + f[1]*h[i+1]
where h[i] is the original histogram.
The Gaussian filter is calculated using the following formula:
f[i] = exp(-i*i/sigma)/norm, i from -2*d to 2*d
where
d = ( (int)w + 1 )/2
sigma = -w * w * 0.25 / log(0.5)
norm = sum of exp( -i * i / sigma), i from -2*d to 2*d
The parameters of the filter are such that the width of the Gaussian curve at half the height equals to
the specified filter width. Please note that for the Gaussian filter, width of the filter (w) can be a noninteger. For example, w can be 3.5.
See Matlab Options and Excel Options for more information on NeuroExplorer post-processing
capabilities.
Page 47
Page 50

2.4. Matlab Options
NeuroExplorer can interact with Matlab via COM interface using Matlab as a powerful postprocessing engine. Immediately after calculating histograms, NeuroExplorer can send the resulting
matrix of histograms to Matlab and then ask Matlab to execute any series of Matlab commands.
Use the Matlab tab in the Analysis parameters dialog to specify the matrix name and the Matlab
command string.
2.5. Excel Options
NeuroExplorer can send numerical results directly to Excel. There are two ways to send the results to
Excel:
•
Use Send to Excel button on the toolbar or menu command File | Save Numerical Results |
Send Results to Excel
•
Use Excel tab in the Analysis Parameters dialog to specify what to transfer to Excel and the
location of the top-left cell for the data from NeuroExplorer.
Page 48
Page 51

2.6. Confidence Limits for Perievent Histograms
If the total time interval (experimental session) is T (seconds) and we have N spikes in the interval,
then the neuron frequency is:
F = N/T
Several options how to calculate neuron frequency F are available. See Options below.
Then if the spike train is a Poisson train, the probability of the neuron to fire in the small bin of the size
b (seconds) is
P = F*b
The expected bin count for the perievent histogram is then:
C = P*NRef, where NRef is the number of the reference events.
The value C is used for drawing the Mean Frequency in the Perievent Histograms and Cross- and
Autocorrelograms.
The confidence limits for C are calculated using the assumption that C has a Poisson distribution.
Assume that a random variable S has a Poisson distribution with parameter C. Then the 99%
confidence limits are calculated as follows:
Low Conf. = x such that Prob(S < x) = 0.005
High Conf. = y such that Prob(S > y) = 0.005
If C < 30, NeuroExplorer uses the actual Poisson distribution
Prob(S = K) = exp(-C) * (C^K) / K!, where C^K is C to the power of K,
to calculate the confidence limits.
If C>= 30., the Gaussian approximation is used. For example, for 99% confidence limits:
Low Conf. = C - 2.58*sqrt(C);
High Conf.= C + 2.58*sqrt(C);
Reference
Abeles M. Quantification, smoothing, and confidence limits for single-units histograms. Journal of
Neuroscience Methods. 5(4):317-25, 1982
Options
The following options to calculate neuron frequency F are available:
•
Use all file data. Here T is the total length of experimental session, N is the total number of
spikes for a given neuron.
•
Use selected time range and interval filter. T is the length of all the time intervals used in
analysis, N is the number of spikes within these intervals.
•
Use time intervals corresponding to prereference bins. This option only works for a stimulationtype data. For example, if you stimulate every second and calculate PSTH with XMin=-0.2,
XMax=0.2, NeuroExplorer can easily distinguish the spikes before and after the stimulus.
However, if you stimulate every 200 ms, the spikes before the second stimulus are also the
spikes after the first stimulus, so you cannot distinguish the spikes that should be used to
calculate the mean firing rate. The algorithm: for each reference event timestamp r, a time
interval (r+XMin, r) is created (where XMin is PSTH or crosscorrelogram time axis minimum
parameter; XMin should be negative). T is the length of all (r+XMin, r) intervals that do not
overlap, N is the number of spikes in these intervals. If more than 5% of intervals overlap, F is
set to zero.
Page 49
Page 52

2.7. Cumulative Sum Graphs
Here is the algorithm that is used to draw optional cumulative sum graphs above the histograms.
Suppose we have a histogram with bin counts bc[i], i=1,...,N. Cumulative Sum Graph displays the
following values cs[i]:
for bin 1: cs[1] = bc[1] - A
for bin 2: cs[2] = bc[1]+bc[2] - A*2
for bin 3: cs[3] = bc[1]+bc[2]+bc[3] - A*3, etc.
The value of A depends on the selected Cumulative Sum option:
A equals to average of all bc[i] if you select "Use all histogram" option
A equals to average of all bc[i] for bins that are before zero on time scale if you select "Use preref as
base" option.
If you use "Use all histogram" option, the value of the cumulative sum for the last bin is always zero:
sc[N] = bc[1]+bc[2]+...bc[N] - A*N, where A = (bc[1]+bc[2]+...bc[N])/N.
NeuroExplorer displays 99% confidence limits for Cumulative Sum Graphs. Confidence limits for "Use
preref as base" option are proportional to square root of bin number.
Calculations of confidence limits for "Use all histogram" option are not trivial. These calculations are
based on the formulas developed by the author of NeuroExplorer, Alexander Kirillov.
Page 50
Page 53

2.8. Rate Histograms
Rate histogram displays firing rate versus time.
Parameters
Parameter Description
XMin/XMax type An option on how XMin and XMax values are specified.
XMin Time axis minimum in seconds.
XMax Time axis maximum in seconds
Bin Histogram bin size in seconds.
Normalization Histogram units (Counts/Bin, Probability or Spikes/Second). See Algorithm
below.
Select Data If Select Data is From Time Range, only the data from the specified (by
Select Data From and Select Data To parameters) time range will be used
in analysis. See also Data Selection Options.
Select Data From Start of the time range in seconds.
Select Data To End of the time range in seconds.
Smooth histogram An option to smooth the histogram after the calculation. See Post-
Processing Options for details.
Smooth Filter Width The width of the smooth filter. See Post-Processing Options for details.
Add to Results / Bin left An option to add an additional vector (containing a left edge of each bin) to
the matrix of numerical results.
Add to Results / Bin
middle
Add to Results / Bin
right
Send to Matlab An option to send the matrix of numerical results to Matlab. See also
Matrix Name Specifies the name of the results matrix in Matlab workspace.
Matlab command Specifies a Matlab command that is executed after the numerical results
Send to Excel An option to send numerical results or summary of numerical results to
Sheet Name The name of the worksheet in Excel where to copy the numerical results.
An option to add an additional vector (containing a middle point of each
bin) to the matrix of numerical results.
An option to add an additional vector (containing a right edge of each bin)
to the matrix of numerical results.
Matlab Options.
are sent to Matlab.
Excel. See also Excel Options.
TopLeft Specifies the Excel cell where the results are copied. Should be in the form
CR where C is Excel column name, R is the row number. For example, A1
is the top-left cell in the worksheet.
Summary of Numerical Results
The following information is available in the Summary of Numerical Results
Column Description
Page 51
Page 54

Variable Variable name.
YMin Histogram minimum.
YMax Histogram maximum.
Spikes The number of spikes used in calculation.
Filter Length The length of all the intervals of the interval filter (if a filter was used) or the
length or the recording session (in seconds).
Mean Freq. Mean firing rate (Spikes/Filter_Length).
Mean Hist. The mean of the histogram bin values.
St. Dev. Hist. The standard deviation of the histogram bin values.
St. Err. Mean. Hist. The standard error of mean of the histogram bin values.
Algorithm
The time axis is divided into bins. The first bin is [XMin, XMin+Bin). The second bin is [XMin+Bin,
Xmin+Bin*2), etc. The left end is included in each bin, the right end is excluded from the bin.
For each bin, the number of events in this bin is calculated.
For example, for the first bin
bin_count = number of timestamps (ts) such that ts >= XMin and ts < XMin + Bin
If Normalization is Counts/Bin, no further calculations are performed.
If Normalization is Spikes/Sec, bin counts are divided by Bin.
Page 52
Page 55

2.9. Interspike Interval Histograms
Interspike interval histogram shows the conditional probability of a first spike at time t0+t after a spike
at time t0.
Parameters
Parameter Description
Min Interval Minimum interspike interval in seconds.
Max Interval Maximum interspike interval in seconds.
Bin Bin size in seconds.
Normalization Histogram units (Counts/Bin, Probability or Spikes/Second). See Algorithm
below.
Log Bins and X Axis An option to use Log10 interval scale (see Algorithm below for details).
Bins per decade A bin size option if Log option is selected (see Algorithm below for details).
Y Axis Limit (%) An option to "zoom in" small histogram values. This parameter specifies
the maximum of Y axis. If it is 100%, the Y axis maximum equals to the
maximum bin value. If it is, for example, 10%, Y axis maximum is set to
10% of the bin max and the small bin values have better visibility.
Select Data If Select Data is From Time Range, only the data from the specified (by
Select Data From and Select Data To parameters) time range will be used
in analysis. See also Data Selection Options.
Select Data From Start of the time range in seconds.
Select Data To End of the time range in seconds.
Int. filter type Specifies if the analysis will use a single or multiple interval filters.
Interval filter Specifies the interval filter(s) that will be used to preselect data before
analysis. See also Data Selection Options.
Create filter on-the-fly Specifies if a temporary interval filter needs to be created (and used to
preselect data).
Create filter around Specifies an event that will be used to create a temporary filter.
Start offset Offset (in seconds, relative to the event specified in Create filter around
parameter) for the start of interval for the temporary filter.
End offset Offset (in seconds, relative to the event specified in Create filter around
parameter) for the end of interval for the temporary filter.
Fix overlaps An option to automatically merge the overlapping intervals in the temporary
filter.
Overlay Graphs An option to draw several histograms in each graph. This option requires
that Int. filter type specifies that multiple interval filters will be used (either
Table (row) or Table (col)).
Smooth Option to smooth the histogram after the calculation. See Post-Processing
Options for details.
Smooth Filter Width The width of the smooth filter. See Post-Processing Options for details.
Add to Results / Bin left An option to add an additional vector (containing a left edge of each bin) to
the matrix of numerical results.
Page 53
Page 56

Add to Results / Bin
middle
An option to add an additional vector (containing a middle point of each
bin) to the matrix of numerical results.
Add to Results / Bin
right
Send to Matlab An option to send the matrix of numerical results to Matlab. See also
Matrix Name Specifies the name of the results matrix in Matlab workspace.
Matlab command Specifies a Matlab command that is executed after the numerical results
Send to Excel An option to send numerical results or summary of numerical results to
Sheet Name The name of the worksheet in Excel where to copy the numerical results.
TopLeft Specifies the Excel cell where the results are copied. Should be in the form
An option to add an additional vector (containing a right edge of each bin)
to the matrix of numerical results.
Matlab Options.
are sent to Matlab.
Excel. See also Excel Options.
CR where C is Excel column name, R is the row number. For example, A1
is the top-left cell in the worksheet.
Summary of Numerical Results
The following information is available in the Summary of Numerical Results
Column Description
Variable Variable name.
YMin Histogram minimum.
YMax Histogram maximum.
Spikes The number of spikes used in calculation.
Filter Length The length of all the intervals of the interval filter (if a filter was used) or the
length or the recording session (in seconds).
Mean Freq. Mean firing rate (Spikes/Filter_Length).
Mean Hist. The mean of the histogram bin values.
St. Dev. Hist. The standard deviation of the histogram bin values.
St. Err. Mean. Hist. The standard error of mean of the histogram bin values.
Mean ISI The mean of the interspike intervals.
St. Dev. ISI The standard deviation of the interspike intervals.
Coeff. Var. ISI Coefficient of variation of the interspike intervals.
Median ISI Median of the interspike intervals.
Mode ISI Mode of the interspike intervals.
Algorithm
If Use Log Bins and X Axis option is not selected:
The time axis is divided into bins. The first bin is [IntMin, IntMin+Bin). The second bin is
[IntMin+Bin, Intmin+Bin*2), etc. The left end is included in each bin, the right end is excluded from
the bin. For each bin, the number of interspike intervals within this bin is calculated.
Page 54
Page 57

For example, for the first bin
bin_count = number of interspike intervals (isi)
such that isi >= IntMin and isi < IntMin + Bin
If Normalization is Counts/Bin, no further calculations are performed.
If Normalization is Probability, bin counts are divided by the number of interspike intervals in the
spike train.
If Normalization is Spikes/Sec, bin counts are divided by NumInt*Bin, where NumInt is the number
of interspike intervals in the spike train.
If Use Log Bins and X Axis option is selected:
The i-th bin (i=1,2,...) is [IntMin * 10 ^ ((i -1)/D), IntMin * 10 ^ (i/D)), where D is the Number of Bins
Per Decade. For each bin, the number of interspike intervals within this bin is calculated. For
discussion on using logarithm of interspike intervals, see:
Alan D. Dorval, Probability distributions of the logarithm of inter-spike intervals yield accurate entropy
estimates from small datasets. Journal of Neuroscience Methods 173 (2008) 129–139
Page 55
Page 58

2.10. Autocorrelograms
Autocorrelogram shows the conditional probability of a spike at time t0+t on the condition that there is
a spike at time t0.
Parameters
Parameter Description
XMin Time axis minimum in seconds.
XMax Time axis maximum in seconds.
Bin Bin size in seconds.
Normalization Histogram units (Counts/Bin, Probability or Spikes/Second). See Algorithm
below.
Select Data If Select Data is From Time Range, only the data from the specified (by
Select Data From and Select Data To parameters) time range will be used
in analysis. See also Data Selection Options.
Select Data From Start of the time range in seconds.
Select Data To End of the time range in seconds.
Int. filter type Specifies if the analysis will use a single or multiple interval filters.
Interval filter Specifies the interval filter(s) that will be used to preselect data before
analysis. See also Data Selection Options.
Create filter on-the-fly Specifies if a temporary interval filter needs to be created (and used to
preselect data).
Create filter around Specifies an event that will be used to create a temporary filter.
Start offset Offset (in seconds, relative to the event specified in Create filter around
parameter) for the start of interval for the temporary filter.
End offset Offset (in seconds, relative to the event specified in Create filter around
parameter) for the end of interval for the temporary filter.
Fix overlaps An option to automatically merge the overlapping intervals in the temporary
filter.
Overlay Graphs An option to draw several histograms in each graph. This option requires
that Int. filter type specifies that multiple interval filters will be used (either
Table (row) or Table (col)).
Smooth Option to smooth the histogram after the calculation. See Post-Processing
Options for details.
Smooth Filter Width The width of the smooth filter. See Post-Processing Options for details.
Draw confidence limits An option to draw the confidence limits.
Confidence (%) Confidence level (percent). See Confidence Limits for details.
Conf. mean calculation An option that specifies how the mean firing rate (that is used in the
derivation of the confidence limits) is calculated.
There are 2 options: Use data selection and Use all file. See Confidence
Limits for details.
Conf. display An option to draw confidence limits either as horizontal lines or as a
Page 56
Page 59

colored background.
Conf. line style Line style for drawing confidence limits (used when Conf. display is Lines ).
Conf. background color Background color for drawing confidence limits (used when Conf. display is
Colored Background ).
Draw mean freq. An option to draw a horizontal line representing the expected histogram
value for a Poisson spike train. See Confidence Limits for details.
Mean line style Line style for drawing mean frequency.
Add to Results / Bin left An option to add an additional vector (containing a left edge of each bin) to
the matrix of numerical results.
Add to Results / Bin
middle
Add to Results / Bin
right
Send to Matlab An option to send the matrix of numerical results to Matlab. See also
Matrix Name Specifies the name of the results matrix in Matlab workspace.
Matlab command Specifies a Matlab command that is executed after the numerical results
Send to Excel An option to send numerical results or summary of numerical results to
Sheet Name The name of the worksheet in Excel where to copy the numerical results.
TopLeft Specifies the Excel cell where the results are copied. Should be in the form
An option to add an additional vector (containing a middle point of each
bin) to the matrix of numerical results.
An option to add an additional vector (containing a right edge of each bin)
to the matrix of numerical results.
Matlab Options.
are sent to Matlab.
Excel. See also Excel Options.
CR where C is Excel column name, R is the row number. For example, A1
is the top-left cell in the worksheet.
Summary of Numerical Results
The following information is available in the Summary of Numerical Results
Column Description
Variable Variable name.
YMin Histogram minimum.
YMax Histogram maximum.
Spikes The number of spikes used in calculation.
Filter Length The length of all the intervals of the interval filter (if a filter was used) or the
length or the recording session (in seconds).
Mean Freq. Mean firing rate (Spikes/Filter_Length).
Mean Hist. The mean of the histogram bin values.
St. Dev. Hist. The standard deviation of the histogram bin values.
Conf. Low Lower confidence level.
Conf. High Upper confidence level.
Mean Expected mean value of the histogram.
Page 57
Page 60

Norm. Factor Normalization factor. Bin counts are divided by this value. See
Normalization in Algorithm below.
First Min. Time Position of the histogram minimum (in seconds). If there are multiple bins
in the histogram where the bin value equals to the histogram minimum, this
value represents the position of the first such bin.
First Max. Time Position of the histogram maximum (in seconds). If there are multiple bins
in the histogram where the bin value equals to the histogram maximum,
this value represents the position of the first such bin.
Algorithm
In general, the Autocorrelogram shows the conditional probability of a spike in the spike train at time t
on the condition that there is a spike at time zero.
The time axis is divided into bins. The first bin is [XMin, XMin+Bin). The second bin is [XMin+Bin,
Xmin+Bin*2), etc. The left end is included in each bin, the right end is excluded from the bin.
Let ts[i] be the spike train (each ts is the timestamp).
For each timestamp ts[k]:
calculate the distances from this spike to all other spikes in the spike train:
d[i] = ts[i] - ts[k]
for each i except i equal to k:
if d[i] is inside the first bin, increment the bin counter for the first bin:
if d[i] >= XMin and d[i] < XMin + Bin
then bincount[1] = bincount[1] +1
if d[i] is inside the second bin, increment the bin counter for the second bin:
if d[i] >= XMin+Bin and d[i] < XMin + Bin*2
then bincount[2] = bincount[2] +1
and so on... .
If Normalization is Counts/Bin, no further calculations are performed.
If Normalization is Probability, bin counts are divided by the number of spikes in the spike train.
Note that the Probability normalization makes sense only for small values of Bin. For Probability
normalization to be valid (so that the values of probability are between 0 and 1), there should be no
more than one spike in each bin. For example, if the Bin value is large and for each ts[k] above there
are many d[i] values such that d[i] >= XMin and d[i] < XMin + Bin, the bin count for the first bin can
exceed the number of spikes in the spike train. Then, the probability value
(bincount[1]/number_of_spikes) could be larger than 1.
If Normalization is Spikes/Sec, bin counts are divided by NumSpikes*Bin, where NumSpikes is the
number of spikes in the spike train.
Page 58
Page 61

2.11. Perievent Histograms
Perievent Histogram shows the conditional probability of a spike at time t0+t on the condition that
there is a reference event at time t0.
Parameters
Parameter Description
Reference Type Specifies if the analysis will use a single or multiple reference events.
Reference Specifies a reference neuron or event (or a group of reference neurons or
events).
XMin Histogram time axis minimum in seconds.
XMax Histogram time axis maximum in seconds
Bin Bin size in seconds.
Normalization Histogram units (Counts/Bin, Probability, Spikes/Second or Z-score). See
Algorithm below.
No Selfcount An option not to count reference events if the target event is the same as
the reference event (prevents a histogram to have a huge peak at zero
when calculating PSTH versus itself).
Select Data If Select Data is From Time Range, only the data from the specified (by
Select Data From and Select Data To parameters) time range will be used
in analysis. See also Data Selection Options.
Select Data From Start of the time range in seconds.
Select Data To End of the time range in seconds.
Int. filter type Specifies if the analysis will use a single or multiple interval filters.
Interval filter Specifies the interval filter(s) that will be used to preselect data before
analysis. See also Data Selection Options.
Create filter on-the-fly Specifies if a temporary interval filter needs to be created (and used to
preselect data).
Create filter around Specifies an event that will be used to create a temporary filter.
Start offset Offset (in seconds, relative to the event specified in Create filter around
parameter) for the start of interval for the temporary filter.
End offset Offset (in seconds, relative to the event specified in Create filter around
parameter) for the end of interval for the temporary filter.
Fix overlaps An option to automatically merge the overlapping intervals in the temporary
filter.
Overlay Graphs An option to draw several histograms in each graph. This option requires
that Int. filter type specifies that multiple interval filters will be used (either
Table (row) or Table (col)).
Overlay Options Specifies line colors and line styles for overlaid graphs.
Smooth Option to smooth the histogram after the calculation. See Post-Processing
Options for details.
Smooth Filter Width The width of the smooth filter. See Post-Processing Options for details.
Page 59
Page 62

Draw confidence limits An option to draw the confidence limits.
Confidence (%) Confidence level (percent). See Confidence Limits for details.
Conf. mean calculation An option that specifies how the mean firing rate (that is used in the
derivation of the confidence limits) is calculated.
There are 3 options: Use data selection, Use all file and Use pre-ref data.
See Confidence Limits for details.
Note that Use pre-ref data option can only be used for a stimulation type
data, i.e. when the distance between any two consecutive reference
events is larger than XMax-XMin. See Confidence Limits for details.
Conf. display An option to draw confidence limits either as horizontal lines or as a
colored background.
Conf. line style Line style for drawing confidence limits (used when Conf. display is Lines ).
Conf. background color Background color for drawing confidence limits (used when Conf. display is
Colored Background ).
Draw mean freq. An option to draw a horizontal line representing the expected histogram
value for a Poisson spike train. See Confidence Limits for details.
Mean line style Line style for drawing mean frequency.
Draw Cusum An option to draw a cumulative sum graph above the histogram. See
Cumulative Sum Graphs for details.
Add to Results / Bin left An option to add an additional vector (containing a left edge of each bin) to
the matrix of numerical results.
Add to Results / Bin
middle
Add to Results / Bin
right
Background An option on how to calculate the histogram background for the peak and
Peak width Peak width (the number of bins in peak) in the peak and through analysis.
Left shoulder Specifies the left shoulder value (in seconds) in the peak and through
Right shoulder Specifies the right shoulder value (in seconds) in the peak and through
Send to Matlab An option to send the matrix of numerical results to Matlab. See also
Matrix Name Specifies the name of the results matrix in Matlab workspace.
Matlab command Specifies a Matlab command that is executed after the numerical results
An option to add an additional vector (containing a middle point of each
bin) to the matrix of numerical results.
An option to add an additional vector (containing a right edge of each bin)
to the matrix of numerical results.
through analysis. See Peak and Trough Statistics below.
See Peak and Trough Statistics below.
analysis. See Peak and Trough Statistics below.
analysis. See Peak and Trough Statistics below.
Matlab Options.
are sent to Matlab.
Send to Excel An option to send numerical results or summary of numerical results to
Excel. See also Excel Options.
Sheet Name The name of the worksheet in Excel where to copy the numerical results.
TopLeft Specifies the Excel cell where the results are copied. Should be in the form
CR where C is Excel column name, R is the row number. For example, A1
is the top-left cell in the worksheet.
Page 60
Page 63

Summary of Numerical Results
The following information is available in the Summary of Numerical Results
Column Description
Variable Variable name.
Reference Reference variable name.
NumRefEvents The number of reference events used in calculation.
YMin Histogram minimum.
YMax Histogram maximum.
Spikes The number of spikes used in calculation.
Filter Length The length of all the intervals of the interval filter (if a filter was used) or the
length or the recording session (in seconds).
Mean Freq. Mean firing rate (Spikes/Filter_Length).
Mean Hist. The mean of the histogram bin values.
St. Dev. Hist. The standard deviation of the histogram bin values.
St. Err. Mean. Hist. The standard error of mean of the histogram bin values.
Conf. Low Lower confidence level.
Conf. High Upper confidence level.
Mean Expected mean value of the histogram. If Z-score normalization is
specified, this value is zero and the expected mean value of the histogram
with Counts/Bin normalization is shown in Z-score Mean.
Norm. Factor Normalization factor. Bin counts are divided by this value. See
Normalization in Algorithm below.
Z-score mean Expected mean value of the histogram before Z-score normalization (in
other words, confidence mean). See Confidence Limits for details .
Mean Before Ref. Mean of the histogram before the reference event (i.e. for all the bins
before zero on time axis).
Bins Before Ref. The number of bins before the bin that contains zero on time axis.
Zero Bin The index of the bin that contains zero on time axis.
Background Mean The mean of the histogram background for the peak and trough analysis.
See Peak and Through Statistics below.
Background Stdev The standard deviation of the histogram background for the peak and
trough analysis. See Peak and Trough Statistics below.
Peak Z-score Peak Z-score. See Peak and Trough Statistics below.
Peak/Mean Histogram peak value divided by the background mean value.
Peak Position Peak position (in seconds).
Peak Half Height The Y value of the half height of the peak, i.e.
histogram_background_mean + (peak-mean)/2.
Peak Width at Half
Height
Peak width at the peak half height level (in seconds).
Page 61
Page 64

Trough Z-score Trough Z-score. See Peak and Trough Statistics below.
Trough/Mean Histogram trough value divided by the background mean value.
Trough Position Trough position (in seconds).
Trough Half Height The Y value of the half height of the trough, i.e.
histogram_background_mean + (trough-mean)/2.
Trough Width at Half
Trough width at the trough half height level (in seconds).
Height
Algorithm
In general, the Perievent Histogram shows the conditional probability of a spike in the spike train at
time t0+t on the condition that there is a reference event (or reference spike) at time t0.
The time axis is divided into bins. The first bin is [XMin, XMin+Bin). The second bin is [XMin+Bin,
XMin+Bin*2), etc. The left end is included in each bin, the right end is excluded from the bin.
Let ref[i] be the array of timestamps of the reference event,
ts[i] be the spike train (each ts is the timestamp)
For each timestamp ref[k]:
1) calculate the distances from this event (or spike) to all the spikes in the spike train:
d[i] = ts[i] - ref[k]
2) for each i:
if d[i] is inside the first bin, increment the bin counter for the first bin:
if d[i] >= XMin and d[i] < XMin + Bin
then bincount[1] = bincount[1] +1
if d[i] is inside the second bin, increment the bin counter for the second bin:
if d[i] >= XMin+Bin and d[i] < XMin + Bin*2
then bincount[2] = bincount[2] +1
and so on... .
If Normalization is Counts/Bin, no further calculations are performed.
If Normalization is Probability, bin counts are divided by the number of reference events.
Note that the Probability normalization makes sense only for small values of Bin. For Probability
normalization to be valid (so that the values of probability are between 0 and 1), there should be no
more than one spike in each bin. For example, if the Bin value is large and for each ref[k] above there
are many d[i] values such that d[i] >= XMin and d[i] < XMin + Bin, the bin count for the first bin can
exceed the number of reference events. Then, the probability value
(bincount[1]/number_of_reference_events) could be larger than 1.
If Normalization is Spikes/Sec, bin counts are divided by NumRefEvents*Bin, where
NumRefEvents is the number of reference events.
If Normalization is Z-score, bin_value = (bin_count - Confidence_mean)/sqrt(Confidence_mean),
where Confidence_mean is the expected mean bin count calculated according to Conf. mean
calculation parameter. Please note that bin counts are assumed to have Poisson distribution
(therefore, standard deviation is equal to square root of expected mean) and Z-score can be
considered to have Normal distribution only for large values (more than 10) of the Confidence_mean.
Peak and Trough Statistics
Page 62
Page 65

NeuroExplorer calculates the histogram peak statistics the following way:
•
Maximum of the histogram is found
•
If the histogram contains several maxima with the same value, peak statistics are not calculated
•
Otherwise, the center of the bin, where the histogram reaches maximum, is shown as Peak
Position in the Summary of Numerical results
•
The mean M and standard deviation S of the bin values of the histogram background are
calculated: If Background parameter is set as Bins outside peak/trough, bins outside peak and
trough (i.e., bins that are more than PeakWidth/2 away from the bin with the histogram
maximum and the bin with the histogram minimum) are used to calculate M and S If Background
parameter is set as Shoulders, bins that are to the left of Left Shoulder or to the right of Right
Shoulder parameters are used to calculate M and S
•
The value M (mean of the background bin values) is shown as Background Mean in the
Summary of Numerical results
•
The value S (standard deviation of the background bin values) is shown as Background Stdev
in the Summary of Numerical results
•
The value (HistogramMaximum – M)/S is shown as Peak Z-score
•
The value (HistogramMaximum + M)/2 is shown as Peak Half Height
•
Histogram intersects a horizontal line drawn at Peak Half Height at time points TLeft and TRight.
(TRight - TLeft) is shown as Peak Width
Histogram trough statistics are calculated in a similar way. The only difference is that histogram
minimum instead of histogram maximum is analyzed.
Page 63
Page 66

2.12. Crosscorrelograms
Crosscorrelogram shows the conditional probability of a spike at time t0+t on the condition that there
is a reference event at time t0.
Compared to PSTH (that calculates the same probabilities), crosscorrelogram allows to use shiftpredictors.
Parameters
Parameter Description
Reference Type Specifies if the analysis will use a single or multiple reference events.
Reference Specifies a reference neuron or event (or a group of reference neurons or
events).
XMin Histogram time axis minimum in seconds.
XMax Histogram time axis maximum in seconds
Bin Bin size in seconds.
Normalization Histogram units (Counts/Bin, Probability, Spikes/Second or Z-score). See
Algorithm below.
No Selfcount An option not to count reference events if the target event is the same as
the reference event (prevents a histogram to have a huge peak at zero
when calculating crosscorrelogram versus itself).
Select Data If Select Data is From Time Range, only the data from the specified (by
Select Data From and Select Data To parameters) time range will be used
in analysis. See also Data Selection Options.
Select Data From Start of the time range in seconds.
Select Data To End of the time range in seconds.
Int. filter type Specifies if the analysis will use a single or multiple interval filters.
Interval filter Specifies the interval filter(s) that will be used to preselect data before
analysis. See also Data Selection Options.
Count bins in filter An option to normalize each bin individually if the interval filter is used. See
Algorithm section below for detailed discussion.
Create filter on-the-fly Specifies if a temporary interval filter needs to be created (and used to
preselect data).
Create filter around Specifies an event that will be used to create a temporary filter.
Start offset Offset (in seconds, relative to the event specified in Create filter around
parameter) for the start of interval for the temporary filter.
End offset Offset (in seconds, relative to the event specified in Create filter around
parameter) for the end of interval for the temporary filter.
Fix overlaps An option to automatically merge the overlapping intervals in the temporary
filter.
Overlay Graphs An option to draw several histograms in each graph. This option requires
that Int. filter type specifies that multiple interval filters will be used (either
Table (row) or Table (col)).
Overlay Options Specifies line colors and line styles for overlaid graphs.
Page 64
Page 67

Smooth Option to smooth the histogram after the calculation. See Post-Processing
Options for details.
Smooth Filter Width The width of the smooth filter. See Post-Processing Options for details.
Shift-predictor An option to use shift-predictor. Shift-predictor requires that an interval
filter is used in the analysis. See Using Shift-Predictor for details.
Shift times How many times to calculate shift predictor. See Using Shift-Predictor for
details.
Subtract predictor An option to subtract shift-predictor from crosscorrelogram. See Using
Shift-Predictor for details.
Predictor line style Specifies the line used to draw shift-predictor (if subtract predictor is not
selected).
Draw confidence limits An option to draw the confidence limits.
Confidence (%) Confidence level (percent). See Confidence Limits for details.
Conf. mean calculation An option that specifies how the mean firing rate (that is used in the
derivation of the confidence limits) is calculated.
There are 3 options: Use data selection, Use all file and Use pre-ref data.
See Confidence Limits for details.
Note that Use pre-ref data option can only be used for a stimulation type
data, i.e. when the distance between any two consecutive reference
events is larger than XMax-XMin. See Confidence Limits for details.
Conf. display An option to draw confidence limits either as horizontal lines or as a
colored background.
Conf. line style Line style for drawing confidence limits (used when Conf. display is Lines ).
Conf. background color Background color for drawing confidence limits (used when Conf. display is
Colored Background ).
Draw mean freq. An option to draw a horizontal line representing the expected histogram
value for a Poisson spike train. See Confidence Limits for details.
Mean line style Line style for drawing mean frequency.
Draw Cusum An option to draw a cumulative sum graph above the histogram. See
Cumulative Sum Graphs for details.
Add to Results / Bin left An option to add an additional vector (containing a left edge of each bin) to
the matrix of numerical results.
Add to Results / Bin
middle
Add to Results / Bin
right
Background An option on how to calculate the histogram background for the peak and
An option to add an additional vector (containing a middle point of each
bin) to the matrix of numerical results.
An option to add an additional vector (containing a right edge of each bin)
to the matrix of numerical results.
through analysis. See Peak and Trough Statistics below.
Peak width Peak width (the number of bins in peak) in the peak and through analysis.
See Peak and Trough Statistics below.
Left shoulder Specifies the left shoulder value (in seconds) in the peak and through
analysis. See Peak and Trough Statistics below.
Right shoulder Specifies the right shoulder value (in seconds) in the peak and through
analysis. See Peak and Trough Statistics below.
Page 65
Page 68

Send to Matlab An option to send the matrix of numerical results to Matlab. See also
Matlab Options.
Matrix Name Specifies the name of the results matrix in Matlab workspace.
Matlab command Specifies a Matlab command that is executed after the numerical results
are sent to Matlab.
Send to Excel An option to send numerical results or summary of numerical results to
Excel. See also Excel Options.
Sheet Name The name of the worksheet in Excel where to copy the numerical results.
TopLeft Specifies the Excel cell where the results are copied. Should be in the form
CR where C is Excel column name, R is the row number. For example, A1
is the top-left cell in the worksheet.
Summary of Numerical Results
The following information is available in the Summary of Numerical Results
Column Description
Variable Variable name.
Reference Reference variable name.
NumRefEvents The number of reference events used in calculation.
YMin Histogram minimum.
YMax Histogram maximum.
Spikes The number of spikes used in calculation.
Filter Length The length of all the intervals of the interval filter (if a filter was used) or the
length or the recording session (in seconds).
Mean Freq. Mean firing rate (Spikes/Filter_Length).
Mean Hist. The mean of the histogram bin values.
St. Dev. Hist. The standard deviation of the histogram bin values.
St. Err. Mean. Hist. The standard error of mean of the histogram bin values.
Conf. Low Lower confidence level.
Conf. High Upper confidence level.
Mean Expected mean value of the histogram. If Z-score normalization is
specified, this value is zero and the expected mean value of the histogram
with Counts/Bin normalization is shown in Z-score Mean.
Norm. Factor Normalization factor. Bin counts are divided by this value. See
Normalization in Algorithm below.
Z-score mean Expected mean value of the histogram before Z-score normalization (in
other words, confidence mean). See Confidence Limits for details .
Mean Before Ref. Mean of the histogram before the reference event (i.e. for all the bins
before zero on time axis).
Bins Before Ref. The number of bins before the bin that contains zero on time axis.
Zero Bin The index of the bin that contains zero on time axis.
Page 66
Page 69

Background Mean The mean of the histogram background for the peak and trough analysis.
See Peak and Through Statistics below.
Background Stdev The standard deviation of the histogram background for the peak and
trough analysis. See Peak and Trough Statistics below.
Peak Z-score Peak Z-score. See Peak and Trough Statistics below.
Peak/Mean Histogram peak value divided by the background mean value.
Peak Position Peak position (in seconds).
Peak Half Height The Y value of the half height of the peak, i.e.
histogram_background_mean + (peak-mean)/2.
Peak Width at Half
Peak width at the peak half height level (in seconds).
Height
Trough Z-score Trough Z-score. See Peak and Trough Statistics below.
Trough/Mean Histogram trough value divided by the background mean value.
Trough Position Trough position (in seconds).
Trough Half Height The Y value of the half height of the trough, i.e.
histogram_background_mean + (trough-mean)/2.
Trough Width at Half
Trough width at the trough half height level (in seconds).
Height
Algorithm
Crosscorrelogram shows the conditional probability of a spike at time t0+t on the condition that there
is a reference event at time t0.
The time axis is divided into bins. The first bin is [XMin, XMin+Bin). The second bin is [XMin+Bin,
XMin+Bin*2), etc. The left end is included in each bin, the right end is excluded from the bin.
Let ref[i] be the array of timestamps of the reference event,
ts[i] be the spike train (each ts is the timestamp).
For each timestamp ref[k]:
1) calculate the distances from this event (or spike) to all the spikes in the spike train:
d[i] = ts[i] - ref[k]
2) for each i:
if d[i] is inside the first bin, increment the bin counter for the first bin:
if d[i] >= XMin and d[i] < XMin + Bin
then bincount[1] = bincount[1] +1
if d[i] is inside the second bin, increment the bin counter for the second bin:
if d[i] >= XMin+Bin and d[i] < XMin + Bin*2
then bincount[2] = bincount[2] +1
and so on... .
If Normalization is Counts/Bin, no further calculations are performed.
If Normalization is Probability, bin counts are divided by the number of reference events.
Note that the Probability normalization makes sense only for small values of Bin. For Probability
normalization to be valid (so that the values of probability are between 0 and 1), there should be no
more than one spike in each bin. For example, if the Bin value is large and for each ref[k] above there
are many d[i] values such that d[i] >= XMin and d[i] < XMin + Bin, the bin count for the first bin can
Page 67
Page 70

exceed the number of reference events. Then, the probability value
(bincount[1]/number_of_reference_events) could be larger than 1.
If Normalization is Spikes/Sec, bin counts are divided by NumRefEvents*Bin, where
NumRefEvents is the number of reference events.
If Normalization is Z-score, bin_value = (bin_count - Confidence_mean)/sqrt(Confidence_mean),
where Confidence_mean is the expected mean bin count calculated according to Conf. mean
calculation parameter. Please note that bin counts are assumed to have Poisson distribution
(therefore, standard deviation is equal to square root of expected mean) and Z-score can be
considered to have Normal distribution only for large values (more than 10) of the Confidence_mean.
If the option Count Bins In Filter is selected, for normalization Spikes/Second, NeuroExplorer will
divide bin count by NumTimesBinWasInFilter*Bin instead of NumRefEvents*Bin. The problem is
that when the interval filter is used, bins close to XMin and to XMax may often (when a reference
event is close to the beginning or to the end of the interval in the interval filter) be positioned outside
the filter and therefore will not be used for many reference events. Hence, the bins close to 0.0 will be
used in analysis more often than the bins close to XMin and XMax. If the option Count Bins In Filter
is selected, NeuroExplorer will count the number of times each bin was used in the calculation and
use this count, NumTimesBinWasInFilter, (instead of the number of reference events) to normalize
the histogram.
Peak and Trough Statistics
NeuroExplorer calculates histogram peak statistics the following way:
•
Maximum of the histogram is found
•
If the histogram contains several maxima with the same value, peak statistics are not calculated
•
Otherwise, the center of the bin, where the histogram reaches maximum, is shown as Peak
Position in the Summary of Numerical results
•
The mean M and standard deviation S of the bin values of the histogram background are
calculated: If Background parameter is set as Bins outside peak/trough, bins outside peak and
trough (i.e., bins that are more than PeakWidth/2 away from the bin with the histogram
maximum and the bin with the histogram minimum) are used to calculate M and S If Background
parameter is set as Shoulders, bins that are to the left of Left Shoulder or to the right of Right
Shoulder parameters are used to calculate M and S
•
The value M (mean of the background bin values) is shown as Background Mean in the
Summary of Numerical results
•
The value S (standard deviation of the background bin values) is shown as Background Stdev
in the Summary of Numerical results
•
The value (HistogramMaximum – M)/S is shown as Peak Z-score
•
The value (HistogramMaximum + M)/2 is shown as Peak Half Height
•
Histogram intersects a horizontal line drawn at Peak Half Height at time points TLeft and TRight.
(TRight - TLeft) is shown as Peak Width
Histogram trough statistics are calculated in a similar way. The only difference is that histogram
minimum instead of histogram maximum is analyzed.
Page 68
Page 71

2.13. Shift-Predictor for Crosscorrelograms
Shift-predictor is defined for a series of trials - you take the spikes of one neuron in trial 1 and
correlate them with the spikes of another neuron in trial 2, etc.
These "trials" are represented in NeuroExplorer as time intervals, i.e. pairs of numbers:
start of interval 1, end of interval 1
start of interval 2, end of interval 2, etc.
To use the shift-predictor, you need to create those "trials" first.
You need an external event that is fired at the beginning of each trial. Suppose Event03 is the event
that happens at the beginning of each trial and each trial lasts 20 seconds.
To create the trial intervals:
•
click on the New Events! button (or select Edit | Operations on Variables menu command)
•
in the operations list, select MakeIntervals
•
in the First Operand, select the external event Event03
•
set Shift Min to 0. and Shift Max to 20.0
•
in the New Var Name, type: Trials
•
press Run the Operation button
•
close the dialog.
Now, click on the Crosscorrelogram button, select Shift-predictor page and select Trials as an interval
filter and specify other shift-predictor parameters.
Shift-Predictor Algorithm
Suppose we have n trial intervals:
[t1a, t1b], [t2a, t2b], ..., [tna, tnb]
and we have 2 spike trains:
reference spike train with N timestamps r1, r2, ... , rN;
and target spike train with M timestamps s1, s2, ... , sM.
Shift-predictor for shift = 1 is calculated like this:
Step 1: find all the timestamps ri that are inside the first trial interval [t1a, t1b]
Step 2: find all the timestamps sj that are inside the second trial interval [t2a, t2b]
Step 3: shift all the timestamps ri so that thay align with the second trial:
calculate new timestamps pi = ri + (t2a-t1a)
Step 4: calculate a crosscorrelogram between pi and sj
Repeat Steps 1 through 4 with interval [t2a, t2b] in Step 1 and interval [t3a, t3b] in Step 2.
...
Repeat Steps 1 through 4 with interval [tna, tnb] in Step 1 and interval [t1a, t1b] in Step 2. Note that
we wrap around the trials: the last trial is correlated with the first one.
Page 69
Page 72

Then, calculate shift-predictors for shift = 2:
Repeat Steps 1 through 4 with interval [t1a, t1b] in Step 1 and interval [t3a, t3b] in Step 2. That is,
shift trials by 2.
...
And, finally, calculate shift-predictors for shift = Number_of_shifts specified in the shift-predictor
options.
More on Time Intervals
These time intervals can be used in other analyses in NeuroExplorer to analyze spikes only from
those intervals.
For example, you may have a pre-drug period in the experiment (say, from 0 to 600 seconds) and
want to analyze the spikes from this pre-drug time period.
To do this, you create a new "interval variable" (let's call it "PreDrug") that contains only one interval
(0., 600.) (press "New Intervals!" button to create a new interval variable).
Then you can calculate, for example, the autocorrelograms for the spikes in the pre-drug period by
specifying PreDrug as an "interval filter" in the Data Selection page of the Autocorrelogram
parameters dialog.
Page 70
Page 73

2.14. Rasters
The raster display shows the spikes as vertical lines (tick) positioned according to the spikes
timestamps. This analysis does not generate numerical results since the raster lines (ticks) are drawn
directly from the data. Some generic analysis statistics are available in the Summary of Numerical
Results (see below).
Parameters
Parameter Description
XMin Time axis minimum in seconds.
XMax Time axis maximum in seconds
Select Data If Select Data is From Time Range, only the data from the specified (by
Select Data From and Select Data To parameters) time range will be used
in analysis. See also Data Selection Options.
Select Data From Start of the time range in seconds.
Select Data To End of the time range in seconds.
Interval filter Specifies the interval filter(s) that will be used to preselect data before
analysis. See also Data Selection Options.
Vert. size The size of a raster tick (in percent of the graph height).
Summary of Numerical Results
The following information is available in the Summary of Numerical Results
Column Description
Variable Variable name.
YMin Y axis minimum (added for compatibility with other analyses, always 0).
YMax Y axis maximum (added for compatibility with other analyses, always 1).
Spikes The number of spikes used in calculation.
Filter Length The length of all the intervals of the interval filter (if a filter was used) or the
length or the recording session (in seconds).
Mean Freq. Mean firing rate (Spikes/Filter_Length).
Algorithm
The raster display shows the spikes as vertical lines positioned according to the spikes timestamps.
Page 71
Page 74

2.15. Perievent Rasters
This analysis shows the timestamps of the selected variable relative to the timestamps of the
reference variable.
Parameters
Parameter Description
Reference Type Specifies if the analysis will use a single or multiple reference events.
Reference Specifies a reference neuron or event (or a group of reference neurons or
events).
XMin Time axis minimum in seconds.
XMax Time axis maximum in seconds
Markers An option to display additional events (as markers - triangles, squares,
etc.) in the perievent raster.
Background Intervals An option to draw intervals in the raster background.
Trial Order This option allows you to specify how the raster lines are drawn. The raster
lines (corresponding to reference events) can be drawn in the order of
reference events (with the first event on top, the last at the bottom of the
perievent raster), or in the reverse order (the first event at the bottom of the
raster display, the last event at the top). This option is ignored if Sort trials
parameter is not None.
Sort trials An option to sort raster lines.
By default, the raster lines (corresponding to reference events) are drawn
in the order specified by the Trial Order parameter. This option allows you
to display reference events in a different order. For example, in behavioral
experiments, you can sort the trials according to the latency of the
response.
Sort event Specifies how to identify the timestamps (in Sort ref. variable) that will be
used in sorting.
Sort ref. Specifies event variable that will be used to sort trials.
Histogram An option to draw a histogram.
Bin Histogram bin size in seconds.
Normalization Histogram units (Counts/Bin, Probability or Spikes/Second). See Algorithm
below.
No Selfcount An option not to count reference events if the target event is the same as
the reference event (prevents a histogram to have a huge peak at zero
when calculating PSTH versus itself).
Select Data If Select Data is From Time Range, only the data from the specified (by
Select Data From and Select Data To parameters) time range will be used
in analysis. See also Data Selection Options.
Select Data From Start of the time range in seconds.
Select Data To End of the time range in seconds.
Int. filter type Specifies if the analysis will use a single or multiple interval filters.
Interval filter Specifies the interval filter(s) that will be used to preselect data before
analysis. See also Data Selection Options.
Page 72
Page 75

Create filter on-the-fly Specifies if a temporary interval filter needs to be created (and used to
preselect data).
Create filter around Specifies an event that will be used to create a temporary filter.
Start offset Offset (in seconds, relative to the event specified in Create filter around
parameter) for the start of interval for the temporary filter.
End offset Offset (in seconds, relative to the event specified in Create filter around
parameter) for the end of interval for the temporary filter.
Fix overlaps An option to automatically merge the overlapping intervals in the temporary
filter.
Smooth histogram An option to smooth the histogram after the calculation. See Post-
Processing Options for details.
Smooth Filter Width The width of the smooth filter. See Post-Processing Options for details.
Summary of Numerical Results
The following information is available in the Summary of Numerical Results
Column Description
Variable Variable name.
Reference Reference variable name.
NumRefEvents The number of reference events used in calculation.
YMin Histogram minimum.
YMax Histogram maximum.
Spikes The number of spikes used in calculation.
Filter Length The length of all the intervals of the interval filter (if a filter was used) or the
length or the recording session (in seconds).
Mean Freq. Mean firing rate (Spikes/Filter_Length).
Mean Hist. The mean of the histogram bin values.
St. Dev. Hist. The standard deviation of the histogram bin values.
St. Err. Mean. Hist. The standard error of mean of the histogram bin values.
Algorithm
For the perievent histogram calculation, see Perievent Histograms.
Let ref[i] be the array of timestamps of the reference event,
sortref[i] be the array of timestamps of the sort event.
If Sort Trials is selected, the following algorithm is used:
1) for each timestamp ref[k], NeuroExplorer finds the smallest sortref[i], such that
sortref[i] > ref[k]
2) the distance between two events is calculated:
dist[k] = sortref[i] - ref[k]
3) the trials ref[k] are sorted array sorted in ascending or descending order of dist[k].
Page 73
Page 76

Continuous variables can also be used in this analysis.
For each ref[k], NeuroExplorer calculates a series of bins. The first bin is:
[ref[k]+XMin, ref[k]+XMin+Bin]
the second bin is [ref[k]+XMin+Bin, ref[k]+XMin+Bin+Bin], etc.
Then, the average value of a continuous variable is calculated within each of the bins and this
average value is displayed using the color scale. If bin does not contain any timestamps of the
continuous variable, the previous value of the continuous variable is used.
Page 74
Page 77

2.16. Joint PSTH
Joint PSTH matrix shows the correlations of the two neurons around the reference events.
Parameters
Parameter Description
XMin Time axis minimum in seconds.
XMax Time axis maximum in seconds
Bin Bin size in seconds.
Reference Specifies a reference neuron or event.
Bottom Neuron The neuron of event shown along the horizontal axis (the vertical axis
shows a neuron selected for analysis).
Diag The width (in seconds) of the area in the scatter matrix around its main
diagonal used to calculate the main diagonal histogram.
Normalization Scatter matrix normalization.
Select Data If Select Data is From Time Range, only the data from the specified (by
Select Data From and Select Data To parameters) time range will be used
in analysis. See also Data Selection Options.
Select Data From Start of the time range in seconds.
Select Data To End of the time range in seconds.
Int. filter type Specifies if the analysis will use a single or multiple interval filters.
Interval filter Specifies the interval filter(s) that will be used to preselect data before
analysis. See also Data Selection Options.
Create filter on-the-fly Specifies if a temporary interval filter needs to be created (and used to
preselect data).
Create filter around Specifies an event that will be used to create a temporary filter.
Start offset Offset (in seconds, relative to the event specified in Create filter around
parameter) for the start of interval for the temporary filter.
End offset Offset (in seconds, relative to the event specified in Create filter around
parameter) for the end of interval for the temporary filter.
Fix overlaps An option to automatically merge the overlapping intervals in the temporary
filter.
Smooth Option to smooth the histograms (not the scatter matrix) after the
calculation. See Post-Processing Options for details.
Smooth Filter Width The width of the smooth filter. See Post-Processing Options for details.
Send to Matlab An option to send the matrix of numerical results to Matlab. See also
Matlab Options.
Matrix Name Specifies the name of the results matrix in Matlab workspace.
Matlab command Specifies a Matlab command that is executed after the numerical results
are sent to Matlab.
Send to Excel An option to send numerical results or summary of numerical results to
Page 75
Page 78

Excel. See also Excel Options.
Sheet Name The name of the worksheet in Excel where to copy the numerical results.
TopLeft Specifies the Excel cell where the results are copied. Should be in the form
CR where C is Excel column name, R is the row number. For example, A1
is the top-left cell in the worksheet.
Matrix Scale An option on how to draw the scatter matrix (color or black and white).
Font size (% matrix) Font size in percent of scatter matrix height.
Summary of Numerical Results
The following information is available in the Summary of Numerical Results
Column Description
Variable Variable name.
YMin Y axis minimum (added for compatibility with other analyses, always 0).
YMax Y axis maximum (added for compatibility with other analyses, always 1).
Spikes The number of spikes of the neuron shown on the vertical axis used in
calculation.
Filter Length The length of all the intervals of the interval filter (if a filter was used) or the
length or the recording session (in seconds).
Mean Freq. Mean firing rate (Spikes/Filter_Length).
Hist. norm. Normalization factor. Histogram bin counts are divided by this value.
Color scale minimum Scatter matrix color scale minimum.
Color scale maximum Scatter matrix color scale maximum.
Algorithm
The main Joint PSTH matrix shows the correlations of the bin counts of two neurons around the
reference events. Histograms to the left of and below the matrix are standard perievent histograms for
two specified neurons. The first histogram to the right of the matrix shows the correlations of nearcoincident spikes around the reference events. The far-right histogram shows correlations of firings of
two neurons around reference events.
See the following paper for details on Joint PSTH analysis:
M. H. J. Aertsen, G. L. Gerstein, M. K. Habib and G. Palm. Dynamics of Neuronal Firing Correlation:
Modulation of "Effective Connectivity" J. Neurophysiol., Vol. 61, pp. 900-917, 1989.
Page 76
Page 79

2.17. Cumulative Activity Graphs
The cumulative activity display shows a stepwise function which makes a jump at the moment of
spike and stays constant between the spikes.
This analysis does not generate numerical results since the stepwise function is drawn directly from
the data. Some generic analysis statistics are available in the Summary of Numerical Results (see
below).
Parameters
Parameter Description
XMin Time axis minimum in seconds.
XMax Time axis maximum in seconds
Summary of Numerical Results
The following information is available in the Summary of Numerical Results
Column Description
Variable Variable name.
YMin Y axis minimum.
YMax Y axis maximum.
Spikes The number of spikes used in calculation.
Mean Freq. Mean firing rate.
Algorithm
The cumulative activity display shows the stepwise function, which makes a jump at the moment of
spike and stays constant between the spikes.
Page 77
Page 80

2.18. Instant Frequency
The instant frequency display shows the instantaneous frequency of the spike train (at the end of
each interspike interval, the inverted interspike interval is drawn as a vertical line).
This analysis does not generate numerical results since the graph is drawn directly from the data.
Some generic analysis statistics are available in the Summary of Numerical Results (see below).
Parameters
Parameter Description
XMin Time axis minimum in seconds.
XMax Time axis maximum in seconds
Graph Style An option to draw vertical lines or points.
Summary of Numerical Results
The following information is available in the Summary of Numerical Results
Column Description
Variable Variable name.
YMin Y axis minimum.
YMax Y axis maximum.
Spikes The number of spikes used in calculation.
Mean Freq. Mean firing rate.
Algorithm
The instant frequency display shows the instantaneous frequency of the spike train.
For each spike that occurred at time t[i] it draws the vertical line
from point ( t[i], 0.) to point (t[i], 1/(t[i] - t[i-1])),
where t[i-1] is the time of the preceding spike.
In other words, at the end of each interspike interval, the inverted interspike interval is drawn as a
vertical line.
Page 78
Page 81

2.19. Interspike Intervals vs. Time
The intervals vs. time graph displays interspike intervals against time (at the end of each interspike
interval, the point is drawn with the Y coordinate equal to the interspike interval).
This analysis does not generate numerical results since the graph is drawn directly from the data.
Some generic analysis statistics are available in the Summary of Numerical Results (see below).
Parameters
Parameter Description
XMin Time axis minimum in seconds.
XMax Time axis maximum in seconds
Summary of Numerical Results
The following information is available in the Summary of Numerical Results
Column Description
Variable Variable name.
YMin Y axis minimum.
YMax Y axis maximum.
Spikes The number of spikes used in calculation.
Mean Freq. Mean firing rate.
Algorithm
The intervals vs. time graph displays interspike intervals against time.
For each spike that occurred at time t[i] it draws the point with the coordinates
(t[i], t[i] - t[i-1]),
where t[i-1] is the time of the preceding spike.
In other words, at the end of each interspike interval, the point is drawn with the Y coordinate equal to
the interspike interval.
Page 79
Page 82

2.20. Poincare Maps
For each spike, a point is displayed where the X coordinate of the point is the current interspike
interval and Y coordinate of the point is the preceding interspike interval.
This analysis does not generate numerical results since the graph is drawn directly from the data.
Some generic analysis statistics are available in the Summary of Numerical Results (see below).
Parameters
Parameter Description
Min Interval Time axis minimum in seconds.
Max Interval Time axis maximum in seconds
Log Axis Scale An option to use Log10 axis scales.
Select Data If Select Data is From Time Range, only the data from the specified (by
Select Data From and Select Data To parameters) time range will be used
in analysis. See also Data Selection Options.
Select Data From Start of the time range in seconds.
Select Data To End of the time range in seconds.
Int. filter type Specifies if the analysis will use a single or multiple interval filters.
Interval filter Specifies the interval filter(s) that will be used to preselect data before
analysis. See also Data Selection Options.
Summary of Numerical Results
The following information is available in the Summary of Numerical Results
Column Description
Variable Variable name.
YMin Y axis minimum.
YMax Y axis maximum.
Spikes The number of spikes used in calculation.
Mean Freq. Mean firing rate.
Algorithm
For each spike that occurred at time t[i], Poincare plot shows the point with the coordinates
(t[i] - t[i-1], t[i-1] - t[i-2]).
That is, the X coordinate of the point is the current interspike interval and Y coordinate of the point is
the preceding interspike interval.
Page 80
Page 83

2.21. Synchrony vs. Time
For each spike, this graph shows the distance from this spike to the closest spike (timestamp) in the
reference event.
This analysis does not generate numerical results since the graph is drawn directly from the data.
Some generic analysis statistics are available in the Summary of Numerical Results (see below).
This analysis is useful in identifying the moments in time when several neurons are in sync with the
reference neuron. To do this, you need to select Inverted Distance. Then, high values will indicate
spikes that are close to each other. Also, it is useful to specify that the graphs are shown in 1 column
(in Num. Columns in Properties panel).
The red arrow in the picture below shows a moment in time where several neurons have almostsynchronous spikes with the reference.
Parameters
Parameter Description
XMin Time axis minimum in seconds.
XMax Time axis maximum in seconds
Reference Reference event.
Distance An option to draw linear or inverted distance.
Summary of Numerical Results
The following information is available in the Summary of Numerical Results
Column Description
Variable Variable name.
Page 81
Page 84

YMin Y axis minimum.
YMax Y axis maximum.
Spikes The number of spikes used in calculation.
Mean Freq. Mean firing rate.
Algorithm
For each spike that occurred at time t[i], this graph shows the distance from this spike to the closest
spike (timestamp) in the reference event:
dist = min(abs(t[i] - ref[j])),
where ref[j] is a timestamp of the reference event
If Distance is Linear, the vertical line is drawn
from point ( t[i], 0.) to point (t[i], dist).
If Distance is Inverted, the vertical line is drawn
from point ( t[i], 0.) to point (t[i], 1/dist).
Page 82
Page 85

2.22. Trial Bin Counts
Trial bin counts analysis computationally is essentially the same as the perievent histogram. The
difference is that the bin counts are saved for each reference event.
Parameters
Parameter Description
Reference Specifies a reference neuron or event.
XMin Histogram time axis minimum in seconds.
XMax Histogram time axis maximum in seconds
Bin Bin size in seconds.
Normalization Histogram units (Counts/Bin, Probability or Spikes/Second). See Algorithm
below.
No Selfcount An option not to count reference events if the target event is the same as
the reference event (prevents a histogram to have a huge peak at zero
when calculating PSTH versus itself).
Select Data If Select Data is From Time Range, only the data from the specified (by
Select Data From and Select Data To parameters) time range will be used
in analysis. See also Data Selection Options.
Select Data From Start of the time range in seconds.
Select Data To End of the time range in seconds.
Interval filter Specifies the interval filter(s) that will be used to preselect data before
analysis. See also Data Selection Options.
Send to Matlab An option to send the matrix of numerical results to Matlab. See also
Matlab Options.
Matrix Name Specifies the name of the results matrix in Matlab workspace.
Matlab command Specifies a Matlab command that is executed after the numerical results
are sent to Matlab.
Send to Excel An option to send numerical results or summary of numerical results to
Excel. See also Excel Options.
Sheet Name The name of the worksheet in Excel where to copy the numerical results.
TopLeft Specifies the Excel cell where the results are copied. Should be in the form
CR where C is Excel column name, R is the row number. For example, A1
is the top-left cell in the worksheet.
Summary of Numerical Results
The following information is available in the Summary of Numerical Results
Column Description
Variable Variable name.
Page 83
Page 86

Color Scale Min Color scale minimum.
Color Scale Max Color scale maximum.
Reference Reverence event.
NumRefEvents Number of reference events.
Bin001Mean Mean of all the values for Bin 1.
Bin001SdDev Standard deviation of all the values for Bin 1.
Algorithm
Trial bin counts analysis computationally is essentially the same as the perievent histogram. The
difference is that the bin counts are saved for each reference event.
The time axis is divided into bins. The first bin is [XMin, XMin+Bin). The second bin is [XMin+Bin,
Xmin+Bin*2), etc. The left end is included in each bin, the right end is excluded from the bin.
Let ref[i] be the array of timestamps of the reference event,
ts[i] be the spike train (each ts is the timestamp)
For each timestamp ref[k]:
Set all bin counts to zero:
bincount[i] = 0 for all i
Calculate the distances from this event (or spike) to all the spikes in the spike train:
d[i] = ts[i] - ref[k]
for each i:
if d[i] is inside the first bin, increment the bin counter for the first bin:
if d[i] >= XMin and d[i] < XMin + Bin
then bincount[1] = bincount[1] +1
if d[i] is inside the second bin, increment the bin counter for the second bin:
if d[i] >= XMin+Bin and d[i] < XMin + Bin*2
then bincount[2] = bincount[2] +1
and so on... .
If Normalization is Counts/Bin, no further calculations are performed.
If Normalization is Spikes/Sec, bin counts are divided by Bin.
Page 84
Page 87

2.23. Power Spectral Densities
This analysis captures the frequency content of continuous variables or neuronal rate histograms.
Parameters
Parameter Description
Max Frequency Maximum frequency of the spectrum in Hz
Number of values Number of values in the spectrum.
Show Frequency From Minimum frequency to be shown in the spectrum graph.
Show Frequency To Maximum frequency to be shown in the spectrum graph.
Normalization Spectrum normalization. See Algorithm below.
Log Frequency Scale An option show Log10 frequency scale.
Smooth Option to smooth the spectrum after the calculation. See Post-Processing
Options for details.
Smooth Filter Width The width of the smooth filter. See Post-Processing Options for details.
Select Data If Select Data is From Time Range, only the data from the specified (by
Select Data From and Select Data To parameters) time range will be used
in analysis. See also Data Selection Options.
Select Data From Start of the time range in seconds.
Select Data To End of the time range in seconds.
Int. filter type Specifies if the analysis will use a single or multiple interval filters.
Interval filter Specifies the interval filter(s) that will be used to preselect data before
analysis. See also Data Selection Options.
Create filter on-the-fly Specifies if a temporary interval filter needs to be created (and used to
preselect data).
Create filter around Specifies an event that will be used to create a temporary filter.
Start offset Offset (in seconds, relative to the event specified in Create filter around
parameter) for the start of interval for the temporary filter.
End offset Offset (in seconds, relative to the event specified in Create filter around
parameter) for the end of interval for the temporary filter.
Fix overlaps An option to automatically merge the overlapping intervals in the temporary
filter.
Overlay Graphs An option to draw several histograms in each graph. This option requires
that Int. filter type specifies that multiple interval filters will be used (either
Table (row) or Table (col)).
Overlay Options Specifies line colors and line styles for overlaid graphs.
Add to Results / Bin left An option to add an additional vector (containing a left edge of each bin) to
the matrix of numerical results.
Add to Results / Bin
middle
Add to Results / Bin An option to add an additional vector (containing a right edge of each bin)
An option to add an additional vector (containing a middle point of each
bin) to the matrix of numerical results.
Page 85
Page 88

right to the matrix of numerical results.
Send to Matlab An option to send the matrix of numerical results to Matlab. See also
Matlab Options.
Matrix Name Specifies the name of the results matrix in Matlab workspace.
Matlab command Specifies a Matlab command that is executed after the numerical results
are sent to Matlab.
Send to Excel An option to send numerical results or summary of numerical results to
Excel. See also Excel Options.
Sheet Name The name of the worksheet in Excel where to copy the numerical results.
TopLeft Specifies the Excel cell where the results are copied. Should be in the form
CR where C is Excel column name, R is the row number. For example, A1
is the top-left cell in the worksheet.
Summary of Numerical Results
The following information is available in the Summary of Numerical Results
Column Description
Variable Variable name.
YMin Spectrum minimum.
YMax Spectrum maximum.
Spikes The number of spikes used in spectrum calculation.
Filter Length The length of all the intervals of the interval filter (if a filter was used) or the
length or the recording session (in seconds).
Mean Freq. Mean firing rate (Spikes/Filter_Length).
First Minimum
Frequency Position
First Maximum
Frequency Position
The position of the minimum of the displayed spectrum (in Hz). If there are
multiple bins in the spectrum where the spectrum value equals to the
spectrum minimum, this value represents the frequency of the first such
bin.
The position of the maximum of the displayed spectrum (in Hz). If there are
multiple bins in the spectrum where the spectrum value equals to the
spectrum maximum, this value represents the frequency of the first such
bin.
Algorithm
Spectral Densities for Spike Trains and Events
NeuroExplorer uses rate histograms to estimate the power spectra of the spike trains. The
parameters of rate histograms are calculated using the following formulas:
Bin = 1./(2.* Maximum_Frequency )
Number_of_Bins = 2* Number_of_Frequency_Values
The time axis is divided into intervals of length Bin*NumberOfBins. Then the power spectrum for each
interval is calculated. The final raw power spectrum is the average of all the spectra for the separate
Page 86
Page 89

intervals.
For each interval:
•
The rate histogram is calculated.
•
The histogram is detrended (its linear regression is subtracted)
•
The Hann window w[i] = (1 - cos(2*PI*i/(NumBer_Of_Bins-1)))/2
is applied to the histogram.
•
FFT of the result is calculated.
•
Raw power spectrum is formed from the FFT.
Spectral Densities for Continuous Variables
If Maximum_Frequency = M and Number_of_Frequency_Values = N, we need to calculate the
spectrum for frequency values 0, M/N, 2*M/N, ..., M.
For a continuous variable with digitizing frequency F, a standard power spectrum with K values in the
FFT transform would produce the spectrum values for frequencies 0, F/K, 2*F/K, ..., F/2.
If a continuous variable digitizing frequency F equals to Maximum_Frequency*2, we can set K =
Number_of_Frequency_Values*2 and calculate the standard power spectrum to get the spectrum
values for the desired frequencies. That is, in the algorithm described above, instead of rate
histogram we simply use continuous variable values in milliVolts.
If a continuous variable digitizing frequency is not equal to Maximum_Frequency*2, we cannot
directly calculate the spectrum for the specified frequencies. Instead, we find a value K such that the
frequency step for continuous variable spectrum is less than the specified step (i.e. F/K < M/N, so that
we should get a more detailed spectrum than specified) and calculate the spectrum for this value of
K. Then, we resample the resulting spectrum to get the spectrum values for the specified frequencies.
Normalization
If Normalization is Raw PSD, the power spectrum is normalized so that the sum of all the spectrum
values equals to the mean squared value of the rate histogram.
If Normalization is % of Total PSD, the power spectrum is normalized so that the sum of all the
spectrum values equals 100.
If Normalization is Log of PSD, the power spectrum is calculated using the formula:
power_spectrum[i] = 10.*log10(raw_spectrum[i])
Page 87
Page 90

2.24. Burst Analysis
This analysis identifies bursts in spike trains. For each spike train (neuron variable), a new interval
variable is created that contains the time intervals corresponding to bursts.
Parameters
Parameter Description
Algorithm Selection of MaxInterval or Surprise algorithms.
Max Interval Maximum interspike interval to start the burst (in seconds, MaxInterval
method).
Max End Interval Maximum interspike interval to end the burst (in seconds, MaxInterval
method).
Min Interburst Interval Minimum interval between bursts (in seconds, MaxInterval method).
Min Burst Duration Minimum burst duration (in seconds, MaxInterval method)
Min Number of Spikes Minimum number of spikes in the burst (MaxInterval method).
Min Surprise Minimum surprise of the burst (Surprise method).
Rate Histogram Bin The value of the bin for the burst rate histogram (in seconds).
Rate Histogram Units Histogram units (Counts/Bin, Probability or Spikes/Second). See Rate
Histograms for details.
Select Data If Select Data is From Time Range, only the data from the specified (by
Select Data From and Select Data To parameters) time range will be used
in analysis. See also Data Selection Options.
Select Data From Start of the time range in seconds.
Select Data To End of the time range in seconds.
Interval filter Specifies the interval filter(s) that will be used to preselect data before
analysis. See also Data Selection Options.
Send to Excel An option to send numerical results or summary of numerical results to
Excel. See also Excel Options.
Sheet Name The name of the worksheet in Excel where to copy the numerical results.
TopLeft Specifies the Excel cell where the results are copied. Should be in the form
CR where C is Excel column name, R is the row number. For example, A1
is the top-left cell in the worksheet.
Page 88
Page 91

Summary of Numerical Results
The following information is available in the Summary of Numerical Results
Column Description
Variable Variable name.
YMin Burst rate histogram minimum.
YMax Burst rate histogram maximum.
Spikes The number of spikes used in spectrum calculation.
Filter Length The length of all the intervals of the interval filter (if a filter was used) or the
length or the recording session (in seconds).
Mean Frequency Mean firing rate (Spikes/Filter_Length).
Num Bursts Number of bursts.
Bursts Per Second Burst rate in bursts per second.
Bursts Per Minute Burst rate in bursts per minute.
% of Spikes in Bursts Percent of spikes in bursts.
Mean Burst Duration Mean duration of burst (in seconds).
St. Dev. of Burst
Duration
Mean Spikes in Burst Average number of spikes in burst.
St. Dev. of Spikes in
Burst
Mean ISI Burst Mean interspike interval in burst.
St. Dev of ISI in Burst Standard deviation of interspike intervals in burst.
Mean Freq. in Burst Mean firing rate (1/interspike_interval) in burst.
St. Dev. of Freq. in
Burst
Mean Peak Frequency
in Burst
St. Dev. Peak
Frequency in Burst
Standard deviation of burst duration.
Standard deviation of the number of spikes in burst.
Standard deviation of (1/interspike_interval) in burst.
Mean of (1/min_interspike_interval), where min_interspike_interval is the
minimum interspike interval in a burst.
Standard deviation of (1/min_interspike_interval), where
min_interspike_interval is the minimum interspike interval in a burst.
Mean Interburst Interval Average length (in seconds) of interburst interval. Interburst interval is the
interval from the end of the previous burst to the start of the current burst.
St. Dev. of Interburst
Interval
Mean Burst Surprise Average burst surprise.
St. Dev. Burst Surprise Standard deviation of burst surprise.
Standard deviation of the interburst intervals.
Page 89
Page 92

Algorithm
For each spike train, NeuroExplorer creates a new Interval variable and stores all the burst intervals
in this variable.
MaxInterval method
Find all the bursts using the following algorithm:
•
Scan the spike train until an interspike interval is found that is less than or equal to Max. Interval
.
•
While the interspike intervals are less than Max. End Interval, they are included in the burst.
•
If the interspike interval is more than Max. End Interval, the burst ends.
•
Merge all the bursts that are less than Min. Interval Between Bursts apart.
•
Remove the bursts that have duration less than Min. Duration of Burst or have fewer spikes
than Min. Number of Spikes.
Surprise method
1) First the mean firing rate ( Freq ) and mean interspike interval ( MeanISI ) of the neuron are
calculated.
Freq = NumberOfSpikes / (FileEndTime – FileStartTime)
MeanISI = 1 / Freq
2) NeuroExplorer scans the spike train until it finds two sequential ISI's so that each of those ISIs is
less than MeanISI/2. The surprise of the resulting 3-spike sequence is calculated:
If we assume that a random variable P has a Poisson distribution with parameter Freq and we also
assume that the burst has N spikes and the distance from the first to the last spike of the burst is T,
then the surprise of the burst is:
S = - log10 (Probability that P has at least N points in a time interval of length T )
3) NeuroExplorer adds the spikes to the end of the burst until the first ISI that is more than MeanISI
and calculates surprise for each of the bursts (with 3 initial spikes, 4 spikes, 5 spikes, etc.) The burst
with maximum surprise Smax is then selected.
4) NeuroExplorer removes the spikes from the beginning of the burst and calculates the surprise for
each of the reduced bursts. The burst with maximum surprise Smax is then selected.
5) If Smax is more than MinSurprise and the number of spikes in the burst is more than 3,
NeuroExplorer adds the burst to the result.
Reference
Legendy C.R. and Salcman M. (1985): Bursts and recurrences of bursts in the spike trains of
spontaneously active striate cortex neurons. J. Neurophysiology, 53(4):926-39.
Page 90
Page 93

2.25. Principal Component Analysis
This analysis calculates eigenvalues and eigenvectors (principal components) of the matrix of
correlations of rate histograms. The analysis creates population vectors corresponding to the principal
components.
Parameters
Parameter Description
Bin Bin size in seconds.
Vectors Prefix A string specifying how the population vector names will be generated. For
example, if prefix is pca1, the vector names will be pca1_01, pca1_02,
etc.
Select Data If Select Data is From Time Range, only the data from the specified (by
Select Data From and Select Data To parameters) time range will be used
in analysis. See also Data Selection Options.
Select Data From Start of the time range in seconds.
Select Data To End of the time range in seconds.
Interval filter Specifies the interval filter that will be used to preselect data before
analysis. See also Data Selection Options.
Send to Matlab An option to send the matrix of numerical results to Matlab. See also
Matlab Options.
Matrix Name Specifies the name of the results matrix in Matlab workspace.
Matlab command Specifies a Matlab command that is executed after the numerical results
are sent to Matlab.
Send to Excel An option to send numerical results or summary of numerical results to
Excel. See also Excel Options.
Sheet Name The name of the worksheet in Excel where to copy the numerical results.
TopLeft Specifies the Excel cell where the results are copied. Should be in the form
CR where C is Excel column name, R is the row number. For example, A1
is the top-left cell in the worksheet.
Summary of Numerical Results
The following information is available in the Summary of Numerical Results
Column Description
Variable Variable name and PCA statistics name.
pca1_N The weight of the variable in the N-th eigenvector. The rows at the bottom
of the table also show Eigenvalue of this eigenvector, % of variance it
explains, and the cumulative percent of the variance explained by this and
preceding eigenvectors.
Corr with VAR Correlation with the specified variable.
Numerical Results
Page 91
Page 94

The Results sheet shows for each neuron the weights this neuron has in all the eigenvectors.
Algorithm
Step 1.
Rate histograms are calculated for each of the selected neurons.
The time axis is divided into bins. The first bin is [0, Bin). The second bin is [Bin, Bin*2), etc. The left
end is included in each bin, the right end is excluded from the bin. For each bin, the number of events
(spikes) in this bin is calculated.
For example, for the first bin
bin_count = number of timestamps (ts) such that ts >= 0 and ts < Bin
Bin counts are calculated in such a way for all the selected variables resulting in a matrix
bin_count[i, j],
where i is the neuron number, j is the bin number.
Step 2.
The matrix of correlations between neurons c[t, s] is calculated:
c[t, s] = correlation between vectors bin_count[t, *] and bin_count[s, *], s, t = 1, ..., number of selected
neurons.
Step 3
The eigenvalues and eigenvectors are calculated for the matrix c[t, s]. The eigenvectors (principal
components) are sorted according to their eigenvalues. The first principal component has the largest
eigenvalue.
Each principal component becomes a new population vector in the data file.
Page 92
Page 95

2.26. PSTH Versus Time
This analysis shows the dynamics of the perievent histogram over time. It calculates multiple PST
histograms using a "sliding window" in time. Each histogram is shown as a vertical stripe with colors
representing the bin counts. Horizontal axis represents the position of the sliding window in time.
Parameters
Parameter Description
Reference Specifies a reference neuron or event.
XMinPSTH Histogram time minimum in seconds.
XMaxPSTH Histogram time maximum in seconds
BinPSTH Histogram bin size in seconds.
Start Start of the first sliding window in seconds.
Duration Duration of the sliding window in seconds.
Shift How much sliding window is shifted each time.
Number of shifts The number of sliding windows to be used.
Normalization Histogram units (Counts/Bin, Probability or Spikes/Second). See Algorithm
below.
No Selfcount An option not to count reference events if the target event is the same as
the reference event (prevents a histogram to have a huge peak at zero
when calculating PSTH versus itself).
Sliding window axis
direction
Sliding window axis
alignment
Smooth Option to smooth the histogram after the calculation. See Post-Processing
Smooth Filter Width The width of the smooth filter. See Post-Processing Options for details.
Send to Matlab An option to send the matrix of numerical results to Matlab. See also
Direction of the sliding window axis (horizontal or vertical).
Allows to specify what values are shown in the sliding window axis. There
are two options: Start of Window and Center of Window.
Suppose your data has the strongest correlation at 50 seconds. This
means that the window that has 50 seconds as its center will show the
highest PST values. If the window width is 20 seconds, it will be the
window [40,60]. If you are using 'Start of window' option, you will see the
peak at 40 instead of 50 seconds. However, with 'Center of window' option,
the peak will be at 50 seconds.
Options for details.
Matlab Options.
Matrix Name Specifies the name of the results matrix in Matlab workspace.
Matlab command Specifies a Matlab command that is executed after the numerical results
are sent to Matlab.
Send to Excel An option to send numerical results or summary of numerical results to
Excel. See also Excel Options.
Sheet Name The name of the worksheet in Excel where to copy the numerical results.
Page 93
Page 96

TopLeft Specifies the Excel cell where the results are copied. Should be in the form
CR where C is Excel column name, R is the row number. For example, A1
is the top-left cell in the worksheet.
Summary of Numerical Results
The following information is available in the Summary of Numerical Results
Column Description
Variable Variable name.
Reference Reference variable name.
YMin PSTH minimum.
YMax PSTH maximum.
Color Scale Min PSTH minimum.
Color Scale Max PSTH maximum.
Spikes The number of spikes in the variable.
Algorithm
NeuroExplorer calculates multiple PST histograms (see Perievent Histogram for more information on
how each PSTH is calculated). Each histogram is calculated for a different interval (window) in time:
[window_start[i], window_end[i]], i = 1,..., Number of Shifts.
That is, for each histogram only the timestamps that are within the window are used.
The following rules are used to calculate the windows:
window_start[1] = Start
window_end[1] = Start + Duration
window_start[2] = Start + Shift
window_end[2] = Start + Shift + Duration
...
Page 94
Page 97

2.27. Correlations with Continuous Variable
This analysis calculates crosscorrelations between a continuously recorded variable and a neuronal
firing rate, or crosscorrelations between a continuously recorded variable and another continuous
variable.
Parameters
Parameter Description
Reference Specifies a reference continuous variable.
XMin Time axis minimum in seconds.
XMax Time axis maximum in seconds.
Bin Type An option to specify how the bin value is determined. If this option is Auto,
NeuroExplorer will use the bin equal to the A/D sampling interval of the
reference variable.
Bin Bin size in seconds. Used if Bin Type is set to User defined.
Select Data If Select Data is From Time Range, only the data from the specified (by
Select Data From and Select Data To parameters) time range will be used
in analysis. See also Data Selection Options.
Select Data From Start of the time range in seconds.
Select Data To End of the time range in seconds.
Interval filter Specifies the interval filter(s) that will be used to preselect data before
analysis. See also Data Selection Options.
Smooth Option to smooth the histogram after the calculation. See Post-Processing
Options for details.
Smooth Filter Width The width of the smooth filter. See Post-Processing Options for details.
Add to Results / Bin left An option to add an additional vector (containing a left edge of each bin) to
the matrix of numerical results.
Add to Results / Bin
middle
Add to Results / Bin
right
Send to Matlab An option to send the matrix of numerical results to Matlab. See also
Matrix Name Specifies the name of the results matrix in Matlab workspace.
An option to add an additional vector (containing a middle point of each
bin) to the matrix of numerical results.
An option to add an additional vector (containing a right edge of each bin)
to the matrix of numerical results.
Matlab Options.
Matlab command Specifies a Matlab command that is executed after the numerical results
are sent to Matlab.
Send to Excel An option to send numerical results or summary of numerical results to
Excel. See also Excel Options.
Sheet Name The name of the worksheet in Excel where to copy the numerical results.
TopLeft Specifies the Excel cell where the results are copied. Should be in the form
CR where C is Excel column name, R is the row number. For example, A1
is the top-left cell in the worksheet.
Page 95
Page 98

Summary of Numerical Results
The following information is available in the Summary of Numerical Results
Column Description
Variable Variable name.
Reference Reference variable name.
YMin Y axis minimum.
YMax Y axis maximum.
Mean Hist. Mean of all the correlation values for this variable.
St. Dev. Hist Standard deviation of all the correlation values for this variable.
St. Eff. Mean Hist Standard error of mean of all the correlation values for this variable.
Algorithm
This analysis calculates standard correlograms for two (continuously recorded) vectors. For spike
trains and events, NeuroExplorer first calculates the rate histograms with the specified bin size, and
then calculates the correlations between the reference variable and the rate histogram.
If x[i], i=1,...N is the reference variable and y[i], i=1,...,N is another continuous variable or spike train
rate histogram, then
c[t] = Pearson correlation between vectors
{ x[1], x[2], …, x[N-t] }
and
{ y[t+1], y[t+2], …, y[N] }
Page 96
Page 99

2.28. Regularity Analysis
This analysis estimates the regularity of neuronal firing after the stimulation.
Parameters
Parameter Description
Reference Specifies a reference event or spike train.
XMin Time axis minimum in seconds.
XMax Time axis maximum in seconds.
Bin Bin size in seconds. Used if Bin Type is set to User defined.
Select Data If Select Data is From Time Range, only the data from the specified (by
Select Data From and Select Data To parameters) time range will be used
in analysis. See also Data Selection Options.
Select Data From Start of the time range in seconds.
Select Data To End of the time range in seconds.
Interval filter Specifies the interval filter that will be used to preselect data before
analysis. See also Data Selection Options.
ISI Graph This parameter determines how the mean ISI values is displayed in the
graph.
SD Graph This parameter determines how the standard deviation of ISI is displayed
in the graph.
CV Graph This parameter determines how the CV (coefficient of variation) values are
displayed in the graph.
CV Graph Max This parameter determines the scale for the CV values in the graph. CV
values are scaled from zero (bottom of the graph) to CV Graph Max (top
of the graph).
Add to Results / Bin left An option to add an additional vector (containing a left edge of each bin) to
the matrix of numerical results.
Add to Results / Bin
middle
Add to Results / Bin
right
An option to add an additional vector (containing a middle point of each
bin) to the matrix of numerical results.
An option to add an additional vector (containing a right edge of each bin)
to the matrix of numerical results.
Summary of Numerical Results
The following information is available in the Summary of Numerical Results
Column Description
Variable Variable name.
NumRefEvents Number of reference events.
YMin Histogram minimum.
YMax Histogram maximum.
Spikes The number of spikes used in calculation.
Page 97
Page 100

Filter Length The length of all the intervals of the interval filter (if a filter was used) or the
length or the recording session (in seconds).
Mean Freq. Mean firing rate (Spikes/Filter_Length).
Mean Hist. The mean of the histogram bin values (ISI values).
St. Dev. Hist. The standard deviation of the histogram bin values.
St. Dev. ISI The mean value of the SD ISI graph.
CV The mean value of the CV ISI graph.
Algorithm
This analysis estimates the regularity of neuronal firing after the stimulation. The time axis is divided
into bins with the first bin starting at the stimulus sync pulse. Interspike intervals are computed and
interval values are placed in time bins according to the latency of the first spike in the interval (if the
end of the ISI is more than XMax, the interval is not used in the analysis). Then the mean and
standard deviation in each bin are calculated. CV is equal to standard deviation for the bin divided by
the mean ISI for the bin.
Let ref[i] be the array of timestamps of the reference event,
ts[i] be the spike train (each ts is the timestamp)
For each timestamp ref[k]:
1a) calculate the distances from this event (or spike) to all the spikes in the spike train:
d[i] = ts[i] - ref[k]
1b) calculate the interspike interval
isi[i] = ts[i+1] - ts[i]
1c) for each i:
if d[i] is inside the first bin ( d[i] >= XMin and d[i] < XMin + Bin )
and d[i] + isi[i] < XMax,
add isi[i] to the series of intervals for the first bin
if d[i] is inside the second bin ( d[i] >= XMin+Bin and d[i] < XMin + Bin*2 )
and d[i] + isi[i] < XMax, add isi[i] to the series of intervals for the second bin
and so on... .
2) for each bin, calculate mean and standard deviation of the series of interspike intervals for this bin.
Reference
E.D. Young, J.-M. Robert and W.P. Shofner. Regularity and latency of units in ventral cochlear
nucleus: implications for unit classification and generation of response properties. J.
Neurophysiology, Vol. 60, 1988, 1-29.
Page 98
 Loading...
Loading...